|
1
|
Imamura M and Maeda S: Genetics of type 2
diabetes: The GWAS era and future perspectives (Review). Endocr J.
58:723–739. 2011. View Article : Google Scholar
|
|
2
|
Yasuda K, Miyake K, Horikawa Y, Hara K,
Osawa H, Furuta H, Hirota Y, Mori H, Jonsson A, Sato Y, et al:
Variants in KCNQ1 are associated with susceptibility to type 2
diabetes mellitus. Nat Genet. 40:1092–1097. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Unoki H, Takahashi A, Kawaguchi T, Hara K,
Horikoshi M, Andersen G, Ng DP, Holmkvist J, Borch-Johnsen K,
Jørgensen T, et al: SNPs in KCNQ1 are associated with
susceptibility to type 2 diabetes in East Asian and European
populations. Nat Genet. 40:1098–1102. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Voight BF, Scott LJ, Steinthorsdottir V,
Morris AP, Dina C, Welch RP, Zeggini E, Huth C, Aulchenko YS,
Thorleifsson G, et al: MAGIC investigators; GIANT Consortium:
Twelve type 2 diabetes susceptibility loci identified through
large-scale association analysis. Nat Genet. 42:579–589. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yamagata K, Senokuchi T, Lu M, Takemoto M,
Fazlul Karim M, Go C, Sato Y, Hatta M, Yoshizawa T, Araki E, et al:
Voltage-gated K+ channel KCNQ1 regulates insulin
secretion in MIN6 β-cell line. Biochem Biophys Res Commun.
407:620–625. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sanyal A, Lajoie BR, Jain G and Dekker J:
The long-range interaction landscape of gene promoters. Nature.
489:109–113. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Maurano MT, Humbert R, Rynes E, Thurman
RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, et
al: Systematic localization of common disease-associated variation
in regulatory DNA. Science. 337:1190–1195. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Smemo S, Tena JJ, Kim KH, Gamazon ER,
Sakabe NJ, Gómez-Marín C, Aneas I, Credidio FL, Sobreira DR,
Wasserman NF, et al: Obesity-associated variants within FTO form
long-range functional connections wit IRX3. Nature. 507:371–375.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pasquali L, Gaulton KJ, Rodríguez-Seguí
SA, Mularoni L, Miguel-Escalada I, Akerman İ, Tena JJ, Morá I,
Gómez-Marín C and van de Bunt M: Pancreatic islet enhancer clusters
enriched in type 2 diabetes risk-associated variants. Nat Genet.
46:136–143. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kassem SA, Ariel I, Thornton PS, Hussain
K, Smith V, Lindley KJ, Aynsley-Green A and Glaser B: 57 (KIP2)
expression in normal islet cells and in hyperinsulinism of infancy.
Diabetes. 50:2763–2769. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Avrahami D, Li C, Yu M, Jiao Y, Zhang J,
Naji A, Ziaie S, Glaser B and Kaestner KH: Targeting the cell cycle
inhibitor 57Kip2 promotes adult human β cell replication. J Clin
Invest. 124:670–674. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Horike S, Mitsuya K, Meguro M, Kotobuki N,
Kashiwagi A, Notsu T, Schulz TC, Shirayoshi Y and Oshimura M:
Targeted disruption of the human LIT1 locus defines a putative
imprinting control element playing an essential role in
Beckwith-Wiedemann syndrome. Hum Mol Genet. 9:2075–2083. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fitzpatrick GV, Soloway PD and Higgins MJ:
Regional loss of imprinting and growth deficiency in mice with a
targeted deletion of KvDMR1. Nat Genet. 32:426–431. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Arima T, Kamikihara T, Hayashida T, Kato
K, Inoue T, Shirayoshi Y, Oshimura M, Soejima H, Mukai T and Wake
N: ZAC, LIT1 (KCNQ1OT1) and 57KIP2 (CDKN1C) are in an imprinted
gene network that may play a role in Beckwith-Wiedemann syndrome.
Nucleic Acids Res. 33:2650–2660. 2005. View Article : Google Scholar :
|
|
15
|
Asahara S, Etoh H, Inoue H, Teruyama K,
Shibutani Y, Ihara Y, Kawada Y, Bartolome A, Hashimoto N, Matsuda
T, et al: Paternal allelic mutation at the Kcnq1 locus reduces
pancreatic β-cell mass by epigenetic modification oCdkn1c. Proc
Natl Acad Sci USA. 112:8332–8337. 2015. View Article : Google Scholar
|
|
16
|
Hiramoto M, Udagawa H, Watanabe A,
Miyazawa K, Ishibashi N, Kawaguchi M, Uebanso T, Nishimura W, Nammo
T and Yasuda K: Comparative analysis of type 2 diabetes-associated
SNP alleles identifies allele-specific DNA-binding proteins for the
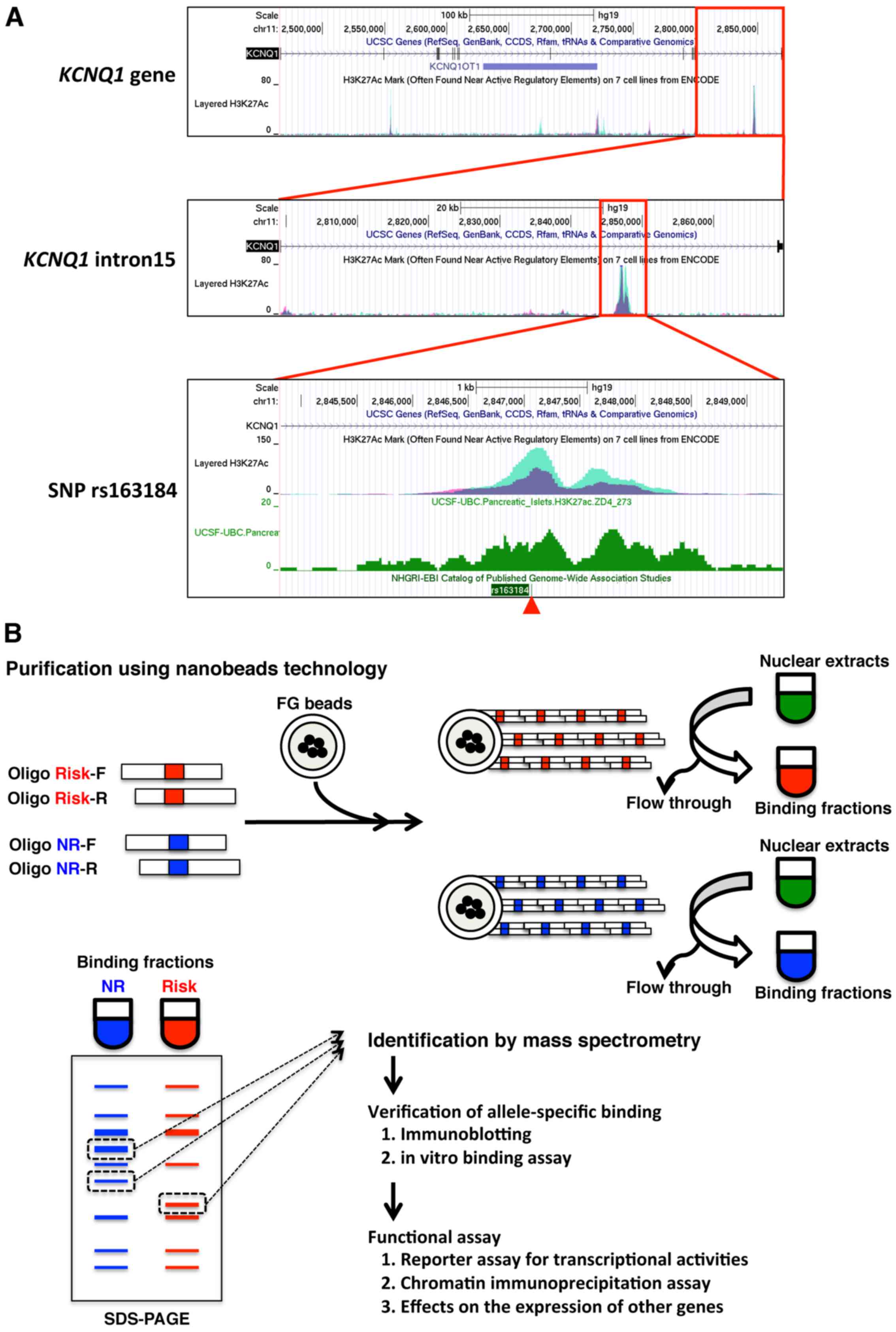
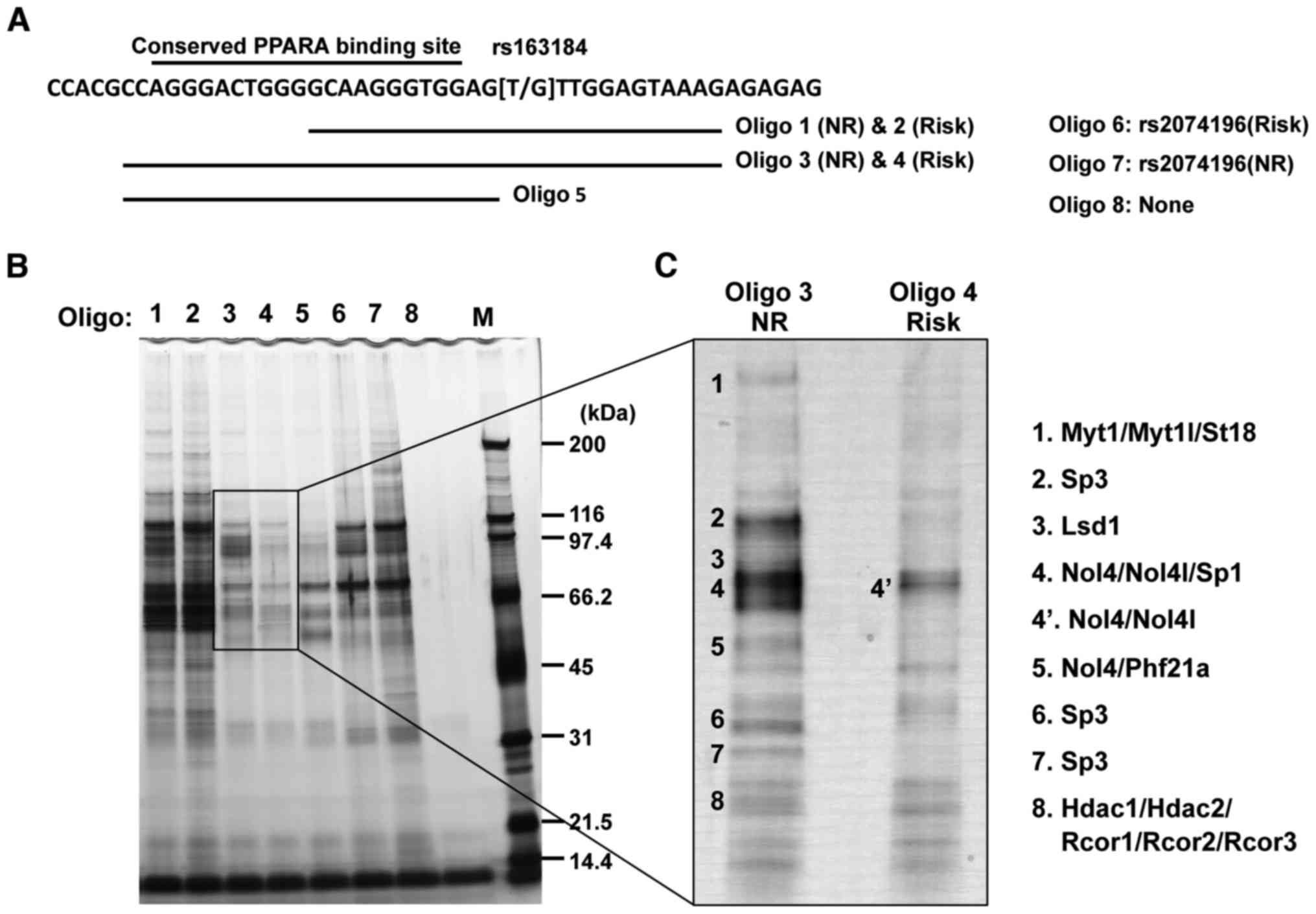
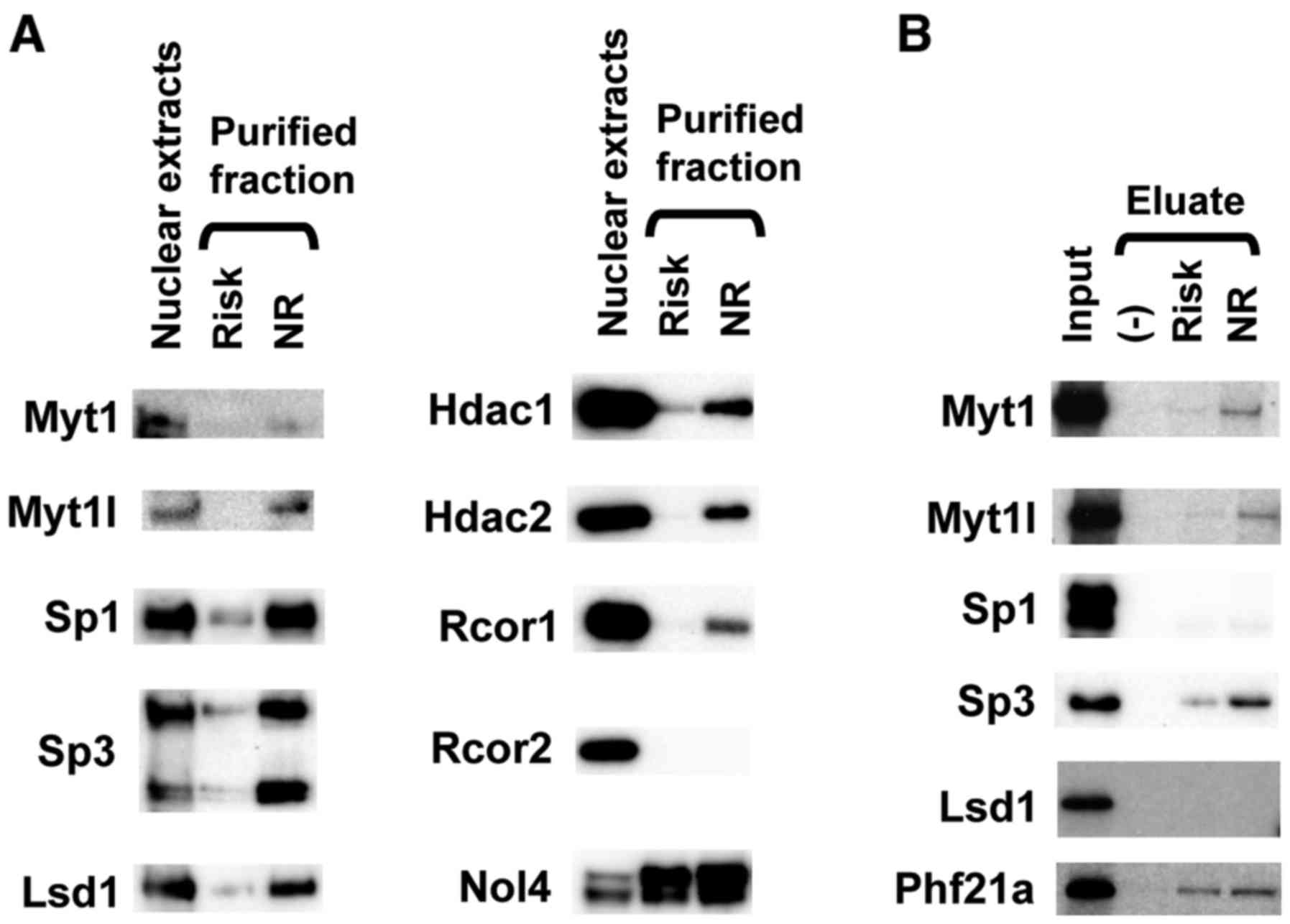
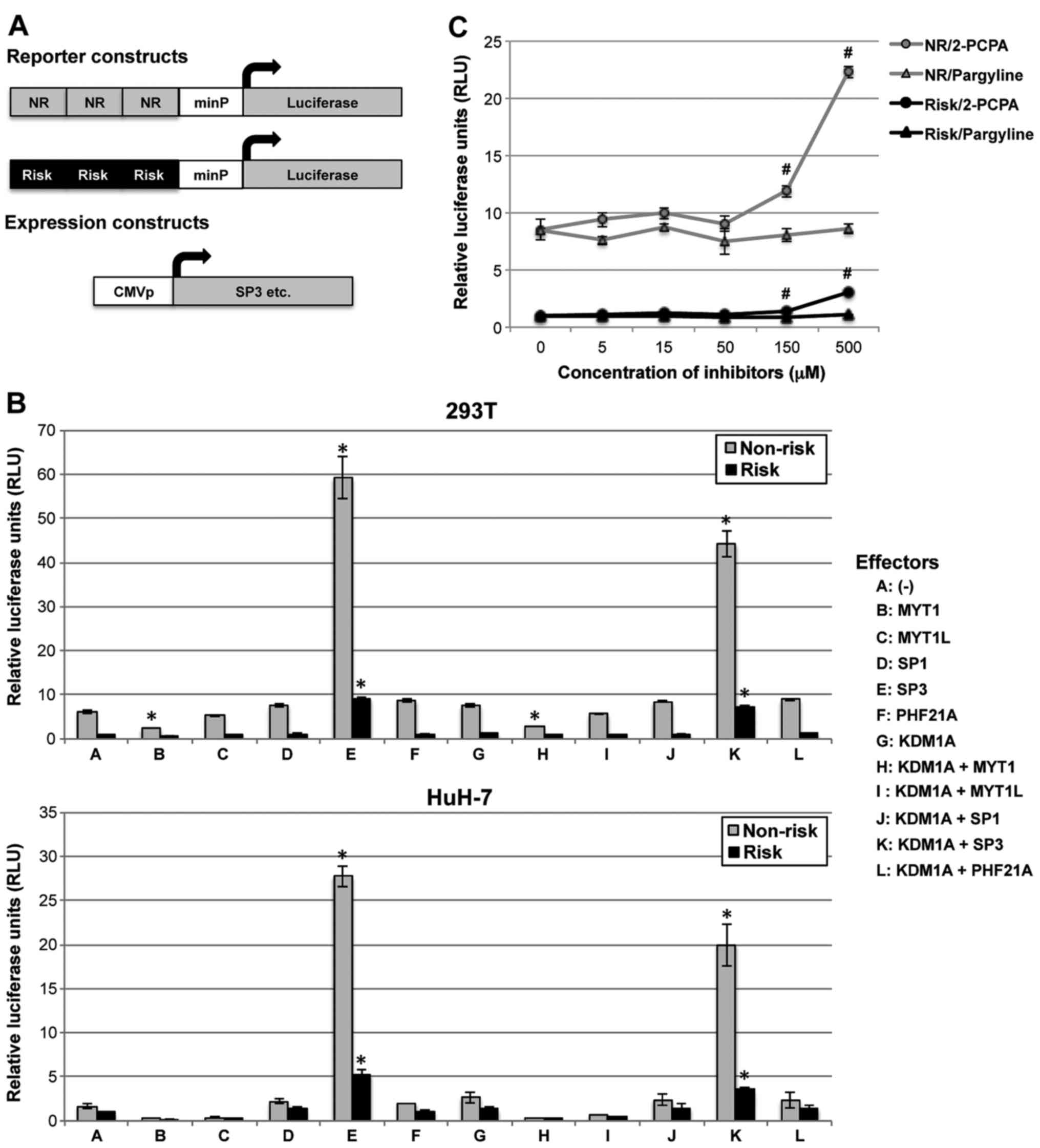
KCNQ1 locus. Int J Mol Med. 36:222–230. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dunham I, Kundaje A, Aldred SF, Collins
PJ, Davis CA, Doyle F, Epstein CB, Frietze S, Harrow J, Kaul R, et
al: ENCODE Project Consortium: An integrated encyclopedia of DNA
elements in the human genome. Nature. 489:57–74. 2012. View Article : Google Scholar
|
|
18
|
Bhaumik SR, Smith E and Shilatifard A:
Covalent modifications of histones during development and disease
pathogenesis. Nat Struct Mol Biol. 14:1008–1016. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Asfari M, Janjic D, Meda P, Li G, Halban
PA and Wollheim CB: Establishment of 2-mercaptoethanol-dependent
differentiated insulin-secreting cell lines. Endocrinology.
130:167–178. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dignam JD, Lebovitz RM and Roeder RG:
Accurate transcription initiation by RNA polymerase II in a soluble
extract from isolated mammalian nuclei. Nucleic Acids Res.
11:1475–1489. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shevchenko A, Wilm M, Vorm O and Mann M:
Mass spectrometric sequencing of proteins silver-stained
polyacrylamide gels. Anal Chem. 68:850–858. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hiramoto M, Maekawa N, Kuge T, Ayabe F,
Watanabe A, Masaike Y, Hatakeyama M, Handa H and Imai T:
High-performance affinity chromatography method for identification
of L-arginine interacting factors using magnetic nanobeads. Biomed
Chromatogr. 24:606–612. 2010.
|
|
23
|
Takahashi E, Okumura A, Unoki-Kubota H,
Hirano H, Kasuga M and Kaburagi Y: Differential proteome analysis
of serum proteins associated with the development of type 2
diabetes mellitus in the KK-Ay mouse model using the
iTRAQ technique. J Proteomics. 84:40–51. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kersey PJ, Duarte J, Williams A,
Karavidopoulou Y, Birney E and Apweiler R: The International
Protein Index: An integrated database for proteomics experiments.
Proteomics. 4:1985–1988. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
26
|
Kerns SL, Guevara-Aguirre J, Andrew S,
Geng J, Guevara C, Guevara-Aguirre M, Guo M, Oddoux C, Shen Y,
Zurita A, et al: A novel variant in CDKN1C is associated with
intrauterine growth restriction, short stature, and
early-adulthood-onset diabetes. J Clin Endocrinol Metab.
99:E2117–E2122. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cucciolla V, Borriello A, Criscuolo M,
Sinisi AA, Bencivenga D, Tramontano A, Scudieri AC, Oliva A, Zappia
V and Della Ragione F: Histone deacetylase inhibitors upregulate
57Kip2 level by enhancing its expression through Sp1 transcription
factor. Carcinogenesis. 29:560–567. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Figliola R, Busanello A, Vaccarello G and
Maione R: Regulation of 57(KIP2) during muscle differentiation:
Role of Egr1, Sp1 and DNA hypomethylation. J Mol Biol. 380:265–277.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shi YJ, Matson C, Lan F, Iwase S, Baba T
and Shi Y: Regulation of LSD1 histone demethylase activity by its
associated factors. Mol Cell. 19:857–864. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Vasilatos SN, Katz TA, Oesterreich S, Wan
Y, Davidson NE and Huang Y: Crosstalk between lysine-specific
demethylase 1 (LSD1) and histone deacetylases mediates
antineoplastic efficacy of HDAC inhibitors in human breast cancer
cells. Carcinogenesis. 34:1196–1207. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ross S, Best JL, Zon LI and Gill G: SUMO-1
modification represses Sp3 transcriptional activation and modulates
its subnuclear localization. Mol Cell. 10:831–842. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Rosendorff A, Sakakibara S, Lu S, Kieff E,
Xuan Y, DiBacco A, Shi Y, Shi Y and Gill GX: P-2 association with
SUMO-2 depends on lysines required for transcriptional repression.
Proc Natl Acad Sci USA. 103:5308–5313. 2006. View Article : Google Scholar
|
|
33
|
Ben-Shushan E, Marshak S, Shoshkes M,
Cerasi E and Melloul D: A pancreatic beta-cell-specific enhancer in
the human PDX-1 gene is regulated by hepatocyte nuclear factor
3beta (HNF-3beta), HNF-1alpha, and SPs transcription factors. J
Biol Chem. 276:17533–17540. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Geiger A, Salazar G and Kervran A: Role of
the Sp family of transcription factors on glucagon receptor gene
expression. Biochem Biophys Res Commun. 285:838–844. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hashimoto T, Nakamura T, Maegawa H, Nishio
Y, Egawa K and Kashiwagi A: Regulation of ATP-sensitive potassium
channel subunit Kir6.2 expression in rat intestinal
insulin-producing progenitor cells. J Biol Chem. 280:1893–1900.
2005. View Article : Google Scholar
|
|
36
|
Baldacchino V, Oble S, Décarie PO,
Bourdeau I, Hamet P, Tremblay J and Lacroix A: The Sp transcription
factors are involved in the cellular expression of the human
glucose-dependent insulinotropic polypeptide receptor gene and
overexpressed in adrenals of patients with Cushing's syndrome. J
Mol Endocrinol. 35:61–71. 2005. View Article : Google Scholar : PubMed/NCBI
|