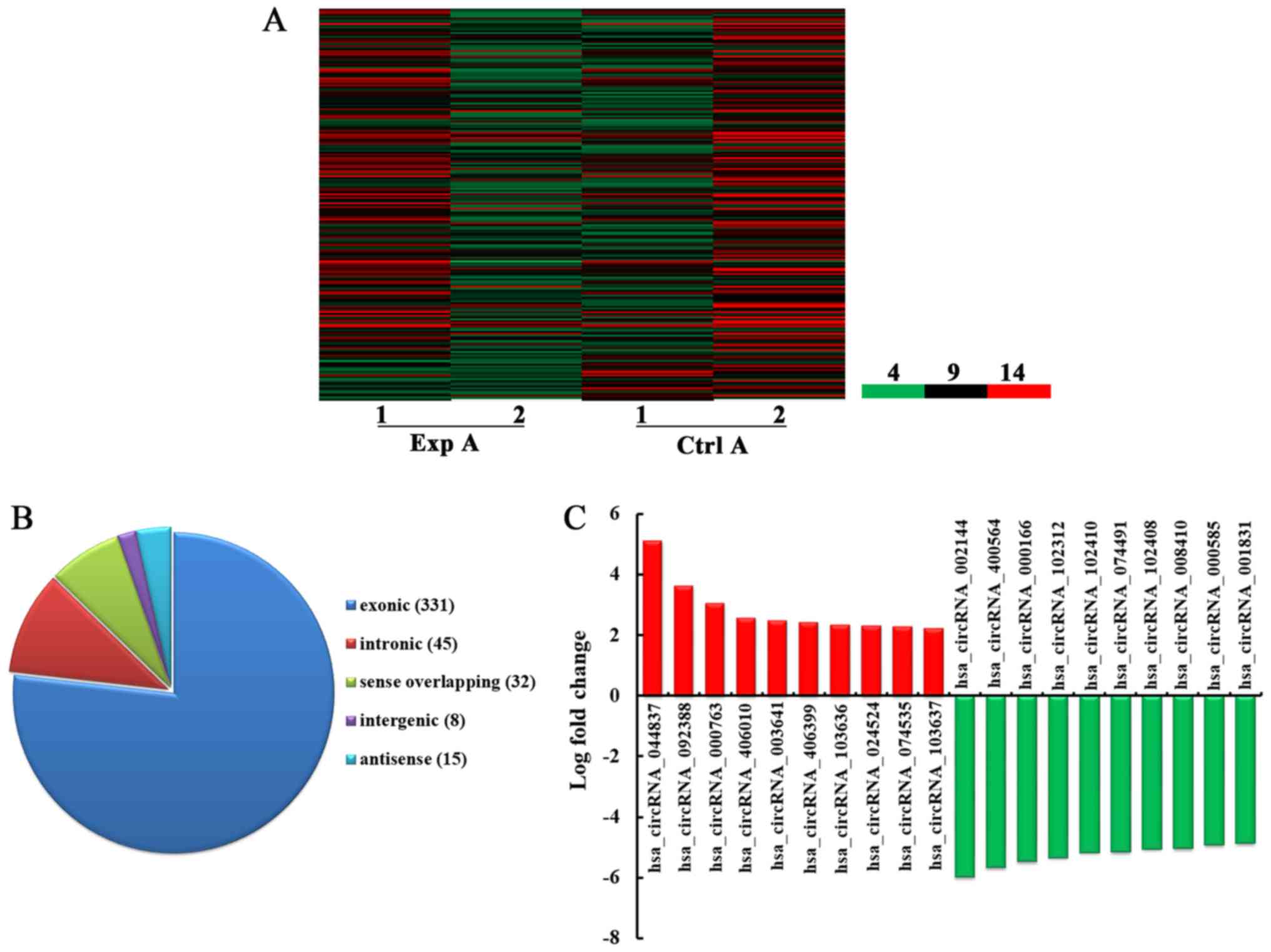
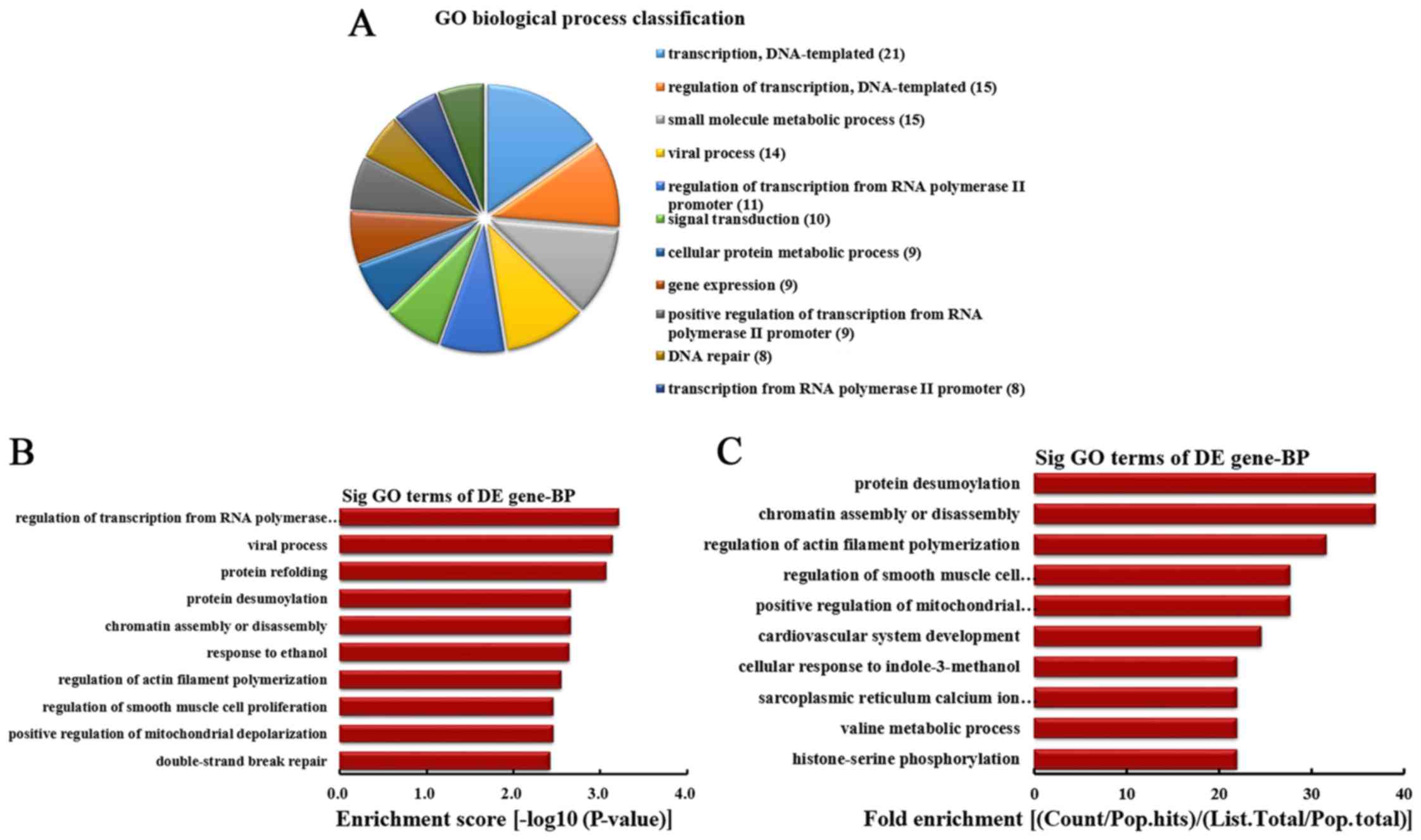
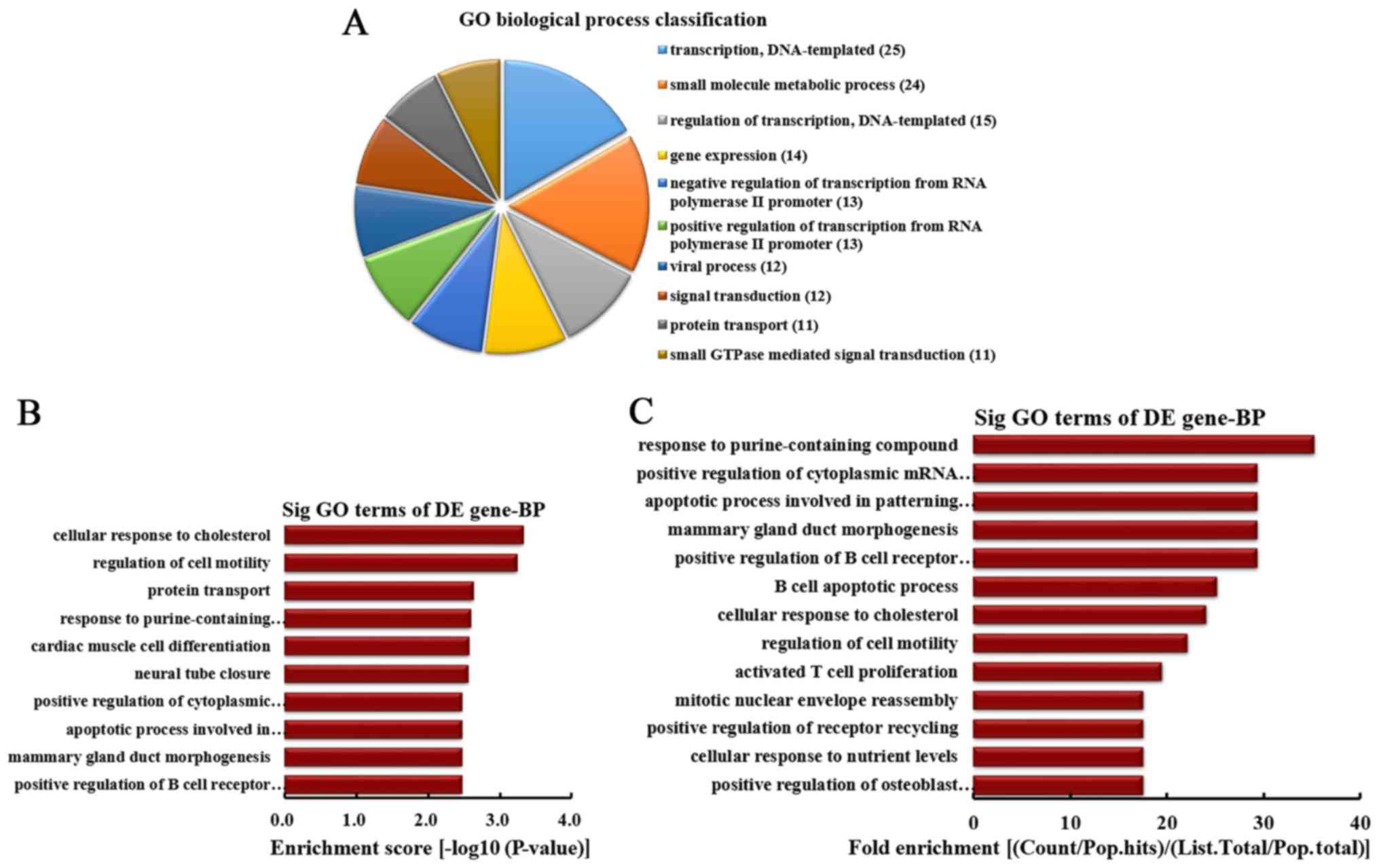
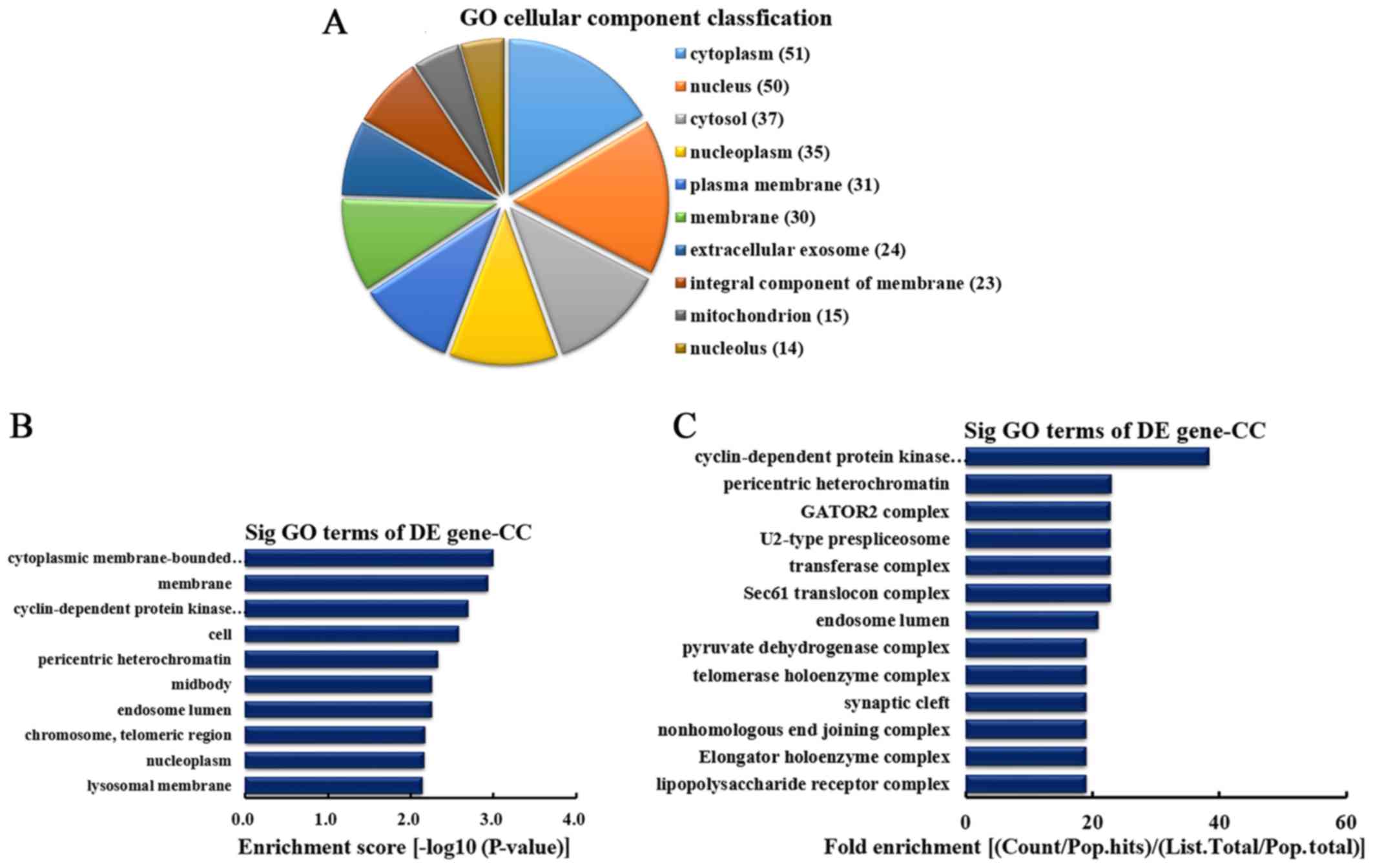
|
1
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar
|
|
2
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel R, Desantis C and Jemal A:
Colorectal cancer statistics, 2014. CA Cancer J Clin. 64:104–117.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shah SA, Haddad R, Al-Sukhni W, Kim RD,
Greig PD, Grant DR, Taylor BR, Langer B, Gallinger S and Wei AC:
Surgical resection of hepatic and pulmonary metastases from
colorectal carcinoma. J Am Coll Surg. 202:468–475. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rotolo N, De Monte L, Imperatori A and
Dominioni L: Pulmonary resections of single metastases from
colorectal cancer. Surg Oncol. 16(Suppl 1): S141–S144. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Goya T, Miyazawa N, Kondo H, Tsuchiya R,
Naruke T and Suemasu K: Surgical resection of pulmonary metastases
from colorectal cancer. 10-year follow-up. Cancer. 64:1418–1421.
1989. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Galandiuk S, Wieand HS, Moertel CG, Cha
SS, Fitzgibbons RJ Jr, Pemberton JH and Wolff BG: Patterns of
recurrence after curative resection of carcinoma of the colon and
rectum. Surg Gynecol Obstet. 174:27–32. 1992.PubMed/NCBI
|
|
8
|
McAuliffe JC, Qadan M and D'Angelica MI:
Hepatic resection, hepatic arterial infusion pump therapy, and
genetic biomarkers in the management of hepatic metastases from
colorectal cancer. J Gastrointest Oncol. 6:699–708. 2015.PubMed/NCBI
|
|
9
|
Chen D, Sun Q, Cheng X, Zhang L, Song W,
Zhou D, Lin J and Wang W: Genome-wide analysis of long noncoding
RNA (lncRNA) expression in colorectal cancer tissues from patients
with liver metastasis. Cancer Med. 5:1629–1639. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mattick JS, Taft RJ and Faulkner GJ: A
global view of genomic information - moving beyond the gene and the
master regulator. Trends Genet. 26:21–28. 2010. View Article : Google Scholar
|
|
11
|
Malouf GG, Zhang J, Yuan Y, Compérat E,
Rouprêt M, Cussenot O, Chen Y, Thompson EJ, Tannir NM, Weinstein
JN, et al: Characterization of long non-coding RNA transcriptome in
clear-cell renal cell carcinoma by next-generation deep sequencing.
Mol Oncol. 9:32–43. 2015. View Article : Google Scholar
|
|
12
|
Li J, Yang J, Zhou P, Le Y, Zhou C, Wang
S, Xu D, Lin HK and Gong Z: Circular RNAs in cancer: Novel insights
into origins, properties, functions and implications. Am J Cancer
Res. 5:472–480. 2015.PubMed/NCBI
|
|
13
|
Sanger HL, Klotz G, Riesner D, Gross HJ
and Kleinschmidt AK: Viroids are single-stranded covalently closed
circular RNA molecules existing as highly base-paired rod-like
structures. Proc Natl Acad Sci USA. 73:3852–3856. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hsu MT and Coca-Prados M: Electron
microscopic evidence for the circular form of RNA in the cytoplasm
of eukaryotic cells. Nature. 280:339–340. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Perkel JM: Assume nothing: The tale of
circular RNA. Biotechniques. 55:55–57. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Salzman J, Gawad C, Wang PL, Lacayo N and
Brown PO: Circular RNAs are the predominant transcript isoform from
hundreds of human genes in diverse cell types. PLoS One.
7:e307332012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Salzman J, Chen RE, Olsen MN, Wang PL and
Brown PO: Cell-type specific features of circular RNA expression.
PLoS Genet. 9:e10037772013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar :
|
|
20
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin
QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long
noncoding RNAs. Mol Cell. 51:792–806. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Guo JU, Agarwal V, Guo H and Bartel DP:
Expanded identification and characterization of mammalian circular
RNAs. Genome Biol. 15:4092014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Qu S, Yang X, Li X, Wang J, Gao Y, Shang
R, Sun W, Dou K and Li H: Circular RNA: A new star of noncoding
RNAs. Cancer Lett. 365:141–148. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bachmayr-Heyda A, Reiner AT, Auer K,
Sukhbaatar N, Aust S, Bachleitner-Hofmann T, Mesteri I, Grunt TW,
Zeillinger R and Pils D: Correlation of circular RNA abundance with
proliferation - exemplified with colorectal and ovarian cancer,
idiopathic lung fibrosis, and normal human tissues. Sci Rep.
5:80572015. View Article : Google Scholar
|
|
26
|
Thorson AG, Christensen MA and Davis SJ:
The role of colonoscopy in the assessment of patients with
colorectal cancer. Dis Colon Rectum. 29:306–311. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jemal A, Thun MJ, Ries LA, Howe HL, Weir
HK, Center MM, Ward E, Wu XC, Eheman C, Anderson R, et al: Annual
report to the nation on the status of cancer, 1975–2005, featuring
trends in lung cancer, tobacco use, and tobacco control. J Natl
Cancer Inst. 100:1672–1694. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yiu HY, Whittemore AS and Shibata A:
Increasing colorectal cancer incidence rates in Japan. Int J
Cancer. 109:777–781. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lin PC, Lin JK, Lin CC, Wang HS, Yang SH,
Jiang JK, Lan YT, Lin TC, Li AF, Chen WS, et al: Carbohydrate
antigen 19-9 is a valuable prognostic factor in colorectal cancer
patients with normal levels of carcinoembryonic antigen and may
help predict lung metastasis. Int J Colorectal Dis. 27:1333–1338.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xie H, Ren X, Xin S, Lan X, Lu G, Lin Y,
Yang S, Zeng Z, Liao W, Ding YQ, et al: Emerging roles of
circRNA_001569 targeting miR-145 in the proliferation and invasion
of colorectal cancer. Oncotarget. 7:26680–26691. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Huang G, Zhu H, Shi Y, Wu W, Cai H and
Chen X: cir-ITCH plays an inhibitory role in colorectal cancer by
regulating the Wnt/β-catenin pathway. PLoS One. 10:e01312252015.
View Article : Google Scholar
|
|
32
|
Ahmed I, Karedath T, Andrews SS, Al-Azwani
IK, Mohamoud YA, Querleu D, Rafii A and Malek JA: Altered
expression pattern of circular RNAs in primary and metastatic sites
of epithelial ovarian carcinoma. Oncotarget. 7:36366–36381. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Huang M, Zhong Z, Lv M, Shu J, Tian Q and
Chen J: Comprehensive analysis of differentially expressed profiles
of lncRNAs and circRNAs with associated co-expression and ceRNA
networks in bladder carcinoma. Oncotarget. 7:47186–47200. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Karam RA, Al Jiffry BO, Al Saeed M, Abd El
Rahman TM, Hatem M and Amer MG: DNA repair genes polymorphisms and
risk of colorectal cancer in Saudi patients. Arab J Gastroenterol.
17:117–120. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
de Freitas IN, de Campos FG, Alves VA,
Cavalcante JM, Carraro D, Coudry Rde A, Diniz MA, Nahas SC and
Ribeiro U Jr: Proficiency of DNA repair genes and microsatellite
instability in operated colorectal cancer patients with clinical
suspicion of lynch syndrome. J Gastrointest Oncol. 6:628–637.
2015.PubMed/NCBI
|
|
36
|
Scarbrough PM, Weber RP, Iversen ES,
Brhane Y, Amos CI, Kraft P, Hung RJ, Sellers TA, Witte JS, Pharoah
P, et al: A cross-cancer genetic association analysis of the DNA
repair and DNA damage signaling pathways for lung, ovary, prostate,
breast, and colorectal cancer. Cancer Epidemiol Biomarkers Prev.
25:193–200. 2016. View Article : Google Scholar
|
|
37
|
Dimberg J, Skarstedt M, Slind Olsen R,
Andersson RE and Matussek A: Gene polymorphism in DNA repair genes
XRCC1 and XRCC6 and association with colorectal cancer in Swedish
patients. APMIS. 124:736–740. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Győrffy B, Stelniec-Klotz I, Sigler C,
Kasack K, Redmer T, Qian Y and Schäfer R: Effects of RAL signal
transduction in KRAS- and BRAF-mutated cells and prognostic
potential of the RAL signature in colorectal cancer. Oncotarget.
6:13334–13346. 2015. View Article : Google Scholar
|
|
39
|
Lu X, Li C, Wang YK, Jiang K and Gai XD:
Sorbitol induces apoptosis of human colorectal cancer cells via p38
MAPK signal transduction. Oncol Lett. 7:1992–1996. 2014.PubMed/NCBI
|
|
40
|
Ji H, Greening DW, Barnes TW, Lim JW,
Tauro BJ, Rai A, Xu R, Adda C, Mathivanan S, Zhao W, et al:
Proteome profiling of exosomes derived from human primary and
metastatic colorectal cancer cells reveal differential expression
of key metastatic factors and signal transduction components.
Proteomics. 13:1672–1686. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Nowakowska-Zajdel E, Mazurek U, Stachowicz
M, Niedworok E, Fatyga E and Muc-Wierzgoń M: Cellular signal
transduction pathways by leptin in colorectal cancer tissue:
Preliminary results. ISRN Endocrinol. 2011:5753972011. View Article : Google Scholar
|
|
42
|
Marte BM and Downward J: PKB/Akt:
Connecting phosphoinositide 3-kinase to cell survival and beyond.
Trends Biochem Sci. 22:355–358. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Mayo MW and Baldwin AS: The transcription
factor NF-kappaB: Control of oncogenesis and cancer therapy
resistance. Biochim Biophys Acta. 1470:M55–M62. 2000.PubMed/NCBI
|
|
44
|
Hidalgo M and Rowinsky EK: The
rapamycin-sensitive signal transduction pathway as a target for
cancer therapy. Oncogene. 19:6680–6686. 2000. View Article : Google Scholar
|
|
45
|
Qi J, Yu Y, Akilli Öztürk Ö, Holland JD,
Besser D, Fritzmann J, Wulf-Goldenberg A, Eckert K, Fichtner I and
Birchmeier W: New Wnt/β-catenin target genes promote experimental
metastasis and migration of colorectal cancer cells through
different signals. Gut. 65:1690–1701. 2016. View Article : Google Scholar
|
|
46
|
Xu K, Chen Z, Qin C and Song X: miR-7
inhibits colorectal cancer cell proliferation and induces apoptosis
by targeting XRCC2. Onco Targets Ther. 7:325–332. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang Y and Zheng T: Screening of hub genes
and pathways in colorectal cancer with microarray technology.
Pathol Oncol Res. 20:611–618. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Slaby O, Sachlova M, Brezkova V, Hezova R,
Kovarikova A, Bischofová S, Sevcikova S, Bienertova-Vasku J, Vasku
A, Svoboda M, et al: Identification of microRNAs regulated by
isothiocyanates and association of polymorphisms inside their
target sites with risk of sporadic colorectal cancer. Nutr Cancer.
65:247–254. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Huhn S, Bevier M, Pardini B, Naccarati A,
Vodickova L, Novotny J, Vodicka P, Hemminki K and Försti A:
Colorectal cancer risk and patients' survival: Influence of
polymorphisms in genes somatically mutated in colorectal tumors.
Cancer Causes Control. 25:759–769. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yuanming L, Lineng Z, Baorong S, Junjie P
and Sanjun C: BRCA1 and ERCC1 mRNA levels are associated with lymph
node metastasis in Chinese patients with colorectal cancer. BMC
Cancer. 13:1032013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Huang MY, Wang JY, Chang HJ, Kuo CW, Tok
TS and Lin SR: CDC25A, VAV1, TP73, BRCA1 and ZAP70 gene
overexpression correlates with radiation response in colorectal
cancer. Oncol Rep. 25:1297–1306. 2011.PubMed/NCBI
|
|
52
|
Sopik V, Phelan C, Cybulski C and Narod
SA: BRCA1 and BRCA2 mutations and the risk for colorectal cancer.
Clin Genet. 87:411–418. 2015. View Article : Google Scholar
|
|
53
|
Kobayashi M, Funayama R, Ohnuma S, Unno M
and Nakayama K: Wnt-β-catenin signaling regulates ABCC3 (MRP3)
transporter expression in colorectal cancer. Cancer Sci.
107:1776–1784. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Liu C, Zhang A, Cheng L and Gao Y: miR 410
regulates apoptosis by targeting Bak1 in human colorectal cancer
cells. Mol Med Rep. 14:467–473. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Chen B, Liu Y, Jin X, Lu W, Liu J, Xia Z,
Yuan Q, Zhao X, Xu N and Liang S: MicroRNA-26a regulates glucose
metabolism by direct targeting PDHX in colorectal cancer cells. BMC
Cancer. 14:4432014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zhou Y, Wan G, Spizzo R, Ivan C, Mathur R,
Hu X, Ye X, Lu J, Fan F, Xia L, et al: miR-203 induces oxaliplatin
resistance in colorectal cancer cells by negatively regulating ATM
kinase. Mol Oncol. 8:83–92. 2014. View Article : Google Scholar
|