Introduction
Major depressive disorder (MDD) is associated with a
high degree of morbidity and mortality (1), and is predicted to be the leading
cause of disease burden by the year 2030 (2). MDD occurs about twice as often in
women than in men (3) and sex
difference is the most obvious characteristics for MDD (3,4).
An increasing number of sex‑specific biomarkers for MDD have been
identified (5). Moreover, male
and female mice have demonstrated fundamentally different
transcriptional and post‑transcriptional responses to stress, and
sex‑specific molecular processes occurred following stress
(6). Notably, males demonstrated
a very robust transcriptional and post-transcriptional response to
stress (6). These findings
revealed that the pathogenesis of MDD is markedly different in
males and females, and transcriptional and post-transcriptional
dysregulation may lead to occurrence of MDD in males. Although
these changes were found to correlate with MDD in males, the
molecular mechanism remains poorly understood.
MicroRNAs (miRNAs or miRs), a prominent class of
small non-coding RNAs, generally exert their functions by binding
the 3′ untranslated regions of target mRNA to suppress target gene
expression in post-transcriptional regulation and control various
biological processes (7). The
majority of known mammalian miRNA are expressed in a
tissue‑specific manner, cause translational repression and may
individually target hundreds of genes (8). miRNAs are highly expressed in
neurons, where they regulate the expression of a large number of
target mRNA. Neuronal miRNA pathways may create an extremely
powerful mechanism to dynamically adjust the protein content of
neuronal compartments, even without the requirement for new gene
transcription (9,10). Dysregulation of specific miRNA has
been observed in patients and animal models of neuro-psychiatric
disorders (11–14). In addition, a critical component
necessary for miRNA biogenesis, Dicer1, has also been implicated in
posttraumatic stress and anxiolytic responses (15,16). With regard to MDD, the evidence
supporting miRNA involvement in the pathophysiology and the
treatment of the disorder is increasing (17). In addition, numerous studies have
implied that miRNA may provide a link between environmental
stressors and gene expression (18), and be an important factor involved
in the mechanism of depression (19–22). In vertebrates, more distinct
miRNAs are expressed in the brain than in any other tissues
(23,24). These studies indicate that miRNA
may serve an important role in the brain of patients with MDD by
acting as the master regulator of gene expression at a
post-transcriptional level (25–27). Previous investigations have
suggested that miRNA have been involved in several pathways that
may contribute to the MDD pathomechanism (27–29). However, how these miRNA contribute
to this illness is not well understood.
The present study identified that miR‑124‑3p, an
abundant miRNA in the brain, was significantly downregulated in the
post-mortem brain BA44 area of males with MDD. The miR-124-3p
target gene, DNA damage-inducible transcript 4 (DDIT4), a regulator
of the tuberous sclerosis proteins 1/2 (TSC1/2) complex, was
associated with MDD pathogenesis and antidepressant effect.
Meanwhile, the expression of miR-124-3p was negatively correlated
with expression of DDIT4 and DDIT4 transcription factor,
specificity protein 1 (SP1), respectively. miR-124-3p regulated the
mammalian target of rapamycin (mTOR) pathway significantly and this
regulation was dependent on the TSC1/2 complex. In addition,
miR-124-3p was downregulated by reduction of its transcription
factor, aryl hydrocarbon receptor nuclear translocator (ARNT), in
males with MDD. These results may help to elucidate the
pathogenesis of MDD and provide novel insight into antidepressant
treatment.
Materials and methods
Gene Expression Omnibus (GEO) dataset
collection and differential expression analysis
Microarray data were obtained from five datasets.
The five series were accessed at the National Centers for
Biotechnology Information GEO database (ncbi.nlm.nih.gov/geo/), which served as a public
repository for gene expression datasets, and the accession numbers
were GSE58105, GSE63377, GSE43261, GSE64119 and GSE22909,
respectively. Differentially expressed genes and miRNAs were
obtained using GEO2R (ncbi.nlm.nih.gov/geo/geo2r/). GEO2R is an interactive
web tool that compares two groups of samples under the same
experimental conditions and is able to analyze almost any GEO
series (30).
miRNA tissue‑specific expression
analysis
Expression levels of miR-124-3p across human tissues
were obtained from the YM500 database (http://ngs.ym.edu.tw/ym500v2/index.php).
Prediction of target genes of miRNA
The miRWalk 2.0 database (zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/)
(31) was used to identify genes
that are directly targeted by miR-124-3p in the validated target
module.
The Cancer Genome Atlas (TCGA) data
exploratory analyses
To determine whether the expression level of DDIT4
was correlated with miR-124-3p in brain tissues, TCGA data about
miRNA and mRNA (RNA Seq v2) expression levels in patients with
low-grade glioma were obtained from synapse. org/ for correlation
analysis in a large data set (n=528). The miR-124 level, DDIT4 mRNA
level, SP1 mRNA level and whole transcriptome data were used in the
present study.
Transcription factor analysis and target
gene binding site prediction
The GeneCards database (genecards.org/), JASPAR database (jaspar.binf.ku.dk/) and PROMO database (alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3)
were used to identify potential transcription factor-gene
interactions. The GeneCards database was used to investigate
whether SP1, a target of miR-124-3p, is a transcription factor of
DDIT4, and whether it regulates DDIT4 expression. The binding
sequence and binding sites were shown in the promoter region of
DDIT4. To investigate the cause of miR-124-3p downregulation,
potential transcription factors, which are also downregulated in
males with MDD according to the results of gene ontology (GO)
analyses, were first screened. Subsequently, the JASPAR and PROMO
databases were used to predict transcription factors of
miR-124.
Gene set enrichment analysis
The association between phenotypes, pathways and
miR-124-3p expression was analyzed using Gene Set Enrichment
Analysis (GSEA v2.2; broad.mit.edu/gsea/). GSEA calculates a gene set
enrichment score (ES) that estimates whether genes from a
pre‑defined gene set (obtained from the Molecular Signatures
Database; http://software.broadinstitute.org/gsea/msigdb/collections.jsp#C2;
MTOR_UP.N4.V1_DN, BILANGES_RAPAMYCIN_ SENSITIVE_VIA_TSC1_AND_TSC2)
are enriched among the highest- (or lowest-) ranked genes or
distributed randomly. Default settings were used. Thresholds for
significance were determined by permutation analysis (1,000
permutations). False discovery rate (FDR) was calculated. A gene
set was considered significantly enriched when the FDR score was
<0.05.
GO analyses
A list of differentially expressed genes in males
with MDD was imported into the Database for Annotation,
Visualization, and Integrated Discovery Bioinformatics Resource
(david.abcc.ncifcrf.gov/). The aim was to
detect significantly over‑represented biological processes affected
by changes in the transcriptomes. Functional enrichment was
assessed using the following three annotation databases: Biological
process, molecular function and cellular component.
Statistical analysis
Statistical analyses were performed using SPSS 19.0
(IBM Corp., Armonk, NY, USA), R 3.2.4 software (r-project.org/), and GraphPad Prism 5 (GraphPad
Software, Inc., La Jolla, CA, USA). The data were presented as the
mean ± standard deviation. Group distributions were compared using
the Student's t-test. For multiple comparisons, one-way analysis of
variance was used followed by Dunnett's post hoc test. The
correlation between miR-124-3p and target gene expression level was
analyzed by Pearson correlation analysis. P<0.05 was considered
to indicate a statistically significant difference.
Results
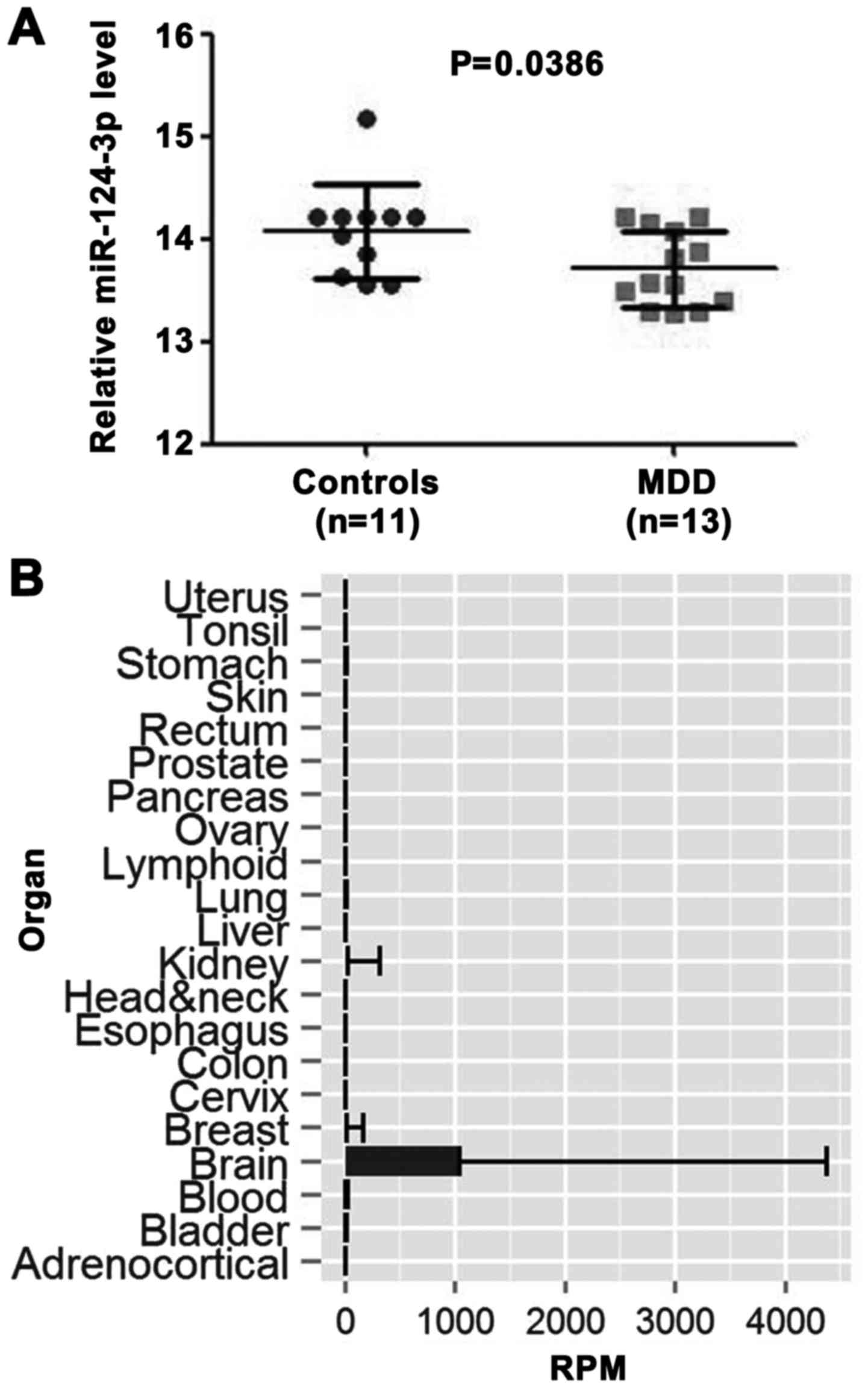
Expression level of miR‑124‑3p is
downregulated in males with MDD
As we know, age and sex are complicated factors when
trying to understand the pathomechanism of MDD (4,32,33). In consideration of this, the data
in the GEO miRNA expression profile (GSE58105) were filtered, and
24 young male samples (MDD, n=13; controls, n=11) were selected,
according to previous studies (32,34). Differential expression analysis
was performed between the prefrontal cortex (PFC) of depressed
males and healthy controls using GEO2R. This result demonstrated
that the miR‑124‑3p expression level was significantly higher in
control samples than in MDD samples (P=0.0386; Fig. 1A). A total of five differentially
expressed miRNA (has-miR-423-5p, -miR-320c, -miR-124, -miR-1825 and
-miR-1281) were screened under the criteria (P<0.05; FC>1.2
or FC<0.8). Subsequently, multiple testing was also performed
and the FDR of candidate differential expression miRNA was
calculated using R 3.2.4. The FDR of miR-124 was also <0.05
(data not shown). According to tissue specificity analysis from the
YM500 database, miR-124-3p was the only miRNA enriched in the brain
(Fig. 1B). Therefore, it was
hypothesized that the downregulation of miR-124-3p may play an
important role in the occurrence of MDD.
DDIT4 and its transcription factor, SP1,
are targets of miR‑124‑3p
To investigate the mechanism of miR-124-3p in the
occurrence of MDD, the validated target genes of miR-124-3p from
the miRWalk 2.0 database were examined. DDIT4 (REDD1), a target of
miR-124-3p (data not shown), was reported to act as a critical
mediator in MDD (35). To further
verify the role of DDIT4, microarray profiles GSE63377 and GSE43261
were used. As demonstrated in Fig.
2A, the DDIT4 mRNA level was upregulated in rats with
depressive-like behavior. In male mice, the DDIT4 mRNA level was
significantly downregulated in fluoxetine sensitivity group
compared with fluoxetine resistance group and control group,
respectively (P=0.011 and P<0.0001, respectively; Fig. 2B). These data revealed the role of
DDIT4 in the occurrence and treatment effect of MDD.
 | Figure 2miR-124-3p inhibits expression of
DDIT4 by directly targeting DDIT4 and its transcription factor,
SP1. (A) Downregulation of the DDIT4 expression level in the PFC of
male rats with depressive-like behavior (GSE63377). (B) Relative
expression level of miR-124-3p in the PFC of male mice. The
expression level of DDIT4 was reduced in the fluoxetine‑S group
compared with the level in the fluoxetine‑R group and control
group, respectively (GSE43261). Significance was determined with
one‑way analysis of variance followed by Dunnett's post hoc test.
(C) miR‑124‑3p expression level is negatively correlated with DDIT4
mRNA expression level, as demonstrated in 528 patients with
low-grade glioma from TCGA dataset. (D) miR-124-3p expression level
is negatively correlated with SP1 mRNA expression level, as
demonstrated in 528 patients with low-grade glioma from TCGA
dataset. (E) SP1 is a potential transcription factor of DDIT4. The
arrow indicates the transcription start site of DDIT4 in the
chromosome position. The vertical line is the SP1 binding site. The
binding position of SP1 on the chromosome and the binding sequence
are shown in the table. (F) MitA, a direct inhibitor of the binding
of SP1 family factors to DNA promoters, suppressed the expression
of DDIT4 (GSE64119). Significance between the two populations was
determined with a two‑tailed t‑test. PFC, prefrontal cortex; ns,
not significant; TCGA, The Cancer Genome Atlas; miR, microRNA;
DDIT4, DNA damage‑inducible transcript 4 protein; MDD, major
depressive disorder; fluoxetine‑S, fluoxetine‑sensitive;
fluoxetine‑R, fluoxetine‑resistant. |
In a previous study, depression also acted as a risk
factor in patients with low-grade glioma (36). It has been proposed that
depression is not only a psychological reaction to the low-grade
glioma, but that the association between these disorders has
biochemical roots (36-38). To further determine whether the
expression level of DDIT4 was regulated by miR-124-3p, correlation
analyses between the expression level of DDIT4 and miR-124-3p were
performed in low-grade glioma patients. This result demonstrated
that the expression level of miR-124-3p was negatively correlated
with the expression level of DDIT4 (Fig. 2C; R2=0.02; P=0.0011).
In addition, the expression level of miR-124-3p was negatively
correlated with the expression level of SP1, the other validated
target gene of miR-124-3p (Fig.
2D; R2=0.13; P<0.001). Notably, SP1, a
transcription factor, could bind to the transcription start site of
DDIT4 (Fig. 2E). To validate
whether SP1 is able to regulate the expression of DDIT4, microarray
profile GSE64119 was used. As indicated in Fig. 2F, DDIT4 was significantly
downregulated by MitA (P<0.05), which is a direct inhibitor of
the binding of SP1 family factors to DNA promoters (39). These data suggested that
miR-124-3p suppresses the expression of DDIT4 by targeting DDIT4
and its transcription factor SP1.
Regulatory effects of miR‑124‑3p on the
mTOR signaling pathway are dependent on the TSC1/2 complex
DDIT4, a target gene of miR-124-3p, is a well-known
antagonist of mTOR signaling in MDD (35). The mTOR signaling pathway serves
an important role in MDD (35),
therefore, to investigate the regulatory effect of miR-124-3p on
mTOR signaling, GSEA was used. The patients with low-grade glioma
from TCGA were divided into miR-124-3p-positive and
miR-124-3p-negative groups, and the association between miR-124-3p
and mTOR signaling was analyzed. As demonstrated in Fig. 3A, the mTOR activation gene set was
significantly enriched in the miR‑124‑3p‑positive group (FDR=0.02).
These data suggested that the miR-124-3p expression level was
positively associated with mTOR signaling. To further verify the
regulation of miR-124-3p on mTOR signaling by targeting DDIT4,
which stabilizes the TSC1/2 complex and inhibits the mTOR
signaling, GSEA was performed and a mTOR pathway activation gene
set was employed, which was obtained from TSC1(−/−) or TSC2(−/−)
mouse embryo fibroblasts. The miR-124-3-positive samples were
divided into high and low miR-124-3p expression groups. As
demonstrated in Fig. 3B, this
mTOR pathway gene set was not significantly enriched in each group.
These results suggested that the regulatory effect of miR-124-3p on
mTOR signaling is dependent on the TSC1/2 complex. Thus, miR-124-3p
may play a role in the regulation of mTOR signaling by targeting
DDIT4.
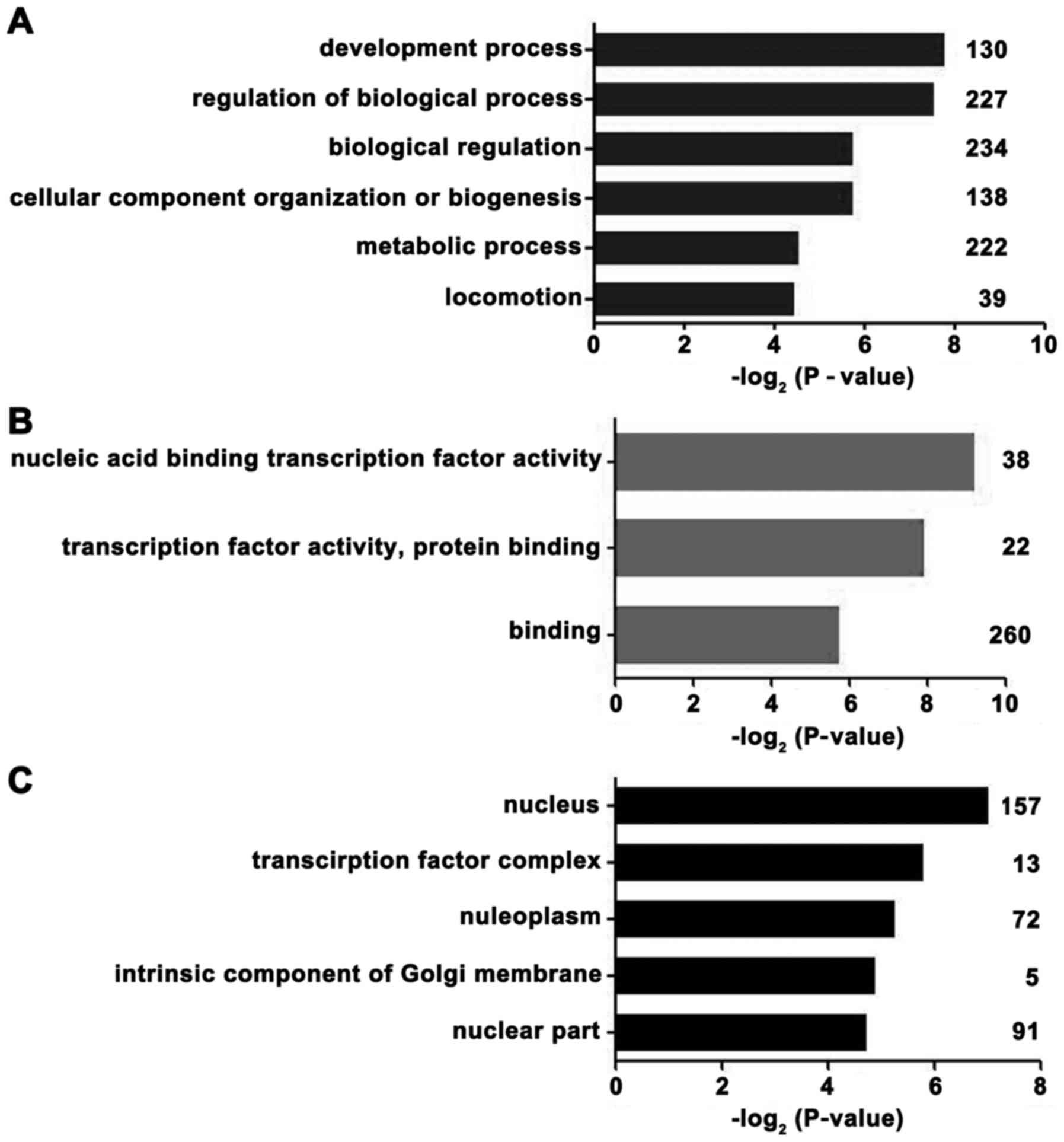
Microarray‑based GO analyses
From the GEO mRNA expression profile (GSE53987),
differentially expressed mRNAs were identified between the PFC of
depressed males and normal control samples. To gain insights into
the biological roles of differently expressed genes, we performed
GO classification enrichment analysis. Genes that demonstrated a
nominal significance level of P<0.01 were selected and were
tested against the background set of all genes with GO annotations.
As indicated in Fig. 4, the most
enriched GO categories of the differentially expressed genes were
'development process', 'nucleic acid binding transcription factor
activity', 'transcription factor complex' and 'nucleus.' According
to these results, the regulation of transcription factors was
associated with the occurrence of MDD in males.
ARNT regulates the expression of
miR‑124‑3p by acting as its transcription factor
According to the GO analysis results, it was
hypothesized that the expression of miR-124-3p was regulated by its
transcription factor in this disease. Among the 38 transcription
factors, 25 were downregulated in males with MDD (data not shown).
Transcription factors of miR-124-3p were predicted from the JASPAR
and PROMO databases. As demonstrated in Fig. 5A, ARNT may be the transcription
factor that has a role in MDD. To further investigate the
regulation of ARNT on miR-124-3p expression, micro-array profile
GSE22909 was used. As indicated in Fig. 5B, miR-124-3p expression was
upregulated by 2,3,7,8-tetrachlo-rodibenzo-p-dioxin, which is an
activator for ARNT (40,41). These results suggested that
miR-124-3p was downregulated by the reduction of ARNT in males with
MDD. As demonstrated in Fig. 6,
ARNT downregulation reduced miR-124-3p expression. miR-124-3p
downregulation may result in mTOR signaling inhibition and
occurrence of MDD in males via regulating DDIT4 expression.
miR-124-3p may regulate DDIT4 expression by targeting DDIT4 and
SP1.
Discussion
Emerging evidences have suggested that miRNA may
serve a key role in the basic mechanisms of brain neuroplasticity,
and they are believed to be involved in the manifestation of
depression and the therapeutic actions of antidepressant drugs
(42). A study by Lopez et
al (25) demonstrated BA44
microarray expression of miRNA in MDD. In the present study, it was
demonstrated that miR-124-3p was one of most significantly
dysregulated miRNA in the BA44 microarray of Lopez et al
(25). To the best of our
knowledge, the present study is the first to report the
downregulation of miR‑124‑3p in males with MDD.
In the present study, the results indicated that
miR-124-3p was downregulated in patients with MDD compared with
healthy controls. To determine the role and mechanism of miR-124-3p
in MDD, bioinformatics analysis was used. As a result, it was
identified that miR‑124‑3p was downregulated in patients with MDD.
DDIT4, a validated target gene of miR-124-3p, was correlated with
the occurrence of MDD and antidepressant effect. miR-124-3p reduced
DDIT4 expression by targeting DDIT4 and DDIT4 transcription factor
SP1, which led to the regulation of the mTOR signaling pathway. In
addition, decreased ARNT levels resulted in reduced miR-124-3p
levels in males with MDD.
Previous studies have reported that miR-124 may
regulate adult neurogenesis, promote neuronal differentiation and
contribute to synaptic plasticity in vivo (5,43).
Although miR-124 has been widely studied in animal models of
depression, few studies have investigated the relationship between
the levels of miR-124 and MDD in clinics. Recent studies have
reported that miR-124-3p was upregulated in the BA46 area of the
brain (44) and in peripheral
blood mononuclear cells in patients with MDD (45). miR-124 expression levels were also
increased in the hippocampus of rat models of depression (46,47). Contrastingly, two previous studies
did not find altered levels of miR-124 (34,48). However, to the best of our
knowledge, the present study is the first to identify that
miR-124-3p was downregulated in the BA44 brain area in patients
with MDD, although the fold-change of miR-124-3p was not
tremendous. Notably, minor fold-change of critical mediators in a
signaling pathway usually have a cascade amplification effect on
occurrence of disease, such as subtle reductions in the dose of
phosphatase and tensin homolog predispose to tumorigenesis
(49). In the present study, the
downregulation of miR-124-3p contributed to the inhibition of the
mTOR signaling pathway, which serves an important role in MDD.
Therefore, miR‑124‑3p may have a cascade amplification effect in
the occurrence of MDD by regulating DDIT4 expression and mTOR
signaling, and so the fold-change is not the only guide for gene
function in the occurrence of disease. The minor downregulation of
miR-124-3p may have stronger biological effects than other notable
differentially expressed genes. In addition, the search for the
main genetic effects in MDD so far has not revealed consistent or
replicated significant findings (50), implying that it is difficult to
identify the main causative genes in MDD, and so the weak
expression differences may be an important feature of MDD.
In the present study, there were limited numbers of
controls and MDD subjects in the GEO database. Perhaps, it is
difficult to collect post-mortem brain tissues. Furthermore, the
incidence of MDD is relatively low in the population (51), which makes it harder to collect
data on patients with MDD. Efforts should be made to collect more
samples in follow-up work.
Previous studies have reported decreased levels of
the mTOR complex 1 (mTORC1) signaling pathway in post-mortem PFC
samples of subjects with MDD (52) and mTOR signaling was correlated
with neuronal brain function in MDD (50). Notably, DDIT4, a critical mediator
of the mTOR signaling pathway in MDD, was a validated target gene
of miR-124-3p. In addition, SP1, which is a transcription factor of
DDIT4, was also a validated target gene of miR-124-3p. miR-124-3p
may regulate expression of DDIT4 by directly targeting DDIT4 and
SP1, which is a transcription factor of DDIT4. To further
investigate the relationship between the mTORC1 signaling pathway
and miR-124-3p, GSEA was performed. The present results indicated
that the miR-124-3p expression level was positively associated with
activation of mTOR pathway. In addition, the regulatory effect of
miR-124-3p on the mTOR signaling pathway is dependent on the TSC1/2
complex. These results revealed that miR-124-3p may regulate the
mTOR signaling pathway by reducing the expression of DDIT4, which
stabilizes the complex formed by TSC1 and TSC2; this complex
subsequently inhibits mTORC1-dependent protein synthesis and cell
growth (50). Research has
demonstrated that miR-124-3p was correlated with cell growth,
proliferation and apoptosis, as well as the mTOR pathway (45). Diagnostically, MDD is associated
with smaller regional brain volumes (53). In addition, the mTOR pathway
serves an important role in neurodevelopment of the cerebral
neocortex and neurodevelopmental diseases (54). Similarly, the results of the
present study indicated that the most aberrant biological process
is the development process in males with MDD. These results
suggested that miR-124-3p may have an important role in these
biological processes.
In a previous study, it was reported that two CpG
islands were identified in the miR-124-3 gene promoter, and that
the promoter methylation level determined the miR-124-3p expression
level in corticosterone-treated rats (44). The present results concerning the
human miR-124-3 gene promoter CpG islands were different to the
finding in this previous study. Similarly, we used MethPrimer
(urogene.org/methprimer/) to predict CpG
islands of miR-124-3 gene promoter in humans. No CpG islands were
identified in the human miR-124-3 gene promoter (data not shown).
According to the present GO analysis results, the majority of
differentially expressed genes were transcription factors or had
transcription factor activity, suggesting that downregulation of
some transcription factors may be one of the prime reasons of
miR-124-3p downregulation. Further bioinformatics analysis revealed
that ARNT was a potential transcription factor that could regulate
the expression of miR-124. To the best of our knowledge, this is a
novel mechanistic study on the regulation of miR-124
expression.
In conclusion, the results of the present study
suggested that miR-124-3p was involved in the pathogenesis and
treatment of MDD in males via regulation of DDIT4 expression and
the mTOR signaling pathway. Reduced expression levels of miR-124-3p
resulted in a reduced ARNT level in MDD. These findings provide
novel insight into the mechanism of MDD occurrence and potential
antidepressant treatment.
Acknowledgments
The present study was supported by the Chinese
National Natural Science Foundation Projects (grant no. 31670774),
Beijing Nova Program (grant no. Z161100004916059) and Support
Project of High-level Teachers in Beijing Municipal Universities in
the Period of 13th Five-year Plan (grant no.
CIT&TCD201704097).
Abbreviations:
|
MDD
|
major depressive disorder
|
|
mTOR
|
mammalian target of rapamycin
|
|
DDIT4
|
DNA damage-inducible transcript 4
protein
|
|
TSC1/2
|
tuberous sclerosis proteins 1/2
|
|
ARNT
|
aryl hydrocarbon receptor nuclear
translocator
|
References
|
1
|
Merikangas KR, Akiskal HS, Angst J,
Greenberg PE, Hirschfeld RM, Petukhova M and Kessler RC: Lifetime
and 12-month prevalence of bipolar spectrum disorder in the
National Comorbidity Survey replication. Arch Gen Psychiatry.
64:543–552. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
World Health Organization: Global burden
of disease 2004 update: Disability weights for diseases and
conditions. WHO; Geneva, Switzerland: 2004, Available at:
http://www.who.int/healthinfo/global_burden_disease/GBD2004_DisabilityWeights.pdf.
|
|
3
|
Seedat S, Scott KM, Angermeyer MC,
Berglund P, Bromet EJ, Brugha TS, Demyttenaere K, de Girolamo G,
Haro JM, Jin R, et al: Cross-national associations between gender
and mental disorders in the World Health Organization World Mental
Health Surveys. Arch Gen Psychiatry. 66:785–795. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Otte C, Gold SM, Penninx BW, Pariante CM,
Etkin A, Fava M, Mohr DC and Schatzberg AF: Major depressive
disorder. Nat Rev Dis Primers. 2:160652016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cheng LC, Pastrana E, Tavazoie M and
Doetsch F: miR-124 regulates adult neurogenesis in the
subventricular zone stem cell niche. Nat Neurosci. 12:399–408.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pfau ML, Purushothaman I, Feng J, Golden
SA, Aleyasin H, Lorsch ZS, Cates HM, Flanigan ME, Menard C,
Heshmati M, et al: Integrative analysis of sex‑specific microRNA
networks following stress in mouse nucleus accumbens. Front Mol
Neurosci. 9:1442016. View Article : Google Scholar
|
|
7
|
Krol J, Loedige I and Filipowicz W: The
widespread regulation of microRNA biogenesis, function and decay.
Nat Rev Genet. 11:597–610. 2010.PubMed/NCBI
|
|
8
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Schratt G: Fine-tuning neural gene
expression with microRNAs. Curr Opin Neurobiol. 19:213–219. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ul Hussain M: Micro-RNAs (miRNAs): Genomic
organisation, biogenesis and mode of action. Cell Tissue Res.
349:405–413. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Forero DA, van der Ven K, Callaerts P and
Del-Favero J: miRNA genes and the brain: Implications for
psychiatric disorders. Hum Mutat. 31:1195–1204. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Miller BH and Wahlestedt C: MicroRNA
dysregulation in psychiatric disease. Brain Res. 1338:89–99. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Moreau MP, Bruse SE, David-Rus R, Buyske S
and Brzustowicz LM: Altered microRNA expression profiles in
postmortem brain samples from individuals with schizophrenia and
bipolar disorder. Biol Psychiatry. 69:188–193. 2011. View Article : Google Scholar :
|
|
14
|
Chan AW and Kocerha J: The Path to
microRNA therapeutics in psychiatric and neurodegenerative
disorders. Front Genet. 3:822012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Dias C, Feng J, Sun H, Shao NY,
Mazei-Robison MS, Damez-Werno D, Scobie K, Bagot R, LaBonté B,
Ribeiro E, et al: β-catenin mediates stress resilience through
Dicer1/microRNA regulation. Nature. 516:51–55. 2014.PubMed/NCBI
|
|
16
|
Wingo AP, Almli LM, Stevens JS, Klengel T,
Uddin M, Li Y, Bustamante AC, Lori A, Koen N and Stein DJ: DICER1
and microRNA regulation in post-traumatic stress disorder with
comorbid depression. Nat Commun. 6:101062015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dwivedi Y: Pathogenetic and therapeutic
applications of microRNAs in major depressive disorder. Prog
Neuropsychopharmacol Biol Psychiatry. 64:341–348. 2016. View Article : Google Scholar
|
|
18
|
Kuss AW and Chen W: MicroRNAs in brain
function and disease. Curr Neurol Neurosci Rep. 8:190–197. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dwivedi Y: Evidence demonstrating role of
microRNAs in the etiopathology of major depression. J Chem
Neuroanat. 42:142–156. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ha TY: MicroRNAs in human diseases: From
autoimmune diseases to skin, psychiatric and neurodegenerative
diseases. Immune Netw. 11:227–244. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mouillet-Richard S, Baudry A, Launay JM
and Kellermann O: MicroRNAs and depression. Neurobiol Dis.
46:272–278. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Smalheiser NR, Lugli G, Rizavi HS, Torvik
VI, Turecki G and Dwivedi Y: MicroRNA expression is down-regulated
and reorganized in prefrontal cortex of depressed suicide subjects.
PLoS One. 7:e332012012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fineberg SK, Kosik KS and Davidson BL:
MicroRNAs potentiate neural development. Neuron. 64:303–309. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lagos-Quintana M, Rauhut R, Yalcin A,
Meyer J, Lendeckel W and Tuschl T: Identification of
tissue‑specific microRNAs from mouse. Curr Biol. 12:735–739. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lopez JP, Lim R, Cruceanu C, Crapper L,
Fasano C, Labonte B, Maussion G, Yang JP, Yerko V, Vigneault E, et
al: miR-1202 is a primate‑specific and brain‑enriched microRNA
involved in major depression and antidepressant treatment. Nat Med.
20:764–768. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xu Y, Liu H, Li F, Sun N, Ren Y, Liu Z,
Cao X, Wang Y, Liu P and Zhang K: A polymorphism in the
microRNA-30e precursor associated with major depressive disorder
risk and P300 waveform. J Affect Disord. 127:332–336. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Saus E, Soria V, Escaramís G, Vivarelli F,
Crespo JM, Kagerbauer B, Menchón JM, Urretavizcaya M, Gratacòs M
and Estivill X: Genetic variants and abnormal processing of
pre-miR-182, a circadian clock modulator, in major depression
patients with late insomnia. Hum Mol Genet. 19:4017–4025. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Meerson A, Cacheaux L, Goosens KA,
Sapolsky RM, Soreq H and Kaufer D: Changes in brain MicroRNAs
contribute to cholinergic stress reactions. Journal of molecular
neuroscience: MN. 40:47–55. 2010. View Article : Google Scholar :
|
|
29
|
Vreugdenhil E, Verissimo CS, Mariman R,
Kamphorst JT, Barbosa JS, Zweers T, Champagne DL, Schouten T,
Meijer OC, de Kloet ER and Fitzsimons CP: MicroRNA 18 and 124a
down-regulate the glucocorticoid receptor: Implications for
glucocorticoid responsiveness in the brain. Endocrinology.
150:2220–2228. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets–update. Nucleic Acids Res. 41:D991–D995. 2013.
View Article : Google Scholar
|
|
31
|
Dweep H, Sticht C, Pandey P and Gretz N:
miRWalk–database: Prediction of possible miRNA binding sites by
'walking' the genes of three genomes. J Biomed Inform. 44:839–847.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kaufman J, Sullivan GM, Yang J, Ogden RT,
Miller JM, Oquendo MA, Mann JJ, Parsey RV and DeLorenzo C:
Quantification of the serotonin 1A receptor using PET:
Identification of a potential biomarker of major depression in
males. Neuropsychopharmacology. 40:1692–1699. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zheng P, Chen JJ, Zhou CJ, Zeng L, Li KW,
Sun L, Liu ML, Zhu D, Liang ZH and Xie P: Identification of
sex-specific urinary biomarkers for major depressive disorder by
combined application of NMR- and GC-MS-based metabonomics. Transl
Psychiatry. 6:e9552016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Belzeaux R, Bergon A, Jeanjean V, Loriod
B, Formisano-Tréziny C, Verrier L, Loundou A, Baumstarck-Barrau K,
Boyer L, Gall V, et al: Responder and nonresponder patients exhibit
different peripheral transcriptional signatures during major
depressive episode. Transl Psychiatry. 2:e1852012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ota KT, Liu RJ, Voleti B, Maldonado-Aviles
JG, Duric V, Iwata M, Dutheil S, Duman C, Boikess S, Lewis DA, et
al: REDD1 is essential for stress-induced synaptic loss and
depressive behavior. Nat Med. 20:531–535. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Mainio A, Tuunanen S, Hakko H, Niemelä A,
Koivukangas J and Räsänen P: Decreased quality of life and
depression as predictors for shorter survival among patients with
low-grade gliomas: A follow-up from 1990 to 2003. Eur Arch
Psychiatry Clin Neurosci. 256:516–521. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Spiegel D and Giese-Davis J: Depression
and cancer: Mechanisms and disease progression. Biol Psychiatry.
54:269–282. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Horrobin DF and Bennett CN: Depression and
bipolar disorder: Relationships to impaired fatty acid and
phospholipid metabolism and to diabetes, cardiovascular disease,
immunological abnormalities, cancer, ageing and osteoporosis.
Possible candidate genes. Prostaglandins Leukot Essent Fatty Acids.
60:217–234. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Seznec J, Silkenstedt B and Naumann U:
Therapeutic effects of the Sp1 inhibitor mithramycin A in
glioblastoma. J Neurooncol. 101:365–377. 2011. View Article : Google Scholar
|
|
40
|
Elyakim E, Sitbon E, Faerman A, Tabak S,
Montia E, Belanis L, Dov A, Marcusson EG, Bennett CF, Chajut A, et
al: hsa-miR-191 is a candidate oncogene target for hepatocellular
carcinoma therapy. Cancer Res. 70:8077–8087. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Nebert DW, Dalton TP, Okey AB and Gonzalez
FJ: Role of aryl hydrocarbon receptor-mediated induction of the
CYP1 enzymes in environmental toxicity and cancer. J Biol Chem.
279:23847–23850. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Song MF, Dong JZ, Wang YW, He J, Ju X,
Zhang L, Zhang YH, Shi JF and Lv YY: CSF miR-16 is decreased in
major depression patients and its neutralization in rats induces
depression-like behaviors via a serotonin transmitter system. J
Affect Disord. 178:25–31. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Makeyev EV, Zhang J, Carrasco MA and
Maniatis T: The MicroRNA miR-124 promotes neuronal differentiation
by triggering brain‑specific alternative pre‑mRNA splicing. Mol
Cell. 27:435–448. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Roy B, Dunbar M, Shelton RC and Dwivedi Y:
Identification of MicroRNA-124-3p as a putative epigenetic
signature of major depressive disorder. Neuropsychopharmacology.
42:864–875. 2017. View Article : Google Scholar
|
|
45
|
Bondarenko EA, Shadrina MI, Grishkina MN,
Druzhkova TA, Akzhigitov RG, Gulyaeva NV, Guekht AB and Slominsky
PA: Genetic analysis of BDNF, GNB3, MTHFR, ACE and APOE variants in
major and recurrent depressive disorders in Russia. Int J Med Sci.
13:977–983. 2016. View Article : Google Scholar :
|
|
46
|
He S, Liu X, Jiang K, Peng D, Hong W, Fang
Y, Qian Y, Yu S and Li H: Alterations of microRNA-124 expression in
peripheral blood mononuclear cells in pre- and post-treatment
patients with major depressive disorder. J Psychiatr Res. 78:65–71.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Bahi A, Chandrasekar V and Dreyer JL:
Selective lentiviral-mediated suppression of microRNA124a in the
hippocampus evokes antidepressants-like effects in rats.
Psychoneuroendocrinology. 46:78–87. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Cao MQ, Chen DH, Zhang CH and Wu ZZ:
Screening of specific microRNA in hippocampus of depression model
rats and intervention effect of Chaihu Shugan San. Zhongguo Zhong
Yao Za Zhi. 38:1585–1589. 2013.In Chinese. PubMed/NCBI
|
|
49
|
Alimonti A, Carracedo A, Clohessy JG,
Trotman LC, Nardella C, Egia A, Salmena L, Sampieri K, Haveman WJ,
Brogi E, et al: Subtle variations in Pten dose determine cancer
susceptibility. Nat Genet. 42:454–458. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Bosker FJ, Hartman CA, Nolte IM, Prins BP,
Terpstra P, Posthuma D, van Veen T, Willemsen G, DeRijk RH, de Geus
EJ, et al: Poor replication of candidate genes for major depressive
disorder using genome-wide association data. Mol Psychiatry.
16:516–532. 2011. View Article : Google Scholar
|
|
51
|
Ferrari AJ, Charlson FJ, Norman RE,
Flaxman AD, Patten SB, Vos T and Whiteford HA: The epidemiological
modelling of major depressive disorder: Application for the Global
Burden of Disease Study 2010. PLoS One. 8:e696372013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Bocchio‑Chiavetto L, Maffioletti E,
Bettinsoli P, et al: Blood microRNA changes in depressed patients
during antidepressant treatment. Eur Neuropsychopharmacol.
23:602–611. 2013. View Article : Google Scholar
|
|
53
|
Kempton MJ, Salvador Z, Munafò MR, Geddes
JR, Simmons A, Frangou S and Williams SC: Structural neuroimaging
studies in major depressive disorder. Meta-analysis and comparison
with bipolar disorder. Arch Gen Psychiatry. 68:675–690. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Fan HM, Sun XY, Guo W, Zhong AF, Niu W,
Zhao L, Dai YH, Guo ZM, Zhang LY and Lu J: Differential expression
of microRNA in peripheral blood mononuclear cells as specific
biomarker for major depressive disorder patients. J Psychiatr Res.
59:45–52. 2014. View Article : Google Scholar : PubMed/NCBI
|