Introduction
Lung cancer is one of the most devastating
malignancies as well as the leading cause of cancer-related
morbidity and mortality globally (1). Non-small cell lung cancer (NSCLC)
accounts for up to 85% of all lung cancers and has a 5-year
survival rate of ~15% (2).
Despite multiple advances in therapeutic options over the years, at
present, platinum-based chemotherapy remains the mainstay of
adjuvant or first-line chemotherapy in NSCLC treatment (3,4).
Cisplatin is the most widely used platinum agent due to its
therapeutic advantages. Compared with carboplatin-based
chemotherapy, cisplatin-based chemotherapy is slightly superior in
terms of response rate and in prolonging survival without being
associated with an increase in severe toxic effects (5). Nevertheless, the clinical response
in patients is frequently compromised by drug resistance, either
intrinsic or acquired, resulting in chemotherapeutic failure and
tumor relapse (6,7). Although researchers have delved into
the modulation of cancer drug resistance at the molecular level,
multiple mechanisms underlying cisplatin resistance require further
clarification.
MicroRNAs (miRNAs) are endogenous single-stranded
non-coding small RNAs ~22 nucleotides in length, which regulate
gene expression at the post-transcriptional level through binding
to the 3′ untranslated region (3′-UTR) of target messenger RNA
(mRNA) (8). miRNAs often exhibit
aberrant expression in human malignancies and function as either
oncogenes or tumor suppressors depending on the biological roles of
their target genes (9).
Dysregulation of miRNA is involved in a wide range of biological
processes, including cell proliferation, apoptosis and metastasis
(10,11). Recent studies have indicated that
miRNAs play a role in chemoresistance to cancer treatment and are
implicated in various biological and pathological processes of
tumor cells (12,13).
miRNA-133b has been indicated to be downregulated in
several types of tumors, including gastric, prostate and colorectal
cancers (14–16). Furthermore, the loss of miR-133b
expression has been demonstrate to correlate with worse survival
time in patients with lung cancer (17); however, to the best of our
knowledge, there have not yet been any studies investigating its
role in the regulation of the biological functions of
drug-resistant lung cancer cells. In the present study, whether
miR-133b modulates chemotherapy resistance, carcinogenesis and
metastasis in cisplatin-resistant lung cancer cells was explored.
The possible target for miR-133b was also predicted and verified in
order to elucidate the effect of the interaction between this miRNA
and its target gene on the biological activity of lung cancer
cells.
Materials and methods
Cell culture
The human lung cancer cell lines A549 and H1299 were
obtained from the Cell Bank of Type Culture Collection of Chinese
Academy of Sciences (Shanghai, China). Cisplatin-resistant A549
(A549/DDP) cells were purchased from the Academy of Military
Medical Sciences (Beijing, China), and cisplatin-resistant H1299
(H1299/DDP) cells were provided by the Institute of Lung Diseases,
Xinqiao Hospital, Third Military Medical University (Chongqing,
China). Cells were cultured in RPMI-1640 (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal
bovine serum (Gibco; Thermo Fisher Scientific, Inc.) at 37°C in a
humidified atmosphere with 5% CO2. The culture medium
for the A549/DDP and H1299/DDP cells included 1 µg/ml
cisplatin to maintain the drug-resistant phenotype. Two days before
each experiment, the culture medium was replaced by fresh medium
without cisplatin to avoid the influence of the drug.
Drugs and antibodies
Cisplatin was purchased from Selleck Chemicals
(Houston, TX, USA). Antibodies against glutathione-S-transferase P1
(GSTP1; cat. no. 3369; 1:1,000), survivin (cat. no. 2808; 1:1,000),
B-cell lymphoma 2 (Bcl-2; cat. no. 15071; 1:1,000),
Bcl-2-associated X protein (Bax; cat. no. 2774; 1:1,000), matrix
metalloproteinase (MMP)-2 (cat. no. 87809; 1:1,000), MMP-3 (cat.
no. 14351; 1:1,000), MMP-9 (cat. no. 13667; 1:1,000) and β-actin
(cat. no. 3700; 1:2,000) were purchased from Cell Signaling
Technology, Inc. (Danvers, MA, USA). Horseradish peroxidase
(HRP)-conjugated goat anti-rabbit and anti-mouse IgG (cat. nos.
111-035-003 and 115-035-003; each 1:10,000) antibodies were
obtained from Jackson ImmunoResearch Laboratories, Inc. (West
Grove, PA, USA).
Transient transfection
A549/DDP and H1299/DDP cells were seeded in 6-well
plates. At 24 h after plating, cells were transfected with 100 nmol
miR-133b mimic or negative control (NC) mimic (Guangzhou RiboBio
Co., Ltd., Guangzhou, China) using Lipofectamine 2000 (Invitrogen;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the
manufacturer’s protocol. The effects were examined at 24 h
post-transfection. The GSTP1 gene was knocked down using small
interfering RNA (siRNA; Guangzhou RiboBio Co., Ltd.). The sequence
of the GSTP1 siRNA was CCTACACCGTGGTCTATTT. The GSTP1-siRNA or
control-siRNA (cat. no. siN05815122147-1-5) was diluted in Opti-MEM
medium (Gibco, Thermo Fisher Scientific, Inc.) and transfected at a
final concentration of 100 nM using Lipofectamine 2000 in Opti-MEM
medium in a 1:1 ratio. The mixture was incubated for 15 min, then
added to fresh culture medium and incubated with the cells for 48
h. The transfected cells were then processed for subsequent
experiments.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Total cellular RNA was extracted using TRIzol
Reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The RNA
sample (500 ng) was then reverse transcribed into cDNA using a
PrimeScript™ RT Reagent kit (Takara Bio, Inc., Otsu, Japan) with
reaction at 37°C for 15 min, followed by 85°C for 5 sec and then
cooling to 4°C. For mRNA quantification, qPCR was performed using
SYBR Premix Ex Taq (Takara Bio, Inc.) with the Applied Biosystems
Prism 7900HT system (Applied Biosystems; Thermo Fisher Scientific,
Inc.). The thermocycling conditions were 1 cycle of 95°C for 5 min
and 40 cycles of 95°C for 5 sec and 60°C for 34 sec. The GSTP1
primers were as follows: Forward, 5′-CCTGTACCAGTCCAATACCATCCT-3′
and reverse, 5′-TCCTGCTGGTCCTTCCCATA-3′. Primers for miR-133b: RT,
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGAC-3′ (to replace the
RT Primer mix of the PrimeScript RT reagent kit), forward,
5′-CTTTGGTCCCCTTCAACCA-3′ and reverse, 5′-CTTTGGTCCCCTTCAACCA-3′.
Primers for U6: Forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse,
5′-AACGCTTCACGAATTTGCGT-3′. Primers for β-actin: Forward,
5′-GCACCACACCTTCTACAATGAGC-3′ and reverse,
5′-GGATAGCACAGCCTGGATAGCAAC-3′. The levels of mature miRNA-133b
expression were normalized to U6 and those of GSTP1 mRNA were
normalized to β-actin mRNA. The fold change in expression was
calculated using the 2−ΔΔCq method (18).
Dual-luciferase reporter assay
Using the web-based miRNA program TargetScan
(http://www.targetscan.org/), it was
predicted that hsa-miR-133b should bind with the 3′-UTR of GSTP1.
The wild-type (WT) and mutant 3′-UTR of GSTP1 were independently
cloned into a pGL3 luciferase vector (Promega Corporation, Madison,
WI, USA) containing the Renilla luciferase gene. The WT
sequence for the GSTP1 3′-UTR was
GGGTTGGGGGGACTCTGAGCGGGAGGCAGAGTTTGCCTTCCTTTCTCCAGGACCAATAAAATTTCTAAGAGAGCTA,
and the mutant sequence was
GGGTTGGGGGGACTCTGAGCGGGAGGCAGAGTTTGCCTTCCTTTCTCCATACTAGCTAAAATTTCTAAGAGAGCTA.
For the luciferase assay, HEK 293T cells (Cell Bank of Type Culture
Collection of Chinese Academy of Sciences, Shanghai, China) were
cotransfected with the miR-133b mimic or NC mimic and the
luciferase reporter plasmid using Lipofectamine 2000. At 48 h post
transfection, firefly luciferase activity was measured using a
Dual-Luciferase Reporter Assay System (Promega Corporation),
according to the manufacturer’s protocol. Firefly luciferase
activity was normalized to Renilla luciferase activity for
each well.
Colony formation assays
At 24 h after the transient transfection, 500 cells
were re-seeded in 6-well plates in triplicate. Following 10 days of
incubation, the colonies were fixed with 4% paraformaldehyde for 30
min and stained with 1% crystal violet for 2 h at room temperature.
The plates were then washed and dried before photographic images
were captured. The colony numbers were counted and the sizes of
colonies were observed.
Cell viability assays
In the growth inhibition assay, transfected cells
were seeded at a density of 8,000 cells/well in 96-well culture
plates and incubated overnight. Following cell adhesion, cisplatin
was applied at a series of concentrations (1, 2, 4, 8, 16, 32, 64
and 128 µM) and the cells were cultured for a further 48 h.
Cell survival was then assayed using a Cell Counting kit (CCK)-8
assay (Dojindo Molecular Technologies, Inc., Kumamoto, Japan). In
the cell proliferation assay, transfected cells were seeded at a
density of 5,000 cells/well, and CCK-8 assays were performed to
examine the cell viability following 24, 48 and 72 h of
culture.
Cell apoptosis analysis
Cells seeded in 6-well plates
(1×105/well) were transfected with miR-133b mimic or NC
mimic for 24 h, and then treated with 10 µM cisplatin for 48
h. Cell apoptosis was assessed using flow cytometry with staining
of the cells using an Annexin V/propidium iodide (PI) kit (cat. no.
556547; BD Biosciences, Franklin Lakes, NJ, USA). Briefly, cells
were collected and washed twice in ice-cold PBS. The washed cells
(2×105) were resuspended in 100 µl binding buffer
(included in the kit), and stained with 5 µl Annexin V and 5
µl PI. Following incubation for 15 min in the dark, flow
cytometry was performed. A flow cytometer (Cytomics FC 500 MPL;
Beckman Coulter, Inc., Brea, CA, USA) was utilized to evaluate the
apoptotic levels in each sample following the manufacturer’s
protocol.
Cell migration assay
Migration experiments were carried out in a chamber
with a BD Falcon™ Cell Culture insert (pore size, 8 µm; BD
Biosciences). In these experiments, 2×104 cells in 200
µl serum-free RPMI-1640 medium were added to the top chamber
of the insert and 600 µl 10% fetal bovine serum-containing
medium (Gibco; Thermo Fisher Scientific, Inc.) was added to the
lower chamber. Following 24 h incubation at 37°C, the cells that
had migrated to the lower chamber were fixed in 4% paraformaldehyde
for 30 min and stained with 1% crystal violet for 2 h at room
temperature. Stained cells were counted in five different fields in
each well under an inverted microscope at a magnification of ×200
(Olympus Corporation, Tokyo, Japan).
Western blot analysis
Total protein lysates were obtained from cultured
cells with a mixture of radioimmunoprecipitation assay buffer
(Beyotime Institute of Biotechnology, Shanghai, China), phosphatase
inhibitor cocktail and protease inhibitor cocktail (both BioTool
AG, Kirchberg, Switzerland). Protein concentrations were determined
using a BCA protein assay kit (Beyotime Institute of
Biotechnology). Cell extracts (20 µg/well) were separated by
10% sodium dodecyl sulfate polyacrylamide Tris-HCl gel
electrophoresis and transferred onto polyvinylidene fluoride
membranes. The membranes were then blocked with 5% skimmed milk in
Tris-buffered saline with Tween-20 (TBST) for 1 h at room
temperature and probed with primary antibodies against GSTP1,
survivin, Bcl-2, Bax, MMP-2, MMP-3, MMP-9 and β-actin overnight at
4°C. After washing with TBST, the membranes were incubated with
HRP-conjugated secondary antibody for 1 h at room temperature, then
washed three times with TBST. The proteins were visualized with
Pierce™ enhanced chemiluminescence reagents (Thermo Fisher
Scientific, Inc.) and detected using a luminescent image analyzer
(ImageQuant LAS4000 mini; GE Healthcare Life Sciences, Little
Chalfont, UK).
Statistical analysis
Statistical analysis was performed using GraphPad
Prism 5.0 software (GraphPad Software, Inc., La Jolla, CA, USA).
Differences between two groups were analyzed using the Student’s
t-test, one-way analysis of variance was applied for the comparison
of more than two groups, and post hoc tests were conducted using
the Bonferroni correction. P<0.05 was considered to indicate a
statistically significant difference.
Results
Expression of miR-133b and GSTP1 in
A549/DPP and H1299/DDP cells compared with their parental cell
lines
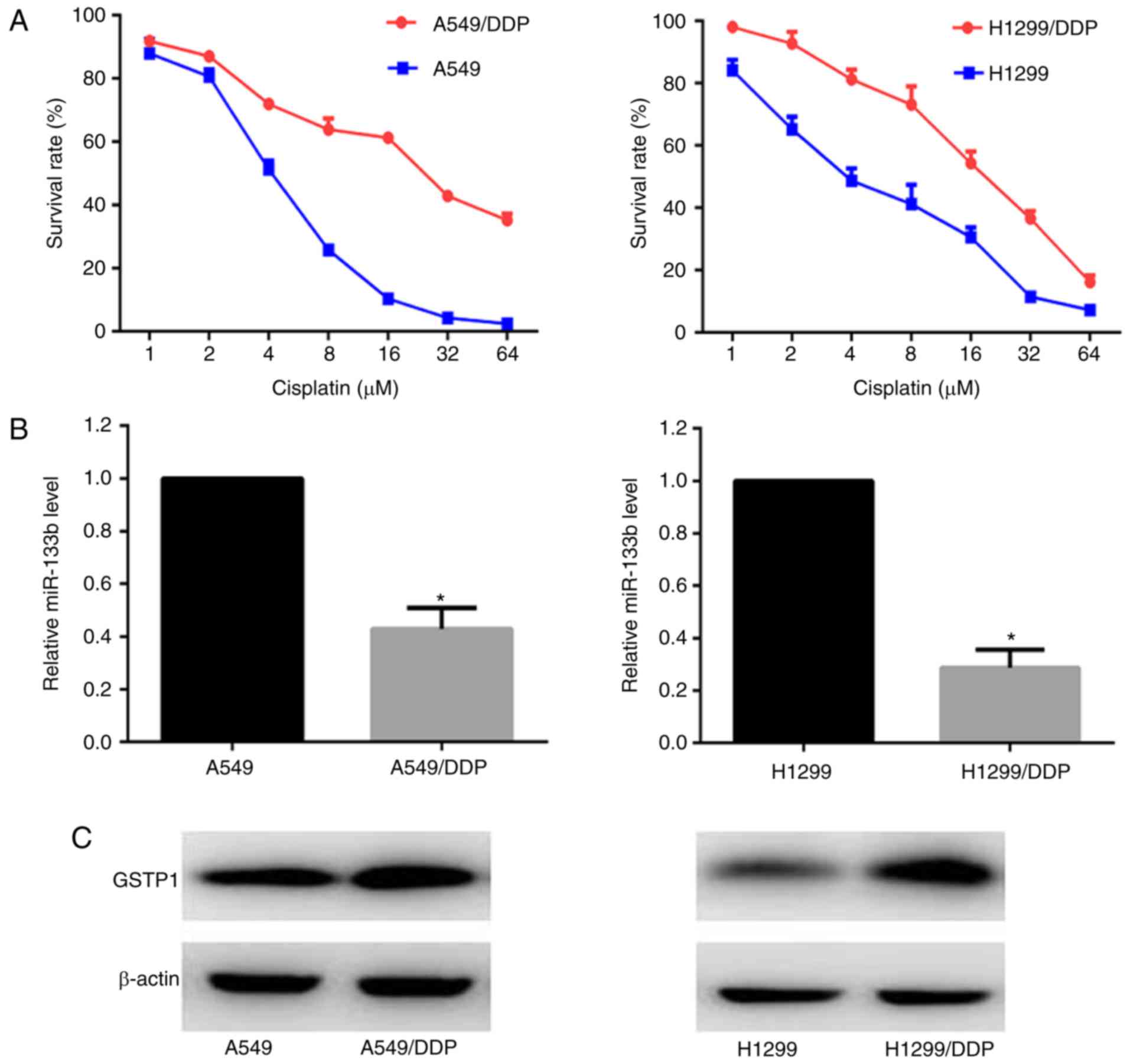
Survival curves of the A549/DDP, A549, H1299/DDP and
H1299 cells in response to various doses of cisplatin were
constructed. CCK-8 growth inhibition assays demonstrated that the
half maximal inhibitory concentration (IC50) of
cisplatin in the A549/DDP cells was ~6 fold higher than that in the
A549 cells, and the H1299/DDP cells exhibited ~4-fold greater
resistance compared with the H1299 cells (Fig. 1A). The expression levels of
miR-133b and GSTP1 protein in the parental and cisplatin-resistant
cells were monitored using RT-qPCR and western blot analysis,
respectively. As shown in Fig.
1B, miR-133b had 2.3- and 3.5-fold lower mean expression levels
in the A549/DPP and H1299/DDP cells compared with the respective
parental cells (P<0.05; Fig.
1B). By contrast, the GSTP1 protein expression was upregulated
in the two cisplatin-resistant cell lines compared with the
parental cell lines (Fig.
1C).
Upregulation of miR-133b increases the
cisplatin chemosensitivity of cisplatin-resistant NSCLC cells
To investigate whether miR-133b modulates the
sensitivity of A549/DPP and H1299/DDP cells to cisplatin, the cells
were transfected with miR-133b mimic or NC mimic. The
IC50 of cisplatin in the A549/DDP cells was
significantly decreased following transfection with miR-133b mimic
compared with those in the blank and NC groups (6.1±0.7 vs.
20.7±1.5 and 20.5±0.5 µM, respectively; both P<0.05;
Fig. 2A). Similarly, the miR-133b
mimic sensitized H1299/DDP cells to cisplatin by ~2.8 fold compared
with the blank and NC groups (IC50 values 4.9±0.1 vs.
14.2±0.5 and 14.0±0.8 µM, respectively; both P<0.05), as
evidenced by the growth inhibition curve (Fig. 2B). To further assess the role of
miR-133b in the apoptosis of cisplatin-resistant cells when exposed
to cisplatin treatment, flow cytometric assays were performed. As
shown in Fig. 2C and D, compared
with the NC and blank groups, the miR-133b mimic significantly
promoted the cisplatin-induced apoptosis of A549/DDP and H1299/DDP
cells. Annexin V/PI-based apoptosis analysis revealed that the
transfection of A549/DDP and H1299/DDP cells with miRNA-133b mimic
increased the percentage of apoptotic cells compared with those in
the blank and NC groups (17.9±0.8 vs. 3.8±0.2 vs. 4.0±0.7%,
respectively, in the A549/DDP cells, and 15.3±0.7 vs. 6.2±0.7 vs.
6.6±1.0%, respectively, in the H1299/DDP cells; all P<0.05).
Western blotting demonstrated decreased expression levels of the
anti-apoptotic proteins Bcl-2 and survivin, and concomitant
activation of the pro-apoptotic protein Bax in the miR-133b mimic
groups compared with the NC and blank groups in the two resistant
cell lines (Fig. 2E). Therefore,
these data suggest that miR-133b enhances the cisplatin
chemosensitivity of cisplatin-resistant NSCLC cells via the
promotion of apoptosis.
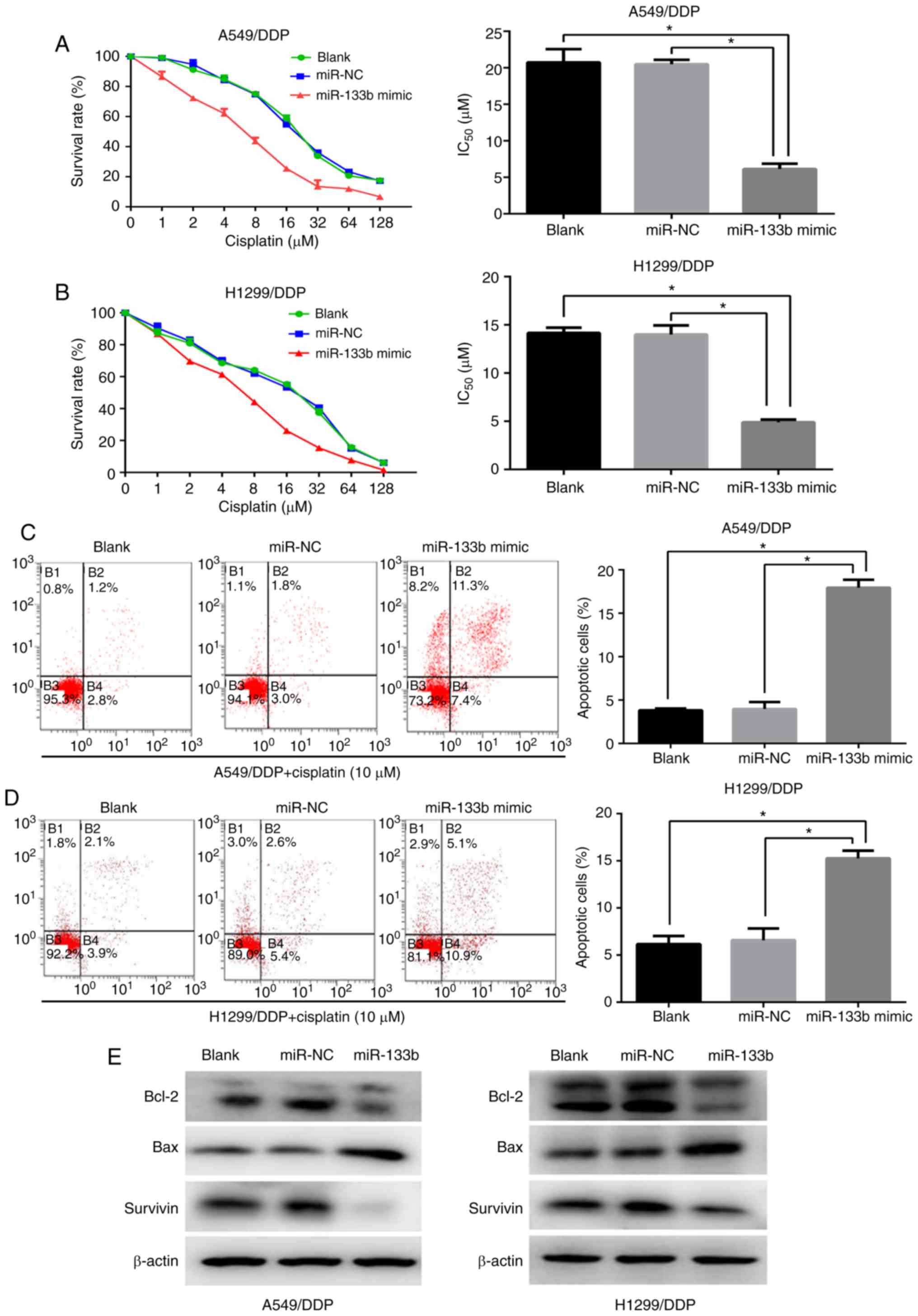 | Figure 2Upregulation of miR-133b increases
the cisplatin chemosensitivity of cisplatin-resistant NSCLC cells.
CCK-8 assays were used to determine the IC50 values of
cisplatin following the transfection of (A) A549/DDP cells and (B)
H1299/DDP cells with miR-133b mimic or negative control. Flow
cytometric assays detected the cell apoptosis of (C) A549/DDP and
(D) H1299/DDP cells when exposed to cisplatin following
transfection with miR-133b mimic or miR-NC. (E) Western blot
detection of Bcl-2, Bax and survivin protein expression following
cisplatin treatment in the blank, miR-NC and miR-133b mimic groups.
Data are the mean ± standard deviation of three separate
experiments. *P<0.05 as indicated. miR, microRNA;
NSCLC, non-small cell lung cancer; CCK, Cell Counting kit;
IC50, half maximal inhibitory concentration A549/DDP,
cisplatin-resistant A549; H1299/DDP, cisplatin-resistant H1299; NC,
negative control; Bcl-2, B-cell lymphoma 2; Bax, Bcl-2-associated X
protein. |
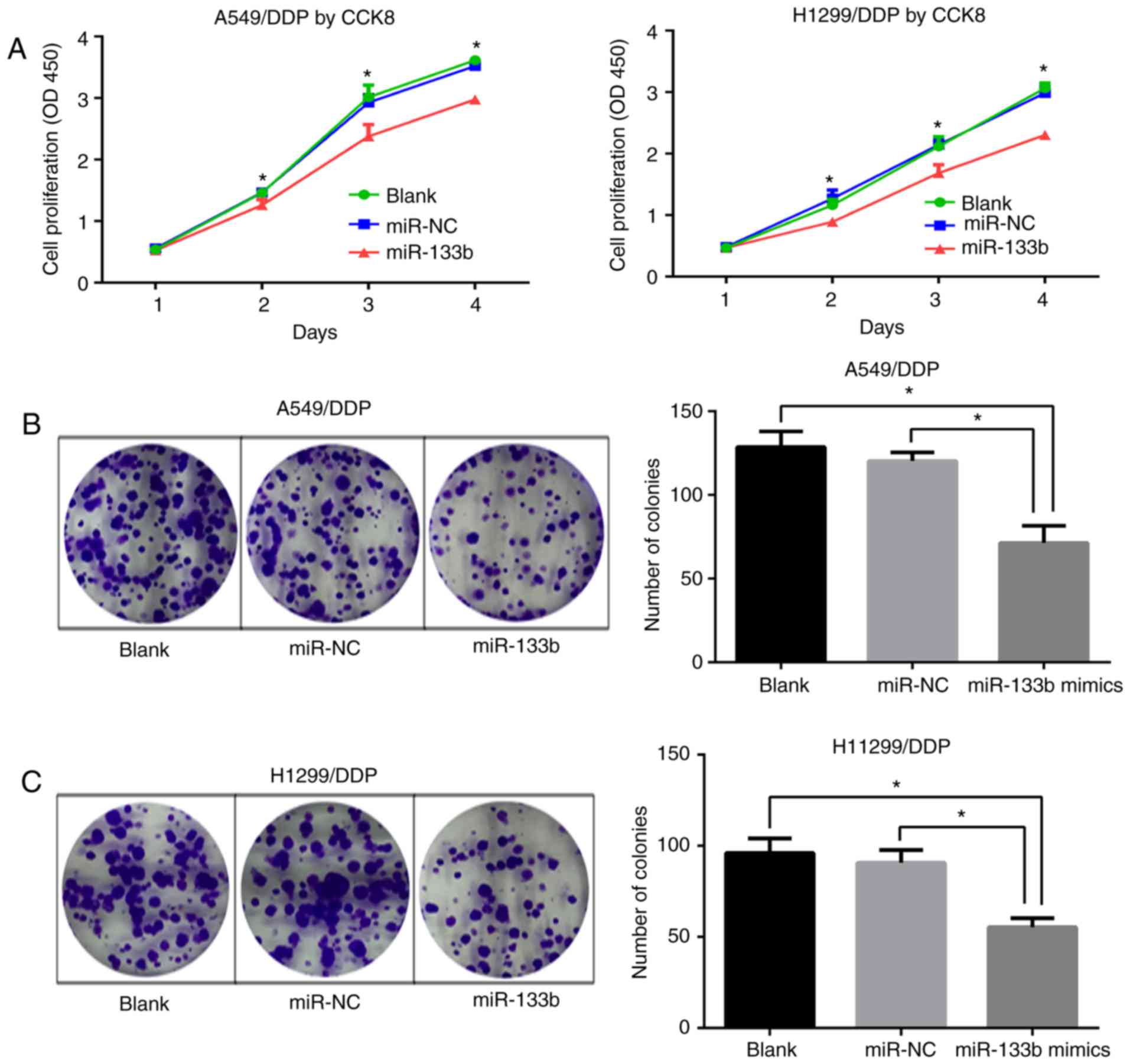
miR-133b inhibits the proliferation and
clonogenesis of A549/DDP and H1299/DDP cells
To ascertain whether miR-133b affects the
proliferative abilities of A549/DDP and H1299/DDP cells, CCK-8
assays were performed. Following transfection with miR-133b mimic,
the proliferation of A549/DDP and H1299/DDP cells was significantly
inhibited at days 2–4 (P<0.05; Fig. 3A). Consistent with this, colony
formation assays demonstrated significantly lower colony-forming
numbers of A549/DDP cells in the miR-133b mimic group compared with
the blank and miR-NC groups (71.3±8.4 vs. 128.7±7.6 and 120.3±4.2,
respectively; both P<0.05; Fig.
3B). Similar results were confirmed in the H1299/DDP cell
groups (55.3±4.1 vs. 96±6.5 and 90.7±5.7, respectively; both
P<0.05; Fig. 3C). These
results indicate that miR-133b impeded the proliferation of
cisplatin-resistant NSCLC cells.
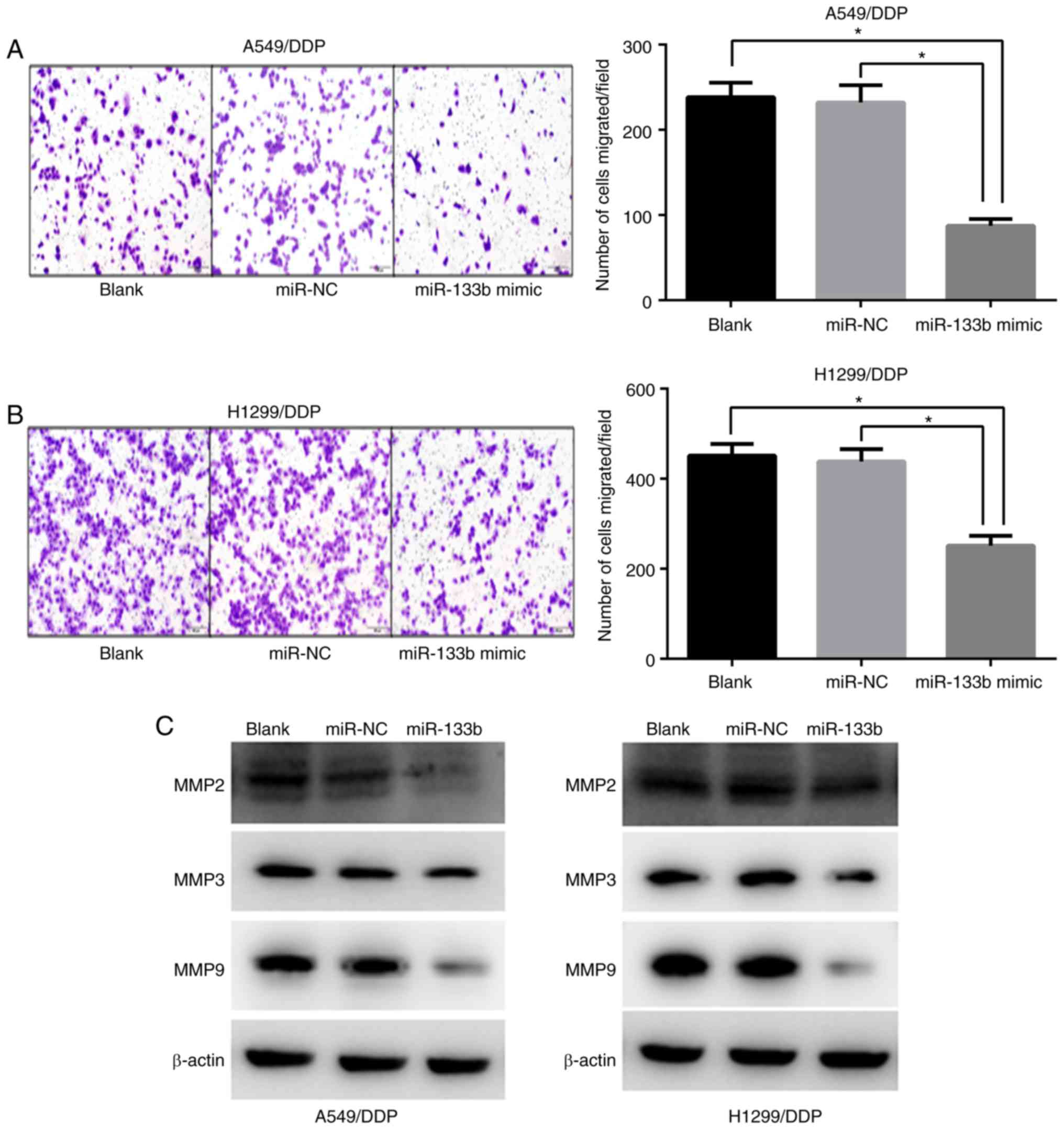
miR-133b attenuates the migration of
A549/DDP and H1299/DDP cells
To further explore the role of miR-133b in the
migration of A549/DDP and H1299/DDP cells, Transwell chamber
migration assays were conducted to assess the effect of miR-133b on
the number of migrating cells. The migratory capacities of the
A549/DDP cells were reduced by ~2.7 fold by the overexpression of
miR-133b compare/d with those of the blank and NC groups (87.3±6.6
vs. 238.3±13.9 and 232.0±16.6, respectively; both P<0.05;
Fig. 4A) and those of the
H1299/DDP cells were reduced by ~1.7 fold (251.3±18.0 vs.
451.3±21.2 and 438.0±22.8, respectively; both P<0.05; Fig. 4B). The expression of
metastasis-associated proteins was then evaluated using western
blotting. Consistent with the lower metastasis capacity following
transfection with miR-133b mimic, the downregulation of MMP2, MMP3
and MMP9 was observed in the miR-133b-transfected cells compared
with the blank and NC groups (Fig.
4C).
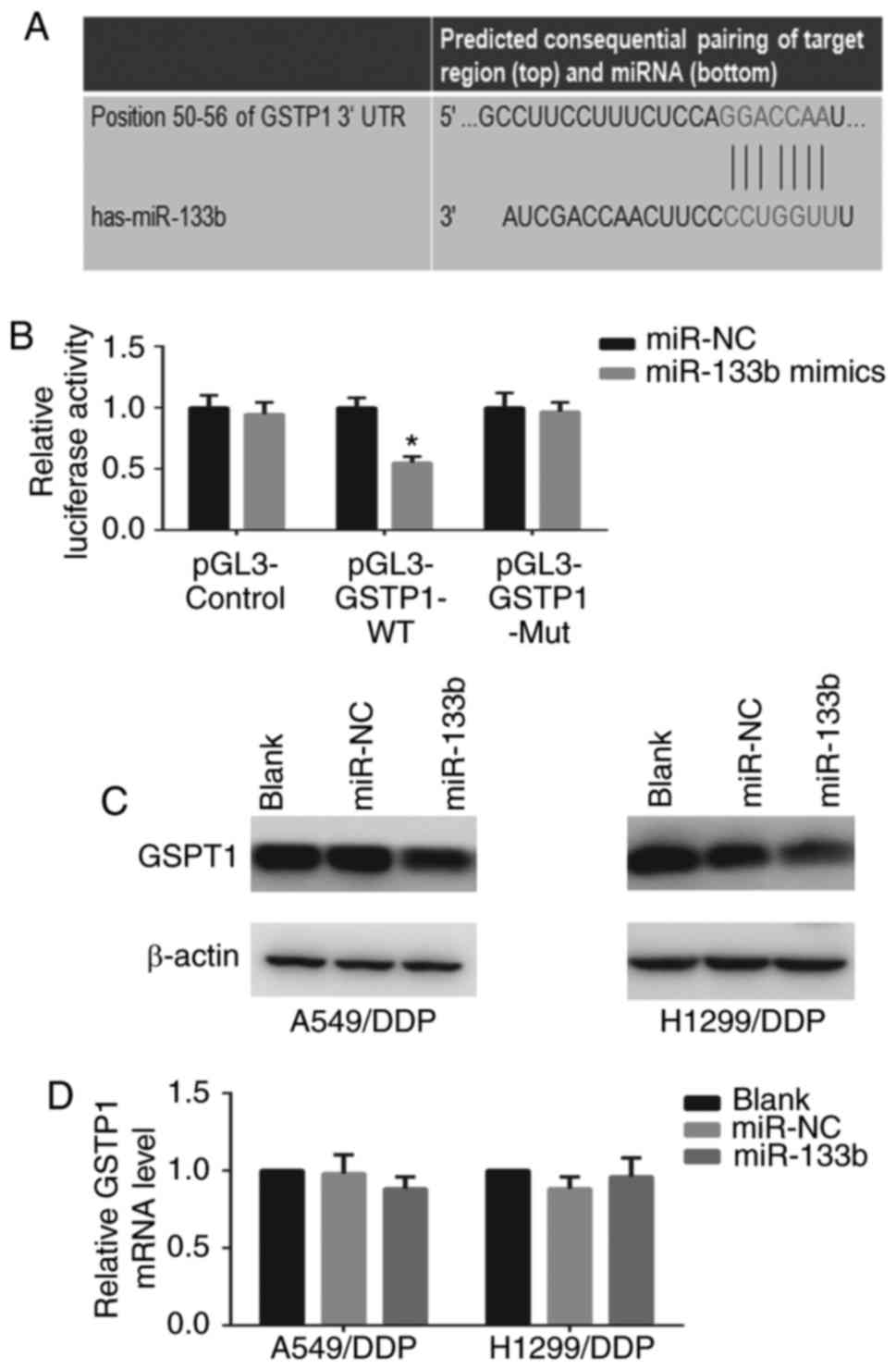
GSTP1 is a direct target of miR-133b
The identification of miRNA-regulated gene targets
is essential to clarify the molecular mechanisms by which miRNAs
mediate cancer progression. The target prediction program
TargetScan Human 7.0 indicated that GSTP1 is a potential target
gene of miR-133b (Fig. 5A). To
confirm that miR-133b regulates GSTP1 expression by directly
binding to the 3′-UTR of GSTP1, the WT or mutant GSTP1 3′-UTR was
cloned into a vector downstream of the luciferase reporter gene and
co-transfected with miR-133b mimic into 293T cells. When the 293T
cells were transfected with the WT GSTP1 3′-UTR, co-transfection
with miR-133b mimic significantly inhibited luciferase activity
(P<0.05; Fig. 5B). By
contrast, the effects of the miR-133b mimic were eliminated in 293T
cells transfected with the mutant type GSTP1 3′-UTR. These results
suggest that miR-133b binds directly to putative GSTP1 3′-UTR
regions, as predicted. Furthermore, the protein expression and mRNA
levels of GSTP1 in the cells following transfection with miR-133b
mimic were examined. Western blotting and RT-qPCR revealed that the
restoration of miR-133b markedly decreased GSTP1 protein (Fig. 5C) but not mRNA expression in
A549/DDP and H1299/DDP cells (data not shown), indicating the
potential post-transcriptional control of GSTP1 expression by
miR-133b. Collectively, these findings confirm the existence of an
inverse correlation between miR-133b and GSTP1 expression.
GSTP1 is critical in the
miR-133b-mediated effects on chemosensitivity to cisplatin, cell
proliferation and migration
The involvement of GSTP1 in the chemosensitivity to
cisplatin, cell proliferation and migration of NSCLC cells
following the regulation of miR-133b expression was investigated.
The GSTP1 siRNA or NC siRNA was transfected into the A549/DDP and
H1299/DDP cells, and the transfected cells were analyzed. The
knockdown of GSTP1 significantly sensitized lung cancer cells to
the induction of apoptosis by cisplatin (Fig. 6A and B) and attenuated the
proliferation and migratory capabilities of the cells compared with
those in the blank and NC groups (Fig. 6C and D). Furthermore, the western
blotting results for GSTP1 silencing were comparable with those for
miR-133b overexpression in the A549/DDP and H1299/DDP cells, and
revealed the downregulation of Bcl-2 and survivin protein
expression and the upregulation of Bax, accompanied by a marked
reduction in MMP expression compared with the respective values in
blank and NC groups (Fig. 6E).
These results indicate that GSTP1 serves a critical role in the
miR-133b-mediated effects on chemosensitivity to cisplatin, cell
proliferation and migration.
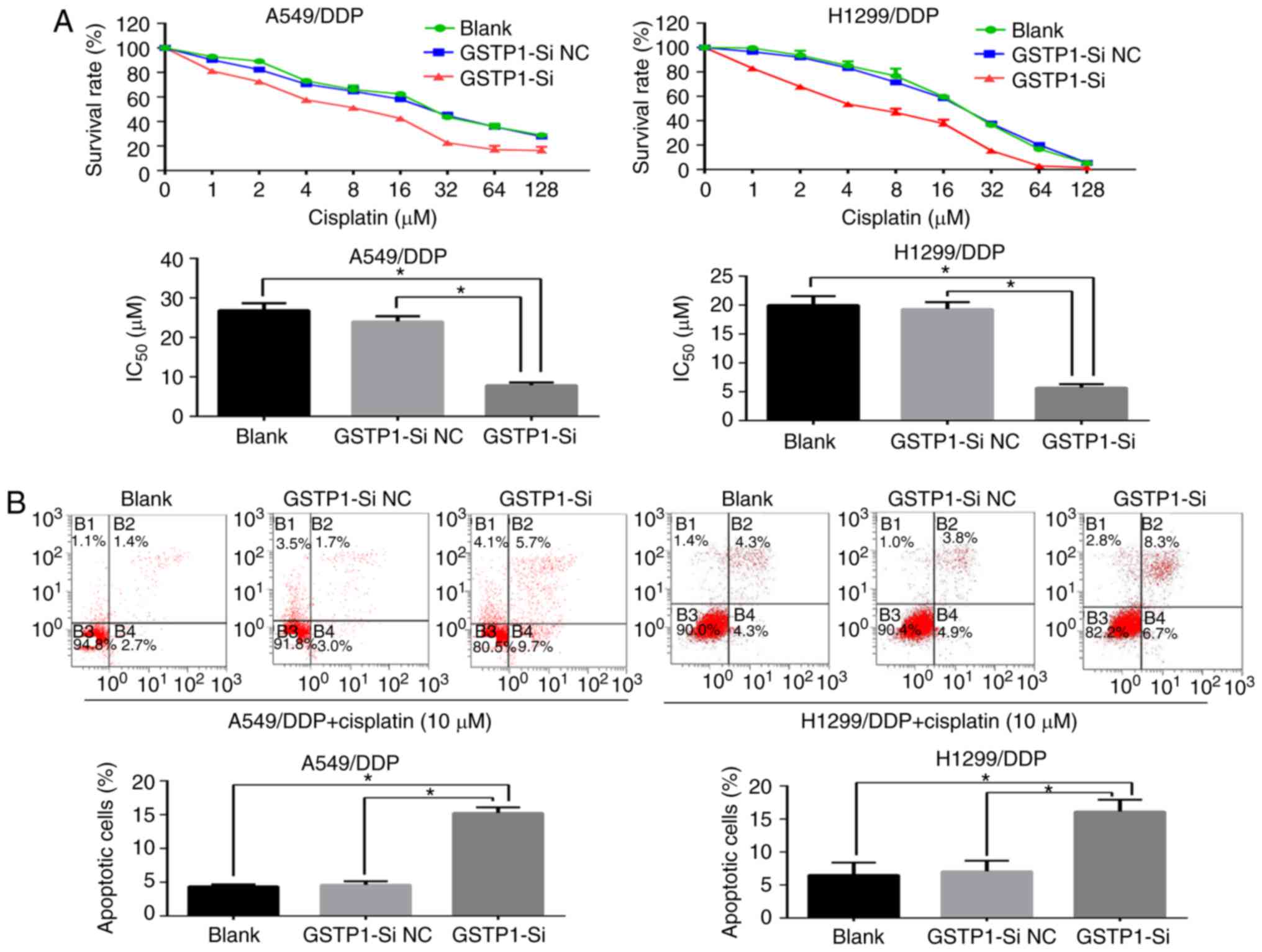 | Figure 6GSTP1 has a critical role in the
miR-133b-mediated chemosensitivity of cisplatin-resistant NSCLC
cells to cisplatin, cell proliferation and migration. (A) Cell
Counting kit-8 and (B) flow cytometric assays demonstrate that the
knockdown of GSTP1 sensitizes lung cancer cells to
cisplatin-induced apoptosis. (C) Cell clone formation and (D)
Transwell migration assays indicate that the knockdown of GSTP1
hampers the proliferation and migratory capabilities of the cells.
(E) Western blot analysis of changes in the expression of apoptotic
and migratory protein following GSTP1 knockdown. Data are the mean
± standard deviation of three separate experiments.
*P<0.05 as indicated. GSTP1,
glutathione-S-transferase P1; miR, microRNA; NSCLC, non-small cell
lung cancer; A549/DDP, cisplatin-resistant A549; H1299/DDP,
cisplatin-resistant H1299; GSTP1-si, small interfering RNA against
GSTP1; NC, negative control; Bcl-2, B-cell lymphoma 2; Bax,
Bcl-2-associated X protein; MMP, matrix metalloproteinase. |
Discussion
In the present study, it was revealed that miR-133b
partly reverses cisplatin-resistance and inhibits the growth and
migration of cisplatin-resistant NSCLC cells. To the best of our
knowledge, this is the first study demonstrating that miR-133b
confers cisplatin sensitivity and inhibits cell proliferation and
migration by targeting GSTP1 in cisplatin-resistant lung cancer
cells.
miRNAs have been indicated to regulate multi-drug
resistance to chemotherapeutic reagents (19). With regard to miR-133b, Chen et
al (20) reported that
miRNA-133b increased the sensitivity of ovarian cancer cells to
chemotherapeutic drugs, including cisplatin and paclitaxel. Zhou
et al (21) demonstrated
that combinational treatment with microRNA-133b and cetuximab
exhibited increased inhibitory effects on the growth and invasion
of colorectal cancer cells compared with either agent used alone.
However, until now, no study has focused on the association between
miR-133b and cisplatin resistance in cisplatin-resistant lung
cancer. In the present study, it was demonstrated that miR-133b was
downregulated in A549/DDP and H1299/DDP cells compared with the
respective parental cells. Additionally, A549/DDP cells displayed
stronger responses to cisplatin following miR-133b mimic
transfection, as did H1299/DDP cells, indicating that miR-133b is a
modulator of cisplatin resistance in NSCLC.
Although the overwhelming majority of studies
support the function of miR-133b as a tumor suppressor in various
cancers, Qin et al (22)
suggested that miR-133b stimulates the progression of cervical
carcinoma, indicating that miR-133b may have disparate effects in
distinct cell environments. Notably, miRNAs are known to play
multiple roles in different tissues depending on the expression of
their target genes, as well as other tissue-specific modulating and
regulatory factors (23,24). In the present study, the ectopic
expression of miR-133b repressed the tumorigenesis and metastasis
of cisplatin-resistant NSCLC cells by attenuating their
proliferation and migratory capabilities, which suggests that
miR-133b acts as a tumor suppressor in lung cancer.
miRNAs are known to regulate the expression of
multiple target genes and affect a variety of cellular pathways.
Nevertheless, the particular pathways affected by miR-133b and the
underlying mechanisms remain unclear. Using TargetScan, an in
silico prediction tool, GSTP1 was identified as a target gene
of miR-133b. GSTP1 belongs to a family of enzymes fulfilling
protective and detoxifying functions in cells (25,26). In addition, GSTP1 is frequently
overexpressed in solid tumors and has been implicated in resistance
against chemotherapy agents (27–29). Previously, Sau et al
(30) reported that targeting
GSTP1 leads to apoptosis in cisplatin-sensitive and -resistant
human osteosarcoma cell lines. Sawers et al (31) found that GSTP1 directly influences
the chemosensitivity of ovarian tumor cell lines to platinum drugs.
In the present study, GSTP1 was validated as a direct target gene
for miR-133b and GSTP1 knockdown was observed to increase the
sensitivity of cisplatin-resistant NSCLC cells to cisplatin.
Furthermore, although GSTP1 protects tumor cells from apoptosis,
little is known about its impact on tumor invasion and migration
(32). The present study provides
evidence that the repression of GSTP1 reduces the migration of
cisplatin-resistant NSCLC cells, and suggests that GSTP1 may be a
promising therapeutic target for the inhibition of tumor
metastasis.
There are certain limitations to the present study.
For example, the signaling pathways by which GSTP1 mediates its
effects have yet to be further clarified. Also, validation in
clinical samples or animal models is necessary to corroborate the
results. However, the findings of the present study provide the
insights that miR-133b partly reverses cisplatin resistance and its
overexpression contributes to the suppression of the malignant
growth and aggressiveness of cisplatin-resistant lung cancer cells
by targeting GSTP1. This could be exploited as a novel therapeutic
strategy to overcome cisplatin resistance.
Abbreviations:
|
miRNA
|
microRNA
|
|
NSCLC
|
non-small cell lung cancer
|
|
MMP
|
matrix metalloproteinase
|
|
3′-UTR
|
3′-untranslated region
|
|
mRNA
|
messenger RNA
|
|
GSTP1
|
glutathione-S-transferase P1
|
|
NC
|
negative control
|
|
siRNA
|
small interfering RNA
|
|
WT
|
wild type
|
|
HRP
|
horseradish peroxidase
|
Acknowledgments
The present study was supported by the National
Natural Science Foundation of China (grant no. 81401896) and the
Shanghai Science and Technology Committee (grant no.
124119a6200).
Notes
[1] Competing
interests
The authors declare that they have no competing
interests.
References
|
1
|
Travis WD, Travis LB and Devesa SS: Lung
cancer. Cancer. 75(1 Suppl): S191–S202. 2015. View Article : Google Scholar
|
|
2
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mitsudomi T, Morita S, Yatabe Y, Negoro S,
Okamoto I, Tsurutani J, Seto T, Satouchi M, Tada H, Hirashima T, et
al: Gefitinib versus cisplatin plus docetaxel in patients with
non-small-cell lung cancer harbouring mutations of the epidermal
growth factor receptor (WJTOG3405): An open label, randomised phase
3 trial. Lancet Oncol. 11:121–128. 2010. View Article : Google Scholar
|
|
4
|
Oliver TG, Mercer KL, Sayles LC, Burke JR,
Mendus D, Lovejoy KS, Cheng MH, Subramanian A, Mu D, Powers S, et
al: Chronic cisplatin treatment promotes enhanced damage repair and
tumor progression in a mouse model of lung cancer. Genes Dev.
24:837–52. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ardizzoni A, Boni L, Tiseo M, Fossella FV,
Schiller JH, Paesmans M, Radosavljevic D, Paccagnella A, Zatloukal
P, Mazzanti P, et al: Cisplatin-versus carboplatin-based
chemotherapy in first-line treatment of advanced non-small-cell
lung cancer: An individual patient data meta-analysis. J Natl
Cancer Inst. 99:847–857. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tan XL, Moyer AM, Fridley BL, Schaid DJ,
Niu N, Batzler AJ, Jenkins GD, Abo RP, Li L, Cunningham JM, et al:
Genetic variation predicting cisplatin cytotoxicity associated with
overall survival in lung cancer patients receiving platinum-based
chemotherapy. Clin Cancer Res. 17:5801–5811. 2010. View Article : Google Scholar
|
|
7
|
Cortés-Sempere M, de Miguel MP, Pernía O,
Rodriguez C, de Castro Carpeño J, Nistal M, Conde E, López-Ríos F,
Belda-Iniesta C, Perona R and Ibanez de Caceres I: IGFBP-3
methylation-derived deficiency mediates the resistance to cisplatin
through the activation of the IGFIR/Akt pathway in non-small cell
lung cancer. Oncogene. 32:1274–1283. 2013. View Article : Google Scholar
|
|
8
|
Orellana EA and Kasinski AL: MicroRNAs in
cancer: A historical perspective on the path from discovery to
therapy. Cancers (Basel). 7:1388–1405. 2015. View Article : Google Scholar
|
|
9
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chitwood DH and Timmermans MC: Small RNAs
are on the move. Nature. 467:415–419. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhai H and Ju J: Implications of microRNAs
in colorectal cancer development, diagnosis, prognosis, and
therapeutics. Front Genet. 2:000782011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhao JJ, Chu ZB, Hu Y, Lin J, Wang Z,
Jiang M, Chen M, Wang X, Kang Y, Zhou Y, et al: Targeting the
miR-221-222/PUMA/BAK/BAX pathway abrogates dexamethasone resistance
in multiple myeloma. Cancer Res. 75:4384–4397. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ma J, Fang B, Zeng F, Ma C, Pang H, Cheng
L, Shi Y, Wang H, Yin B, Xia J, et al: Down-regulation of miR-223
reverses epithelial-mesenchymal transition in gemcitabine-resistant
pancreatic cancer cells. Oncotarget. 6:1740–1749. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao Y, Huang J, Zhang L, Qu Y, Li J, Yu
B, Yan M, Yu Y, Liu B and Zhu Z: MiR-133b is frequently decreased
in gastric cancer and its overexpression reduces the metastatic
potential of gastric cancer cells. BMC Cancer. 14:342014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tao J, Wu D, Xu B, Qian W, Li P, Lu Q, Yin
C and Zhang W: microRNA-133 inhibits cell proliferation, migration
and invasion in prostate cancer cells by targeting the epidermal
growth factor receptor. Oncol Rep. 27:1967–1975. 2012.PubMed/NCBI
|
|
16
|
Akçakaya P, Ekelund S, Kolosenko I,
Caramuta S, Ozata DM, Xie H, Lindforss U, Olivecrona H and Lui WO:
miR-185 and miR-133b deregulation is associated with overall
survival and metastasis in colorectal cancer. Int J Oncol.
9:311–318. 2011.
|
|
17
|
Liu L, Shao X, Gao W, Zhang Z, Liu P, Wang
R, Huang P, Yin Y and Shu Y: MicroRNA-133b inhibits the growth of
non-small-cell lung cancer by targeting the epidermal growth factor
receptor. FEBS J. 279:3800–3812. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
19
|
MacDonagh L, Gray SG, Finn SP, Cuffe S,
O’Byrne KJ and Barr MP: The emerging role of microRNAs in
resistance to lung cancer treatments. Cancer Treat Rev. 41:160–169.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen S, Jiao JW, Sun KX, Zong ZH and Zhao
Y: MicroRNA-133b targets glutathione S-transferase π expression to
increase ovarian cancer cell sensitivity to chemotherapy drugs.
Drug Des Dev Ther. 9:5225–5235. 2015.
|
|
21
|
Zhou J, Lv L, Lin C, Hu G, Guo Y, Wu M,
Tian B and Li X: Combinational treatment with microRNA 133b and
cetuximab has increased inhibitory effects on the growth and
invasion of colorectal cancer cells by regulating EGFR. Mol Med
Rep. 12:5407–5414. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Qin W, Dong P, Ma C, Mitchelson K, Deng T,
Zhang L, Sun Y, Feng X, Ding Y, Lu X, et al: MicroRNA-133b is a key
promoter of cervical carcinoma development through the activation
of the ERK and AKT1 pathways. Oncogene. 31:4067–4075. 2012.
View Article : Google Scholar
|
|
23
|
Mendell JT: MiRiad roles for the miR-17-92
cluster in development and disease. Cell. 133:217–222. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Garzon R, Calin GA and Croce CM: MicroRNAs
in cancer. Annu Rev Med. 60:167–179. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Singh S: Cytoprotective and regulatory
functions of glutathione S-transferases in cancer cell
proliferation and cell death. Cancer Chemother Pharmacol. 75:1–15.
2015. View Article : Google Scholar
|
|
26
|
Schnekenburger M, Karius T and Diederich
M: Regulation of epigenetic traits of the glutathione S-transferase
P1 gene: From detoxification toward cancer prevention and
diagnosis. Front Pharmacol. 5:1702014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Louie SM, Grossman EA, Crawford LA, Ding
L, Camarda R, Huffman TR, Miyamoto DK, Goga A, Weerapana E and
Nomura DK: GSTP1 is a driver of triple-negative breast cancer cell
metabolism and pathogenicity. Cell Chem Biol. 23:567–578. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Townsend DM and Tew KD: The role of
glutathione-S-transferase in anti-cancer drug resistance. Oncogene.
22:7369–7375. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Depeille P, Cuq P, Passagne I, Evrard A
and Vian L: Combined effects of GSTP1 and MRP1 in melanoma drug
resistance. Br J Cancer. 93:216–223. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sau A, Filomeni G, Pezzola S, D’Aguanno S,
Tregno FP, Urbani A, Serra M, Pasello M, Picci P, Federici G and
Caccuri AM: Targeting GSTP1-1 induces JNK activation and leads to
apoptosis in cisplatin-sensitive and -resistant human osteosarcoma
cell lines. Mol Biosyst. 8:994–1006. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sawers L, Ferguson MJ, Ihrig BR, Young HC,
Chakravarty P, Wolf CR and Smith G: Glutathione S-transferase P1
(GSTP1) directly influences platinum drug chemosensitivity in
ovarian tumour cell lines. Br J Cancer. 111:1150–1158. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Holley SL, Fryer AA, Haycock JW, Grubb SE,
Strange RC and Hoban PR: Differential effects of glutathione
S-transferase pi (GSTP1) haplotypes on cell proliferation and
apoptosis. Carcinogenesis. 28:2268–2273. 2007. View Article : Google Scholar : PubMed/NCBI
|