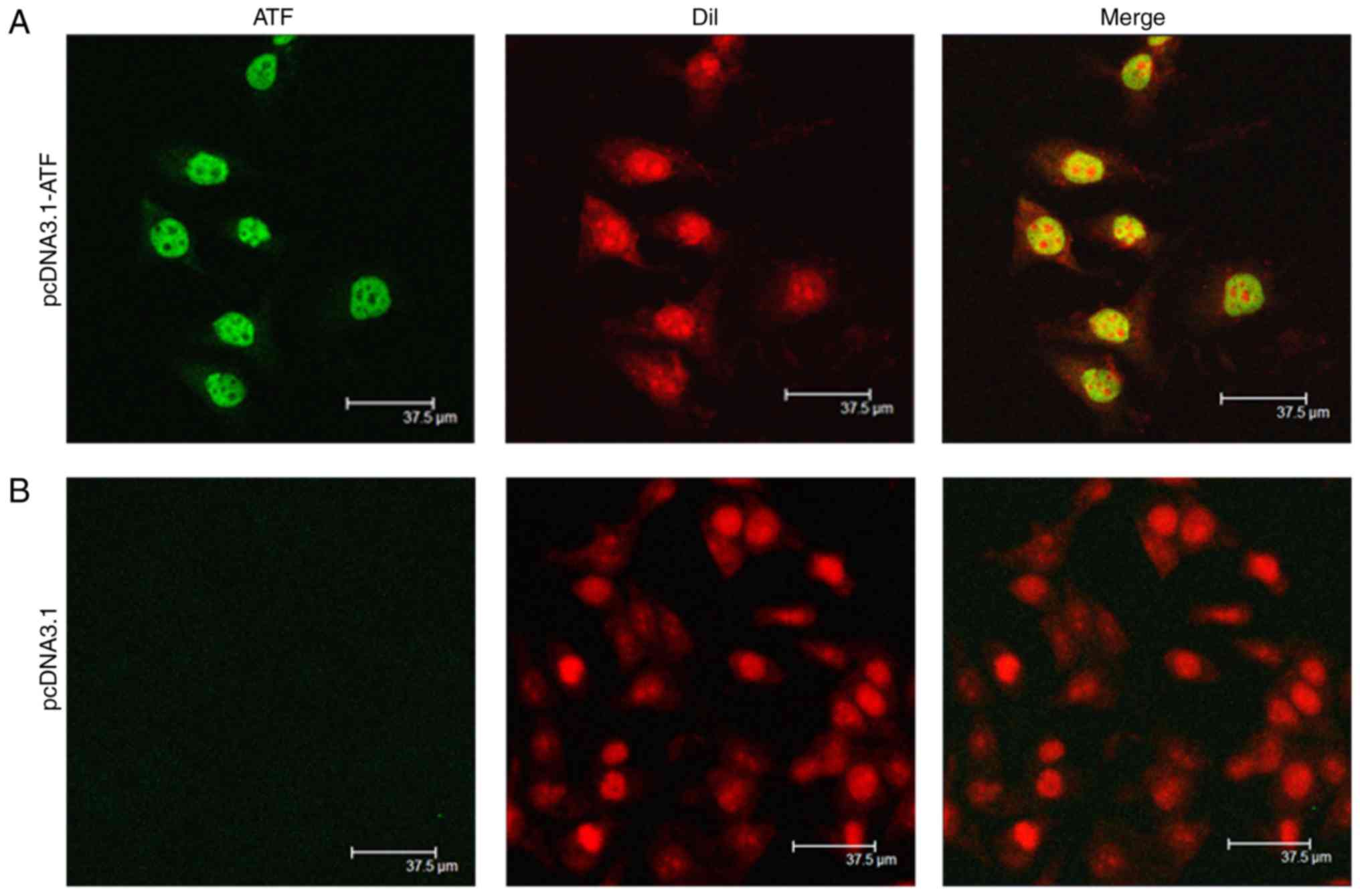
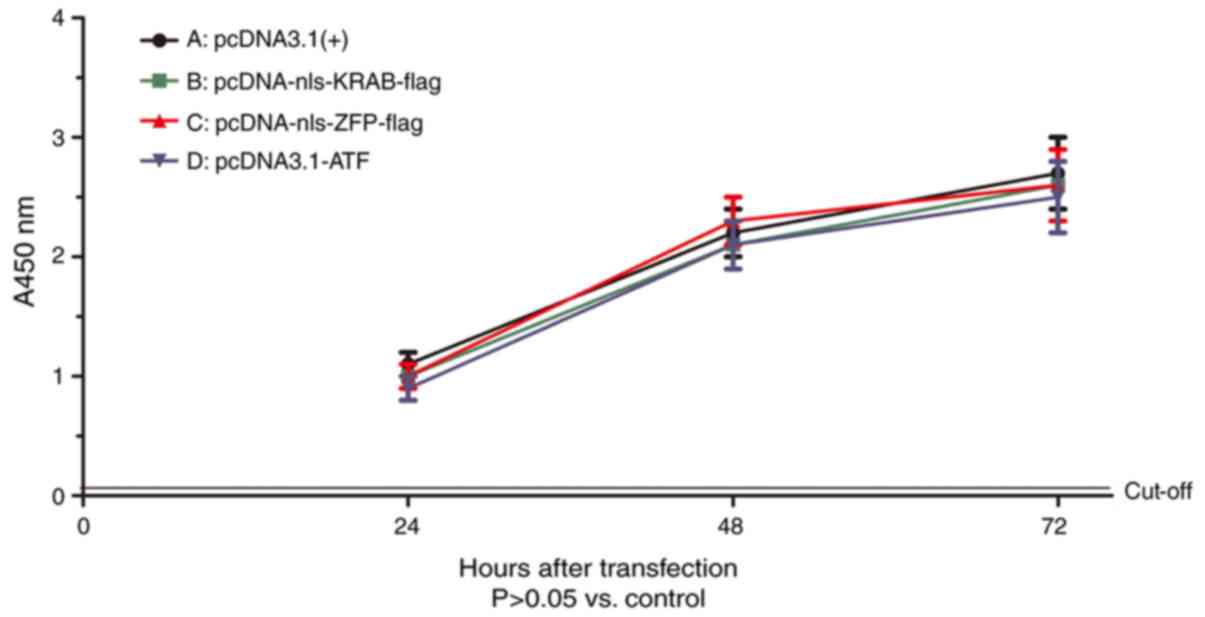
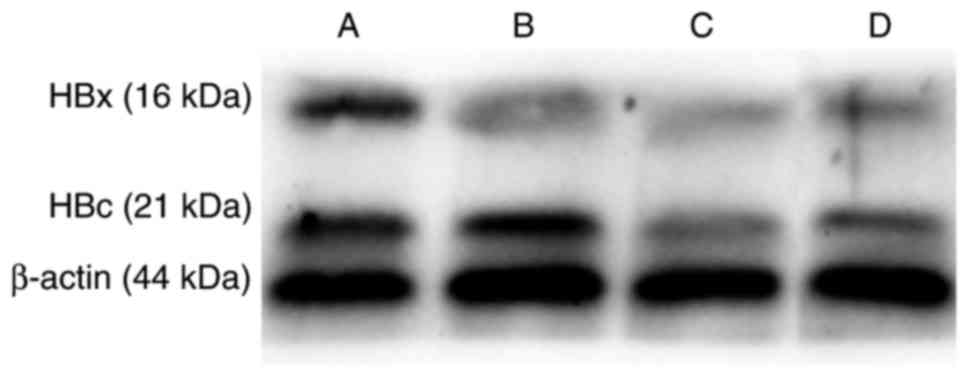
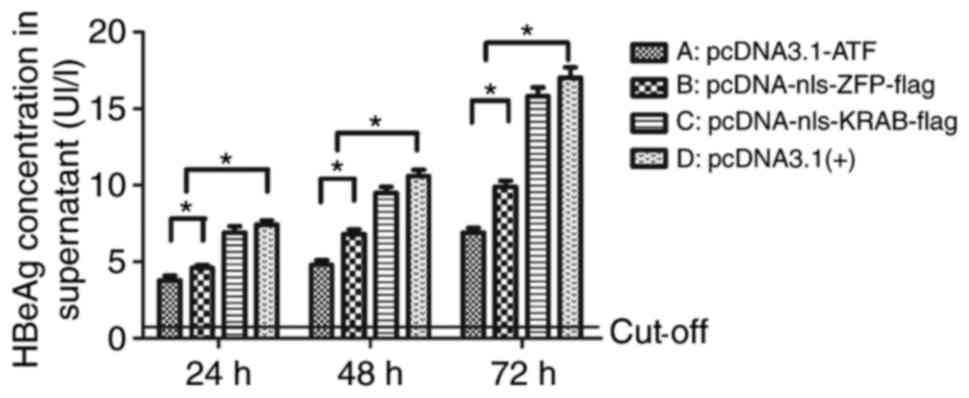
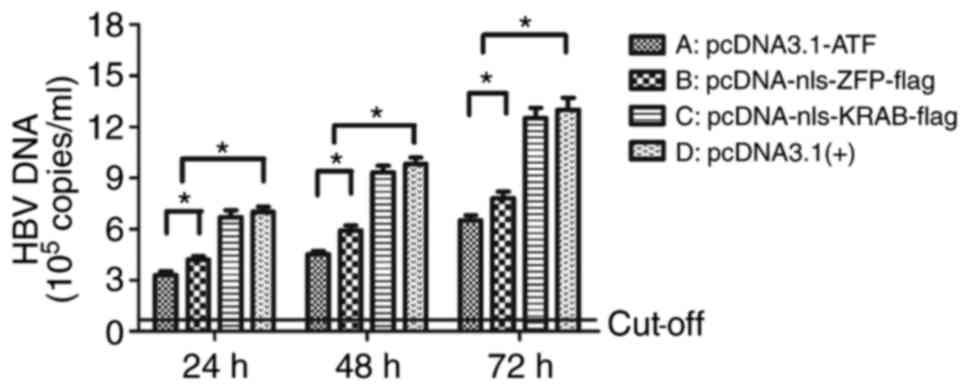
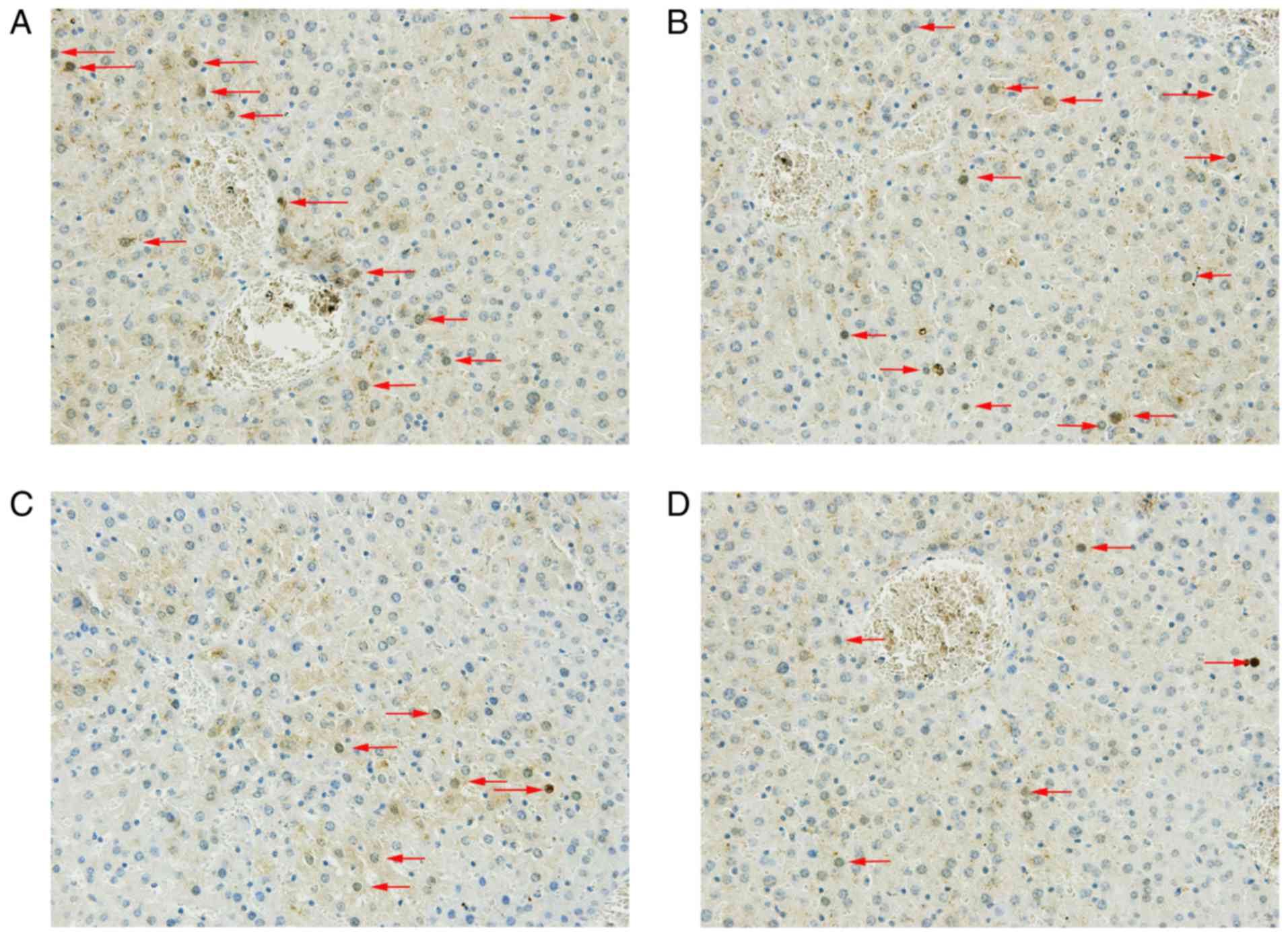
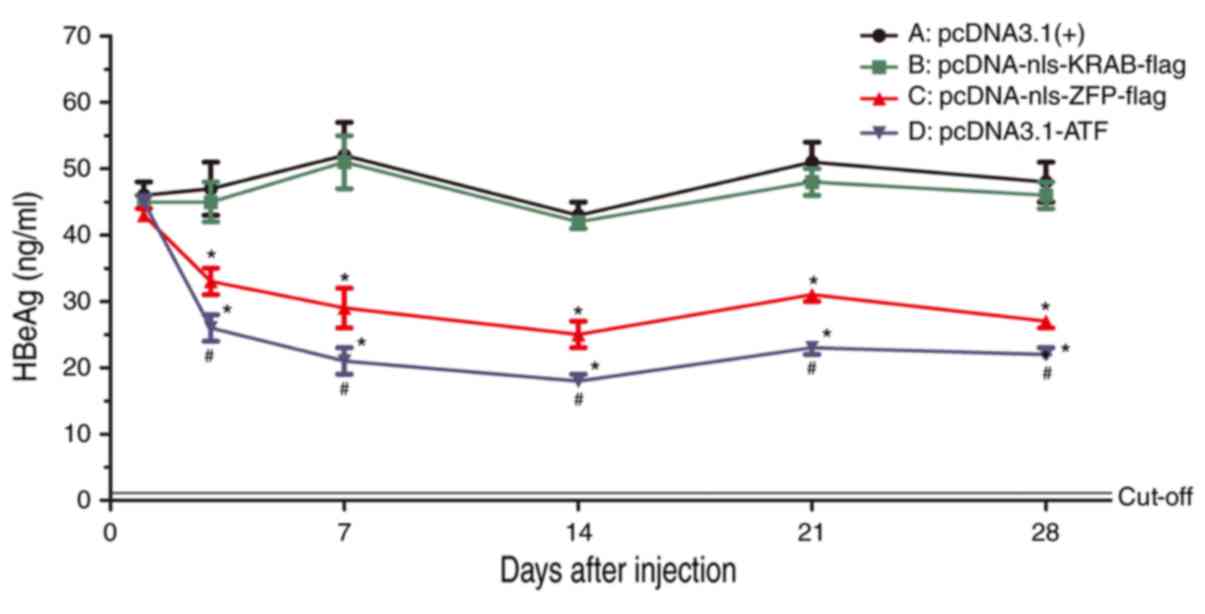
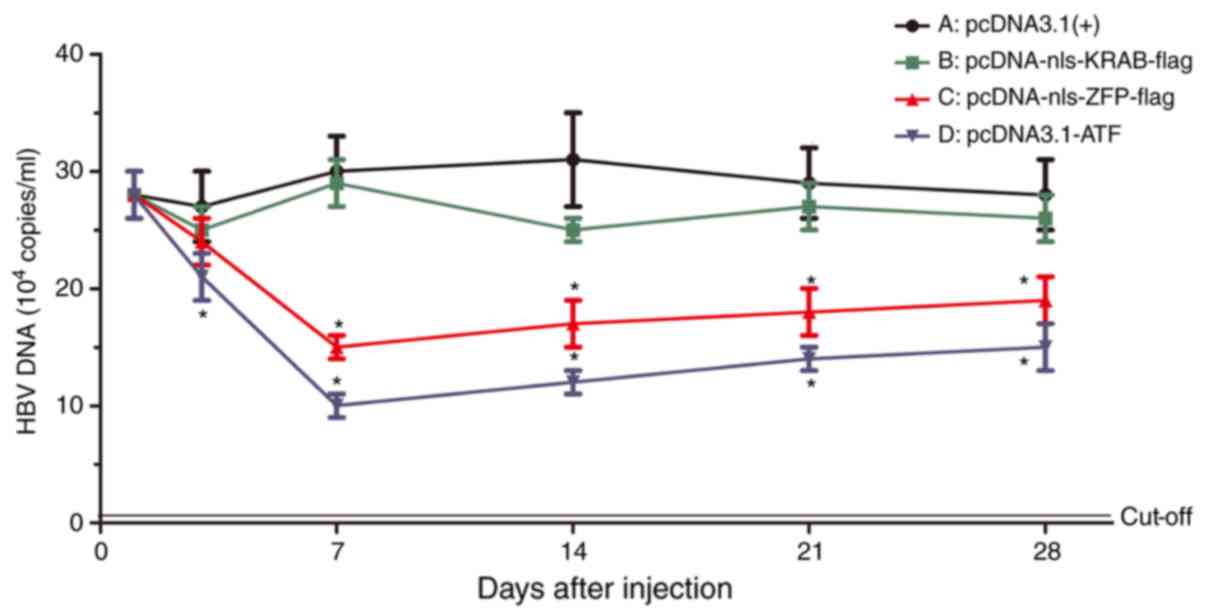
|
1
|
Barker LF, Maynard JE, Purcell RH,
Hoofnagle JH, Berquist KR and London WT: Viral hepatitis, type B,
in experimental animals. Am J Med Sci. 270:189–195. 1975.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Moolla N, Kew M and Arbuthnot P:
Regulatory elements of hepatitis B virus transcription. J Viral
Hepat. 9:323–331. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Beasley RP, Hwang LY, Lin CC and Chien CS:
Hepatocellular carcinoma and hepatitis B virus. A prospective study
of 22, 707 men in Taiwan. Lancet. 2:1129–1133. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ganem D and Varmus HE: The molecular
biology of the hepatitis B viruses. Annu Rev Biochem. 56:651–693.
1987. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ganem D and Prince AM: Hepatitis B virus
infection natural history and clinical consequences. N Engl J Med.
350:1118–1129. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lupberger J and Hildt E: Hepatitis B
virus-induced oncogenesis. World J Gastroenterol. 13:74–81. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chang JJ and Lewin SR: Immunopathogenesis
of hepatitis B virus infection. Immunol Cell Biol. 85:16–23. 2007.
View Article : Google Scholar
|
|
8
|
Pramoolsinsap C: Acute hepatitis A and
acquired immunity to hepatitis A virus in hepatitis B virus (HBV)
carriers and in HBV- or hepatitis C virus-related chronic liver
diseases in Thailand. J Viral Hepat. 7(Suppl 1): S11–S12. 2000.
View Article : Google Scholar
|
|
9
|
Liaw YF, Tsai SL, Sheen IS, Chao M, Yeh
CT, Hsieh SY and Chu CM: Clinical and virologival course of chronic
hepatitis B virus infection with hepatitis C and D virus markers.
Am J Gastroenterol. 93:354–359. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen LP, Zhao J, Du Y, Han YF, Su T, Zhang
HW and Cao GW: Antiviral treatment to prevent chronic hepatitis B
or C-related hepatocellular carcinoma. World J Virol. 12:174–183.
2012. View Article : Google Scholar
|
|
11
|
Dusheiko G: Treatment of HBeAg positive
chronic hepatitis B: Interferon or nucleoside analogues. Liver Int.
33(Suppl 1): S137–S150. 2013. View Article : Google Scholar
|
|
12
|
Craxi A, Antonucci G and Cammà C:
Treatment options in HBV. J Hepatol. 44(Suppl 1): S77–S83. 2006.
View Article : Google Scholar
|
|
13
|
Fung J, Lai CL, Seto WK and Yuen MF:
Nucleoside/nucleotide analogues in the treatment of chronic
hepatitis B. J Antimicrob Chemother. 66:2715–2725. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sera T: Zinc-finger-based artificial
transcription factors and their applications. Adv Drug Deliv Rev.
61:513–526. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Papworth M, Kolasinska P and Minczuk M:
Designer zinc-finger proteins and their applications. Gene.
366:27–38. 2006. View Article : Google Scholar
|
|
16
|
Greisman HA and Pabo CO: A general
strategy for selecting high-affinity zinc finger proteins for
diverse DNA target sites. Science. 275:657–661. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dreier B, Beerli RR, Segal DJ, Flippin JD
and Barbas CF III: Development of zinc finger domains for
recognition of the 5′-ANN-3′ family of DNA sequences and their use
in the construction of artificial transcription factors. J Biol
Chem. 276:29466–29478. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Isalan M, Klug A and Choo Y: A rapid,
generally applicable method to engineer zinc fingers illustrated by
targeting the HIV-1 promoter. Nat Biotechnol. 19:656–660. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu Q, Segal DJ, Ghiara JB and Barbas CF
III: Design of polydactyl zinc-finger proteins for unique
addressing within complex genomes. Proc Natl Acad Sci USA.
94:5525–5530. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kim JS and Pabo CO: Getting a handhold on
DNA: Design of poly-zinc finger proteins with femtomolar
dissociation constants. Proc Natl Acad Sci USA. 95:2812–2817. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Desjarlais JR and Berg JM: Toward rules
relating zinc finger protein sequences and DNA-binding site
preferences. Proc Natl Acad Sci USA. 89:7345–7349. 1992. View Article : Google Scholar
|
|
22
|
Rebar EJ and Pabo CO: Zinc finger phage:
Affinity selection of fingers with new DNA-binding specificities.
Science. 263:671–673. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Laity JH, Lee BM and Wright PE: Zinc
finger proteins: New insights into structural and functional
diversity. Curr Opin Struct Biol. 11:39–46. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Blancafort P, Magnenat L and Barbas CF
III: Scanning the human genome with combinatorial transcription
factor libraries. Nat Biotechnol. 21:269–274. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jamieson AC, Miller JC and Pabo CO: Drug
discovery with engineered zinc-finger proteins. Nat Rev Drug
Discov. 2:361–368. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lund CV, Blancafort P, Popkov M and Barbas
CF III: Promoter-targeted phage display selections with
preassembled synthetic zinc finger libraries for endogenous gene
regulation. J Mol Biol. 340:599–613. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wu H, Yang WP and Barbas CF III: Building
zinc fingers by selection: Toward a therapeutic application. Proc
Natl Acad Sci USA. 92:344–348. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Papworth M, Moore M, Isalan M, Minczuk M,
Choo Y and Klug A: Inhibition of herpes simplex virus 1 gene
expression by designer zinc-finger transcription factors. Proc Natl
Acad Sci USA. 100:1621–1626. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
ZF Tools. http://www.scripps.edu/mb/barbas/zfdesign/zfdesign-home.php.
|
|
30
|
Margolin JF, Friedman JR, Meyer WK,
Vissing H, Thiesen HJ and Rauscher FJ III: Krüppel-associated boxes
are potent transcriptional repression domains. Proc Natl Acad Sci
USA. 91:4509–4513. 1994. View Article : Google Scholar
|
|
31
|
Sadowski I, Ma J, Triezenberg S and
Ptashne M: GAL4-VP16 is an unusually potent transcriptional
activator. Nature. 335:563–564. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
López-Terrada D, Cheung SW, Finegold MJ
and Knowles BB: Hep G2 is a hepatoblastoma-derived cell line. Hum
Pathol. 40:1512–1515. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Walters KA, Joyce MA, Addison WR, Fischer
KP and Tyrrell DL: Superinfection exclusion in duck heaptitis B
virus infection is mediated by the large surface antigen. J Virol.
78:7925–7937. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
35
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Larionov A, Krause A and Miller W: A
standard curve based method for relative real time PCR data
processing. BMC Bioinformatics. 6:622005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Shimizu S, Nomura F, Tomonaga T, Sunaga M,
Noda M, Ebara M and Saisho H: Expression of poly(ADP-ribose)
polymerase in human hepatocellular carcinoma and analysis of biopsy
specimens obtained under sonographic guidance. Oncol Rep.
12:821–825. 2004.PubMed/NCBI
|
|
38
|
Shintani M, Urano M, Takakuwa Y, Kuroda M
and Kamoshida S: Immunohistochemical characterization of pyrimidine
synthetic enzymes, thymidine kinase-1 and thymidylate synthase, in
various types of cancer. Oncol Rep. 23:1345–1350. 2010.PubMed/NCBI
|
|
39
|
Dikstein R, Faktor O and Shaul Y:
Hierarchic and cooperative binding of the rat liver nuclear protein
C/EBP at the hepatitis B virus enhancer. Mol Cell Biol.
10:4427–4430. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Faktor O, Budlovsky S, Ben-Levy R and
Shaul Y: A single element within the hepatitis B virus enhancer
binds multiple proteins and responds to multiple stimuli. J Virol.
64:1861–1863. 1990.PubMed/NCBI
|
|
41
|
Siegrist CA, Durand B, Emery P, David E,
Hearing P, Mach B and Reith W: RFX1 is identical to enhancer factor
C and functions as a transactivator of the hepatitis B virus
enhancer. Mol Cell Biol. 13:6375–6384. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ori A, Zauberman A, Doitsh G, Paran N,
Oren M and Shaul Y: p53 binds and represses the HBV enhancer: An
adjacent enhancer element can reverse the transcription effect of
p53. EMBO J. 17:544–553. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chandra V, Huang P, Potluri N, Wu D, Kim Y
and Rastinejad F: Multidomain integration in the structure of the
HNF-4α nuclear receptor complex. Nature. 495:394–398. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Quasdorff M and Protzer U: Control of
hepatitis B virus at the level of transcription. J Viral Hepat.
17:527–536. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Liu B, Wen X, Huang C and Wei Y:
Unraveling the complexity of hepatitis B virus: From molecular
understanding to therapeutic strategy in 50 years. Int J Biochem
Cell Biol. 45:1987–1996. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Levrero M, Pollicino T, Petersen J,
Belloni L, Raimondo G and Dandri M: Control of cccDNA function in
hepatitis B virus infection. J Hepatol. 51:581–592. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Beck J and Nassal M: Hepatitis B virus
replication. World J Gastroenterol. 13:48–64. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Christine N, Wei Y and Buendia MA:
Mechanisms of HBV-related hepatocarcinogenesis. J Hepatol.
52:594–604. 2010. View Article : Google Scholar
|
|
49
|
Bock CT, Malek NP, Tillmann HL, Manns MP
and Trautwein C: The enhancer I core region contributes to the
replication level of hepatitis B virus in vivo and in vitro. J
Virol. 74:2193–2202. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Antonucci TK and Rutter WJ: Hepatitis B
virus (HBV) promoters are regulated by the HBV enhancer in a
tissue-specific manner. J Virol. 63:579–583. 1989.PubMed/NCBI
|
|
51
|
Doitsh G and Shaul Y: Enhancer I
predominance in hepatitis B virus gene expression. Mol Cell Biol.
24:1799–1808. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Soussan P, Garreau F, Zylberberg H, Ferray
C, Brechot C and Kremsdorf D: In vivo expression of a new hepatitis
B virus protein encoded by a spliced RNA. J Clin Invest. 105:55–60.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Pavletich NP and Pabo CO: Crystal
structure of a five-finger GLI-DNA complex: New perspectives on
zinc fingers. Science. 261:1701–1707. 1993. View Article : Google Scholar
|
|
54
|
Urnov FD and Rebar EJ: Designed
transcription factors as tools the therapeatics and functional
genomics. Biochem Pharmacol. 64:919–923. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ramanan V, Shlomai A, Cox DB, Schwartz RE,
Michailidis E, Bhatta A, Scott DA, Zhang F, Rice CM, Bhatia SN, et
al: CRISPR/Cas9 cleavage of viral DNA efficiently suppresses
hepatitis B virus. Sci Rep. 5:108332015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Karimova M, Beschorner N, Dammermann W,
Chemnitz J, Indenbirken D, Bockmann JH, Grundhoff A, Lüth S,
Buchholz F, Schulze zur Wiesch J and Hauber J: CRISPR/Cas9
nickase-mediated disruption of hepatitis B virus open reading frame
S and X. Sci Rep. 5:137342015. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Mino T, Hatono T, Matsumoto N, Mori T,
Mineta Y, Aoyama Y and Sera T: Inhibition of DNA replication of
human papilloma-virus by artificial zinc finger proteins. J Virol.
80:5405–5412. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zimmerman KA, Fischer KP, Joyce MA and
Tyrrell DL: Zinc finger proteins designed to specifically target
duck hepatitis B virus covalently closed circular DNA inhibit viral
transcription in tissue culture. J Virol. 82:8013–8021. 2008.
View Article : Google Scholar : PubMed/NCBI
|