Introduction
Nonalcoholic fatty liver disease (NAFLD) is a
metabolic-related liver disease associated with obesity, insulin
resistance (IR), type 2 diabetes mellitus (T2DM), and other
components of the metabolic syndrome (MetS). Proportionally, it is
now the most common chronic liver disease worldwide (1). The spectrum of NAFLD ranges from
nonalcoholic fatty liver (NAFL), nonalcoholic steatohepatitis
(NASH), liver cirrhosis and hepatocellular carcinoma (HCC).
Multiple mechanisms are considered to underlie NAFLD, such as
oxidative stress, mitochondrial dysfunction, endoplasmic reticulum
stress, and bacterial endotoxins (1).
It is well known that genetic factors play an
important role in NAFLD. Recently, epigenetic factors (DNA
methylation, histone modifications and noncoding RNA) have also
been uncovered to serve as the molecular basis of NAFLD (2,3).
Of these epigenetic factors, DNA methylation reflects a level of
epigenetic regulation that is closely linked to transcription
factor (TF) binding and chromatin accessibility. Upregulated DNA
methylation at the gene promoter usually leads to transcriptional
repression (4,5). Accumulating proofs have revealed
that DNA methylation takes a critical role in the regulation of IR,
obesity and T2DM, which are major risk factors for NAFLD. Moreover,
methyl-depleted diets are reported to promote NASH in animal
models, whereas methyl-rich diets prevent NASH (6,7).
Locus-specific and global hypomethylation are also associated with
NAFLD (8–10). Thus, DNA methylation may be of
great importance in the initiation and progression of NAFLD.
Despite its accuracy in NAFLD diagnosis, liver
biopsy is limited in clinical application because of its
invasiveness. Ultrasound, another widely used method for NAFLD
diagnosis, displays poor sensitivity in patients with mild
steatosis (<33%) (11,12). Thus, biomarkers for noninvasive,
early diagnosis of NAFLD remained to be explored until now.
Fortunately, DNA methylation of peripheral leukocytes is currently
employed, serving as alternative for that of organic tissue, in the
diagnosis of various diseases (13–16). For example, lowered level of DNA
methylation in peripheral blood reflects early T2DM (13). A nested case-control study showed
that global hypomethylation in leukocytes may be useful biomarker
of HCC susceptibility (16).
Genomic DNA methylation in peripheral leukocytes is associated with
gastric cancer in a population-based, case-control study (15).
We, therefore, enrolled NAFLD patients and normal
controls from Chinese population so as to profile leukocytic DNA
methylation. The association between DNA methylation and hepatic
pathology was then subjected to evaluation. Characteristic sites of
NAFLD-related methylation were filtered, and further assessed for
noninvasive diagnosis.
Patients and methods
Study population
A total of 65 unrelated adults (aged 18–70 years)
were recruited from March, 2012 to May, 2013. Of these, NAFLD
patients were enrolled from Xinhua Hospital, Shanghai, China
(n=14); Tianjin Hospital of Infectious Diseases, Tianjin, China
(n=4) and Zhengxing Hospital, Zhangzhou, Fujian, China (n=17),
respectively. Thirty healthy controls were recruited accordingly.
All subjects were Han Chinese in origin. Each patient underwent
both ultrasound-guided percutaneous liver biopsy and
FibroScan® 502 (Echosens, Paris, France) examination,
and met the diagnostic criteria for NAFLD (1). The exclusion criteria were as
follows: i) excessive alcohol consumption (>30 g/day for males
and >20 g/day for females); ii) other diseases that led to fatty
liver, such as chronic hepatitis C, drug-induced liver injury,
Wilson's disease, total parenteral nutrition, and autoimmune
hepatitis; iii) previous liver transplantation; and iv) other
end-stage disease or malignancy. All control subjects were
confirmed to be free of liver diseases by B-mode ultrasound and
FibroScan examination [controlled attenuation parameter (CAP)
<240 dB/m and liver stiffness measurement (LSM) values <7.0
kPa] (17,18). These subjects demonstrated normal
liver function without evidence of liver injury. The study protocol
was approved by the Ethics Committee of Xinhua Hospital in
accordance with the Declaration of Helsinki. All participants were
enrolled under informed consent.
Clinical and laboratory evaluation
Demographic and anthropometric measurements were
carried out in NAFLD patients and normal controls, including sex,
age, body mass index [BMI, weight (kg)/height (m)2], and waist-to-hip ratio [WHR, waist
circumference (cm)/hip circumference (cm)]. Biochemical tests were
conducted, including fasting blood glucose (FBG), fasting serum
insulin, total cholesterol (TC), triglycerides (TG), high-density
lipoprotein cholesterol (HDL-C), low-density lipoprotein
cholesterol (LDL-C), alanine transaminase (ALT), aspartate
transaminase (AST), γ-glutamyl-transpeptidase (GGT), total
bilirubin (TBIL), direct bilirubin (DBIL), and uric acid (UA). All
biochemical parameters were measured using a conventional automated
analyzer (7600; Hitachi, Tokyo, Japan). Homeostasis model
assessment of insulin resistant (HOMA-IR) was used to evaluate IR:
HOMA-IR = fasting serum insulin (µIU/ml) x fasting plasma
glucose (mmol/l)/22.5.
Liver histology
Percutaneous liver biopsy was performed in all NAFLD
patients under real-time ultrasound guidance. Biopsy specimens were
then formalin-fixed, paraffin-embedded, sectioned, and treated with
hematoxylin and eosin (H&E), Masson's and reticulin staining.
Histological changes were assessed according to Kleiner's
classification, with the NALFD activity score (NAS) based on
steatosis, lobular inflammation and hepatocyte ballooning (19). Patients with NAS <3 were
excluded of NASH. While those with NAS of 3–4 (at least 1 for
ballooning degeneration), and ≥5 were diagnosed to be borderline
NASH, and NASH, respectively. Liver fibrosis was staged as follows:
F0, none; F1, perisinusoidal or portal fibrosis; F2, perisinusoidal
and periportal fibrosis without bridging; F3, bridging fibrosis;
and F4, cirrhosis (20).
DNA extraction and bisulfite
conversion
Genomic DNA was extracted from peripheral blood
samples obtained from each subject using the nucleic acid
extraction kit (Qiagen, Hilden, Germany). The DNA quality was
determined by a NanoDrop 2000c spectrophotometer (Thermo Fisher
Scientific Inc., Wilmington, DE, USA). Bisulfite conversion of DNA
(500 ng/sample) was then performed by the EZ DNA meth-ylation kit
(Zymo Research, Irvine, CA, USA) according to the manufacturer's
protocol, with a modified thermo-cycling procedure as suggested by
Illumina (San Diego, CA, USA). The protocol included 16 cycles of
denaturing at 95°C for 30 sec, incubation at 50°C for 60 min, and a
final holding step at 4°C.
Methylation analysis
The Human Methylation 450K BeadChip (Illumina) was
used to analyze the genome-wide DNA methylation profile across 485
577 loci distributed in promoters, gene bodies, 3′-untranslated
regions (3′-UTR), and intergenic regions, respectively. In detail,
bisulfite-converted DNA (4 µl) was hybridized with the
Methylation 450K BeadChip following the Illumina Infinium HD
Methylation protocol. Illumina's Genome Studio®
Methylation module version 1.0 (Illumina) was employed to calculate
the methylation level at each CpG site by β-value [β=intensity of
the methylated allele (M)/(intensity of the unmethylated allele (U)
+ intensity of the methylated allele (M) + 100)]. The obtained
β-values, ranging from 0 (fully unmethylated) to 1 (fully
methylated), was further normalized by R package (Partek Inc., St.
Louis, MO, USA) (21).
Resultantly, average Δ-β-value was calculated to indicate the
differential methylation between NAFLD patients and controls.
Quality control inclusion valuation depended on hybridization
detection P-values of <0.05. DNA loci with differential
methylation were filtered by Δ-β-value and DiffScore as follows
(21,22): DiffScore=10Sign
(βNAFLD−βcontrols) log10P.
Bioinformatics analysis
The effect of differentially methylated sites on
biological functions was analyzed by gene ontology (GO). Moreover,
KEGG algorithm was used to identify signaling pathways that
significantly related to the differentially methylated sites. The
false discovery rate (FDR) was employed to exclude false positive
results (23,24).
Pyrosequencing
Acyl-CoA synthetase long-chain family member 4
(ACSL4) was filtered for validation using pyrosequencing. In
detail, forward, GTGATGGATTTTGGGGTTTT, reverse,
AAAACTCCCTAACCCTCAATTAC and sequencing primers,
GTATTTAGAGGGTTAGAAGTTAT were designed with PyroMark Assay Design
software (version 2.0; Qiagen). Bisulfite treated DNA was amplified
by PCR using the PyroMark PCR kit (Qiagen). The pyrosequencing
assay was then performed on a PyroMark Q96 instrument using the
PyroMark Gold Q96 kit (both from Qiagen). The sequencing results
were analyzed using the PyroMark CpG software (Qiagen).
Statistical analysis
Continuous variables were determined by the unpaired
Student's t-test (data with normal distribution) or Mann-Whitney U
test (data with skewed distribution). Comparative analyses of
categorical variables were carried out using the Chi-square test.
Fisher's exact test and Chi-square test were employed to classify
the GO category and significant signaling pathways, respectively.
Two-way ANOVA was used to compare groups with two independent
variables (age and sex). Because of its non-normal distribution,
methylation level of CpG sites was analyzed after log2
transformation. The optimal cut-off value for each CpG site was
determined by maximizing the Youden index. CpG sites with
methylated levels ≥ and < optimal cut-off were defined as high,
and low methylation, respectively. Multivariate regression analysis
was then performed to identify independent risk CpG sites for NAFLD
or NASH. In addition, the diagnostic efficiency for risk CpG sites
was analyzed by area under the receiver operating characteristic
curves (AUC) and 95% confidence interval (CI). All statistical
analyses were performed by SPSS 13.0 software (SPSS, Inc., Chicago,
IL, USA). A two-tailed P<0.05 was considered statistically
significant. The statistical methods used in this study were
reviewed by Dr Guang-Yu Chen from Clinical Epidemiology Center,
Shanghai Jiao Tong University.
Results
Clinical characteristics of the study
population
The clinical characteristics of the study population
are outlined in Table I. When
compared to those of control group, levels of BMI, WHR, ALT, GGT,
TG, TC, HDL-C, LDL-C, UA, CAP and LSM were significantly higher in
the NAFLD group (P<0.01–0.05).
 | Table IClinical characteristics of NAFLD
patients and normal controls. |
Table I
Clinical characteristics of NAFLD
patients and normal controls.
|
Characteristics | NAFLD (n=35) | Controls
(n=30) | P-value |
|---|
| Gender (M/F) | 27/8 | 18/12 | 0.118 |
| Age (years) | 37.72±12.25 | 46.63±7.24 | 0.002 |
| WHR | 0.95±0.04 | 0.86±0.06 | <0.0001 |
| BMI
(kg/m2) | 27.58±3.54 | 23.02±2.63 | <0.0001 |
| ALT (U/l) | 71.99±53.46 | 15.60±4.42 | <0.0001 |
| ALP (U/l) | 95.30±41.02 | 90.20±23.46 | 0.885 |
| GGT (U/l) | 81.46±65.23 | 14.67±5.49 | <0.0001 |
| TG (mmol/l) | 2.25±1.55 | 0.82±0.34 | <0.0001 |
| TC (mmol/l) | 4.94±0.91 | 4.34±0.61 | 0.004 |
| HDL-C (mmol/l) | 1.21±0.30 | 1.43±0.25 | 0.003 |
| LDL-C (mmol/) | 2.97±0.96 | 2.29±0.49 | <0.0001 |
| UA
(µmol/l) | 378.62±113.51 | 257.83±62.14 | <0.0001 |
| FBG (mmol/l) | 5.90±2.80 | 5.29±0.39 | 0.595 |
| Insulin
(µU/ml) | 7.29±5.24 | 3.86±2.48 | <0.0001 |
| HOMA-IR | 1.68±1.03 | 0.90±0.56 | <0.0001 |
| CAP (dB/m) | 310 (277–359) | 195 (174–228) | <0.0001 |
| LSM (kPa) | 7.60 (5.40–12) | 4.15
(3.70–4.70) | <0.0001 |
DNA methylation profile of peripheral
leukocytes differentiated NAFLD patients from normal controls
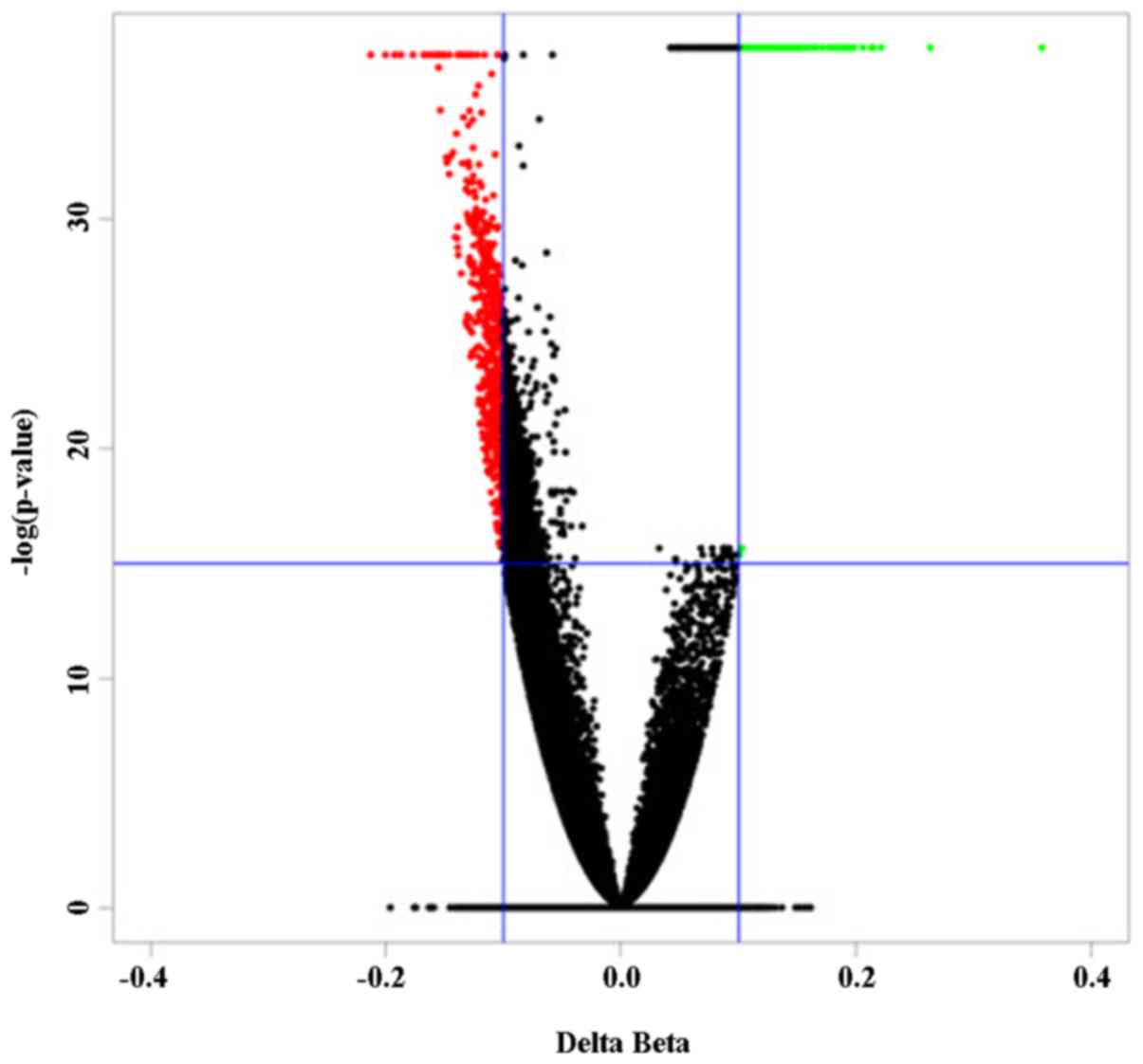
When compared to those of normal controls, a panel
of 863 differentially methylated (DM) CpG sites in peripheral blood
cells characterized the NAFLD patients (Fig. 1). Among these DM sites, 183
(21.2%) were hypermethylated in NAFLD patients, whereas 680 sites
(78.8%) were hypomethylated.
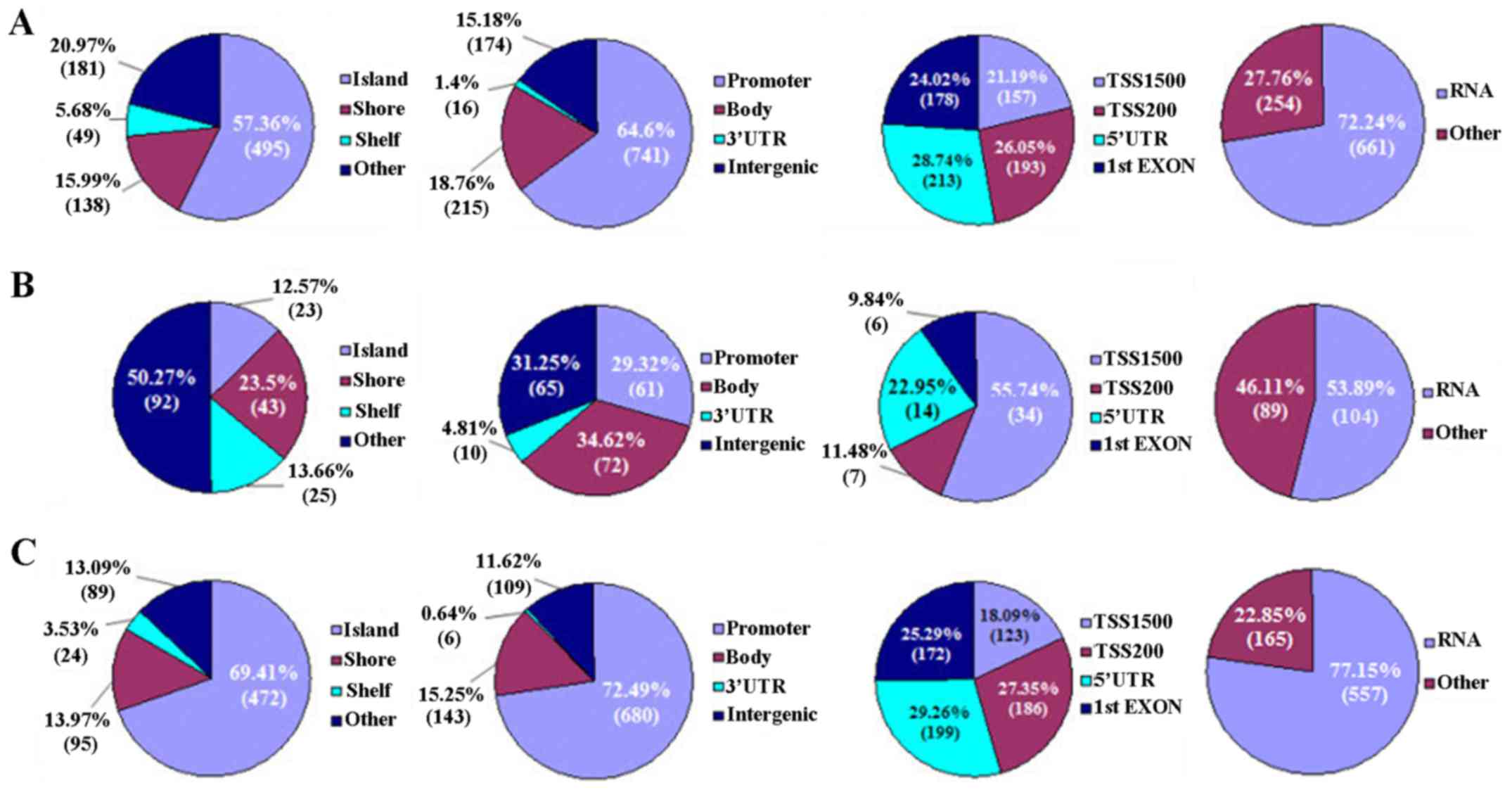
According to genomic location, CpG sites distribute
in CpG islands (CGIs), shores [0–2 kilobase (kb) from CpG islands],
shelves (2–4 kb from CpG islands) and other/open sea, respectively.
On the other hand, the location of CpG sites is classified as
promoter, gene body, 3′-UTR and intergenic, respectively. The
promoter is further divided into surrounding transcription sites
(TSS −200 to −1500 bp, TSS200 and TSS1500), 5′-UTR and 1st exon,
respectively (Fig. 2).
In the present study, most DM CpG sites (57.36%)
localized in CGIs, followed by 20.97 and 15.99% in the other/open
sea and shore regions, respectively. On the contrary, these CpG
sites were enriched in promoter (64.60%), and less likely to be in
the gene body (18.76%), intergenic regions (15.18%), and 3′-UTR
(1.4%). In the promoter, the DM CpG sites usually distributed in
the TSS1500, TSS200, 5′-UTR and 1st exon regions (Fig. 2A).
Methylatively, hypermethylated CpG sites dominated
the other/open sea (50.27%) and gene body (34.62%), respectively.
In the promoter, most hypermethylated CpG sites (55.74%) localized
in TSS500 (Fig. 2B). In contrast,
the hypomethylated CpG sites predominantly located in CGIs (69.41%)
and promoter (72.49%), respectively. In the promoter, these
hypomethylated CpG sites were likely to localize in 5′-UTR (29.26%)
(Fig. 2C). Taken together, the DM
CpG sites (hyper- or hypomethylated sites) were predominantly
distributed in the RNA coding regions of the genome (Fig. 2).
Differential methylation of the
adipocytokine signaling pathway characterized NAFLD patients
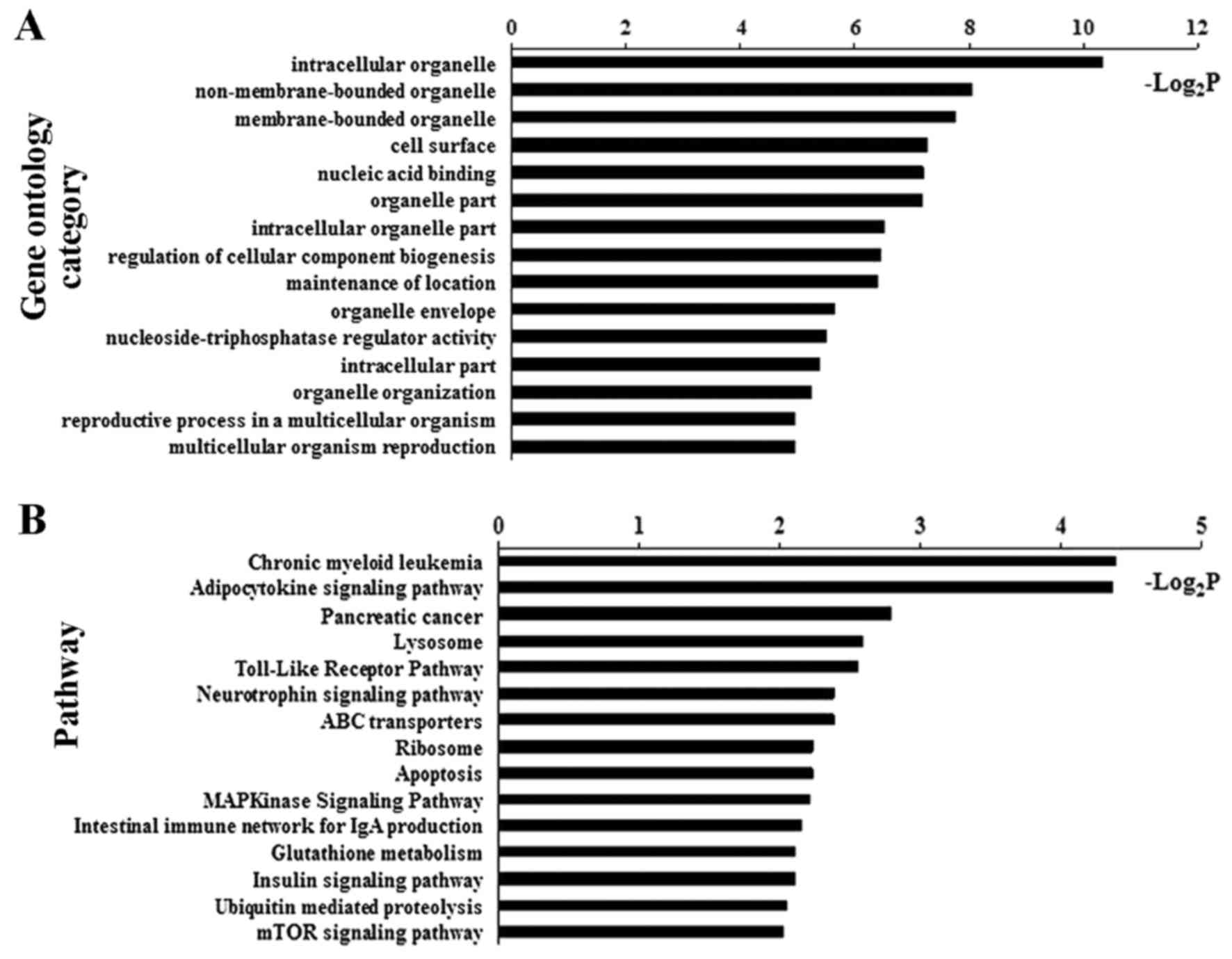
Bioinformatic analysis identified GOs and signaling
pathways significantly associated with the DM CpG sites in NAFLD
patients (Fig. 3 and Table II). Of these, adipocytokine
signaling pathway demonstrated one of the top-ranking pathways.
Four critical genes, including ACSL4, carnitine
palmitoyltransferase 1C (CPT1C), insulin receptor substrate 4
(IRS4) and inhibitor of κ light polypeptide gene enhancer in
B-cells, kinase γ (IKBKG), of the adipocytokine signaling pathway
were verified to be hypomethylated in their promoter (Table II).
 | Table IISignaling pathways related to
differentially methylated genes. |
Table II
Signaling pathways related to
differentially methylated genes.
| Pathways | P-value | Genes |
|---|
| Chronic myeloid
leukemia | 0.047 | GAB2, ARAF, IKBKG,
TGFB3, SMAD3 |
| Adipocytokine
signaling pathway | 0.048 | ACSL4, CPT1C, IRS4,
IKBKG |
| Pancreatic
cancer | 0.144 | ARAF, IKBKG, TGFB3,
SMAD3 |
| Lysosome | 0.165 | LAMP2, AP4E1, IDS,
ATP6AP1, GAA |
| Toll-like receptor
pathway | 0.170 | IRAK1, IKBKG,
ELK1 |
Hypomethylated ACSL4 (cg15536552) and
CPT1C (cg21604803) in adipocytokine signaling pathway conferred
susceptibility to NAFLD
When compared to those in the control group,
β-values of 2 loci in the ACSL4 gene (cg15536552 and cg06822229)
and 1 locus the CPT1C gene (cg21604803) were markedly lower in the
NAFLD group (ACSL4 cg15536552, P=0.009; ACSL4 cg06822229, P=0.023;
CPT1C cg21604803, P=0.004) (Table
III). No significant differences could be found in other loci
between the NAFLD patients and controls.
 | Table IIIDifference in DNA methylation between
NAFLD patients and normal controls. |
Table III
Difference in DNA methylation between
NAFLD patients and normal controls.
| NAFLD (n=35) | Control (n=30) | P-value |
|---|
| ACSL4 | | | |
| cg15536552 | 3.29
(3.09–3.60) | 3.86
(3.37–6.12) | 0.009 |
| cg06822229 | 2.57
(2.23–3.45) | 3.11
(2.75–6.14) | 0.023 |
| cg08855111 | 3.31
(3.12–3.75) | 3.49
(3.18–6.15) | 0.354 |
| cg19635884 | 0.87
(0.08–1.71) | 1.00
(0.46–5.86) | 0.119 |
| cg26119746 | 2.89
(2.70–3.15) | 3.06
(2.57–6.09) | 0.767 |
| cg09091181 | 0.82
(0.34–1.36) | 1.01
(0.32–5.86) | 0.506 |
| cg10721440 | 0.62
(0.13–1.44) | 0.80
(0.17–5.88) | 0.354 |
| CPT1C | | | |
| cg21604803 | 3.22
(2.87–3.53) | 3.64
(3.24–4.11) | 0.004 |
| IRS4 | | | |
| cg06779802 | 3.28
(2.79–4.52) | 3.93
(3.09–6.10) | 0.061 |
| IKBKG | | | |
| cg08417382 | 2.42
(2.06–3.17) | 2.71
(2.15–6.35) | 0.248 |
| cg02869694 | 2.13
(1.79–2.61) | 2.16
(1.75–6.19) | 0.418 |
| cg00813156 | 2.27
(2.08–2.64) | 2.35
(2.10–6.18) | 0.549 |
| cg08560117 | 2.70
(2.55–3.02) | 2.92
(2.60–6.01) | 0.266 |
| cg08873063 | 3.22
(3.09–3.64) | 3.51
(3.17–6.06) | 0.068 |
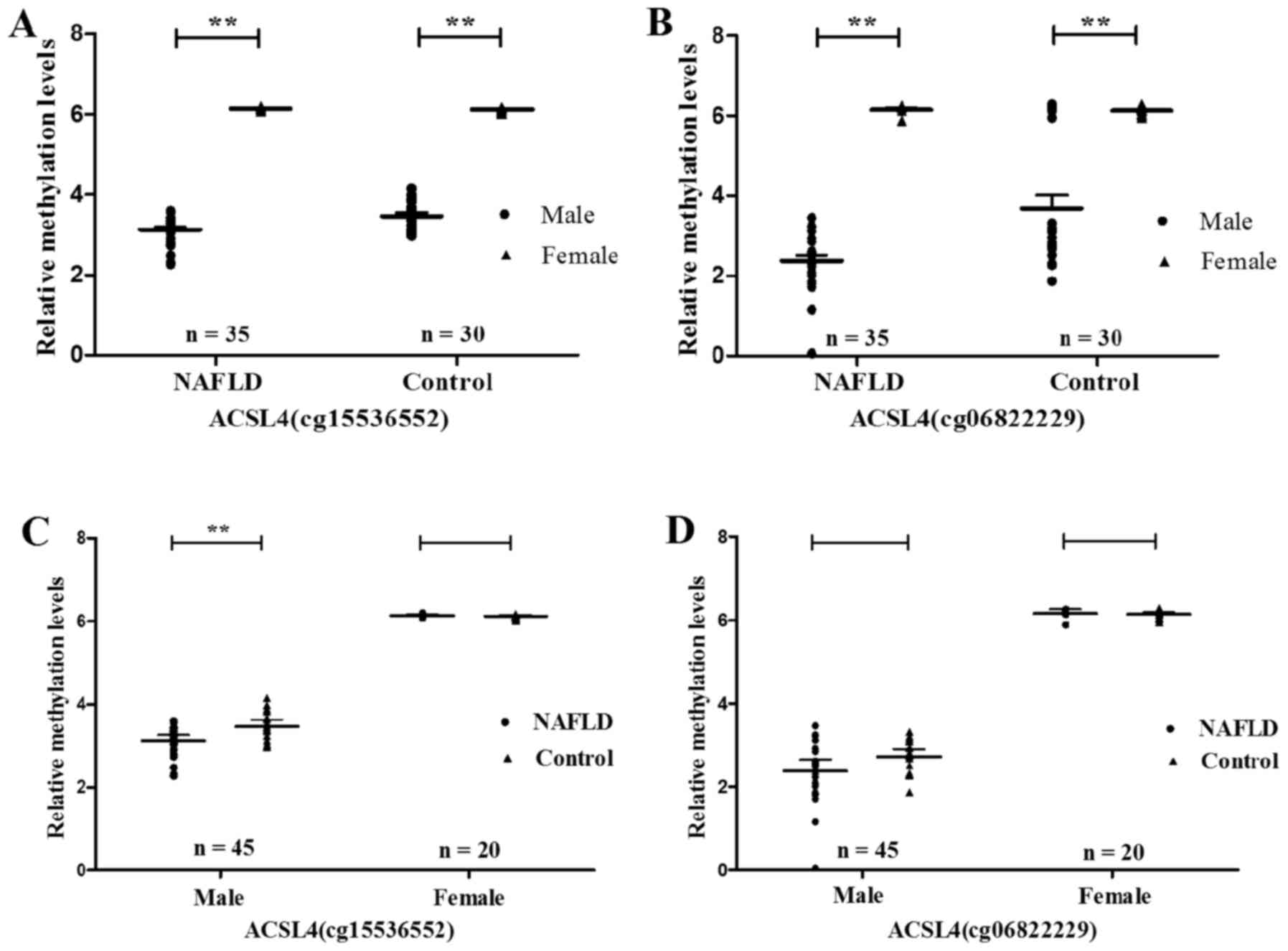
After grouping by sex and age, the ACSL4 (cg15536552
and cg06822229) methylation level was proved to be significantly
higher in females than in males, regardless whether NAFLD patients
(ACSL4 cg15536552, P<0.0001; ACSL4 cg06822229, P<0.0001) or
normal controls (ACSL4 cg15536552, P<0.0001; ACSL4 cg06822229,
P<0.0001) (Fig. 4A and B).
Nevertheless, male NAFLD patients demonstrated methylation levels
of ACSL4 (cg15536552 and cg06822229) much lower than those in the
normal controls correspondingly (ACSL4 cg15536552, P=0.007; ACSL4
cg06822229, P=0.035) (Fig. 4C and
D). As compared to that of normal controls, the methylation
values of ACSL4 (cg15536552) and CPT1C (cg21604803) were also
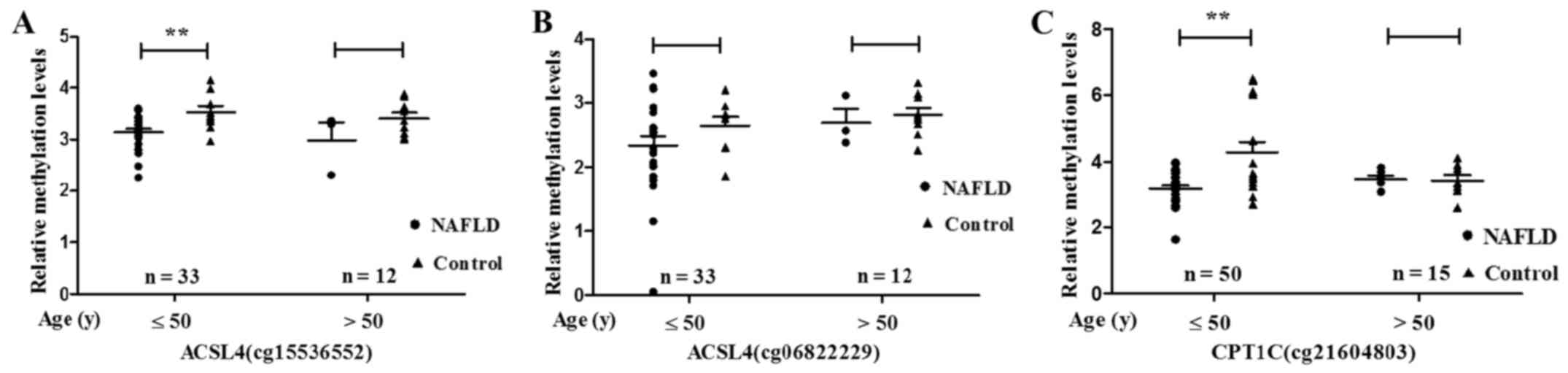
significantly lower in NAFLD patients ≤50 years (ACSL4 cg15536552,
P=0.009; CPT1C cg21604803, P=0.001) (Fig. 5).
Moreover, hypomethylation of ACSL4 (cg15536552) and
CPT1C (cg21604803) was proved to increase the risk of NAFLD (ACSL4
cg15536552, OR: 10.50, 95% CI: 1.70–64.99, P=0.014; CPT1C
cg21604803, OR: 7.67, 95% CI: 2.14–27.49, P=0.001). After adjusting
for BMI and HOMA-IR, the hypomethylated ACSL4 (cg15536552) and
CPT1C (cg21604803) still conferred susceptibility to NAFLD (ACSL4
cg15536552, OR: 11.44, 95% CI: 1.04–125.37, P=0.046; CPT1C
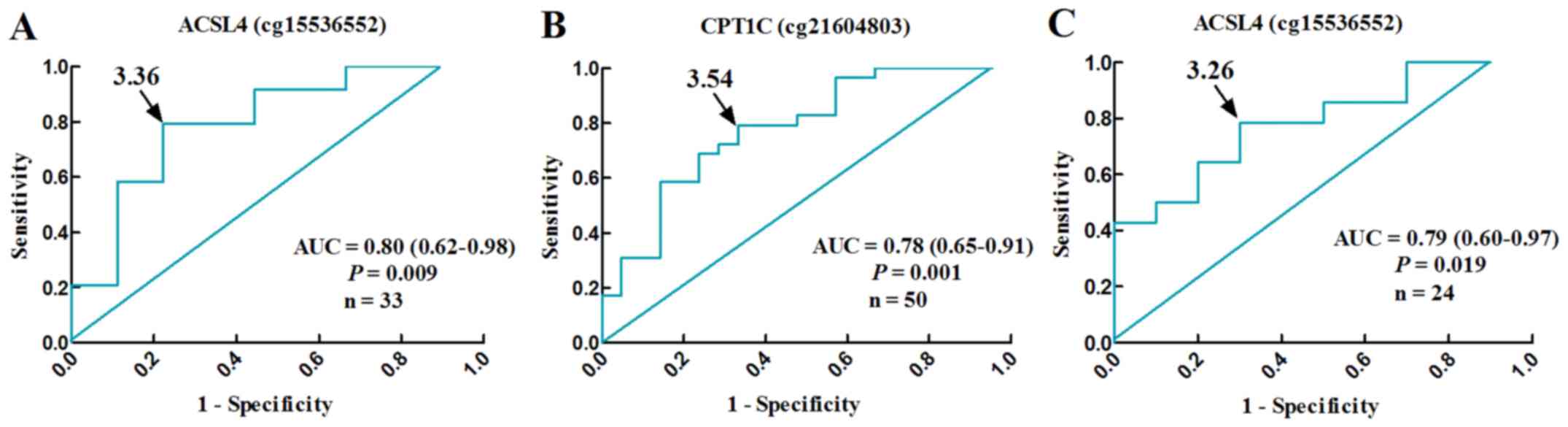
cg21604803, OR: 6.57, 95% CI: 1.02–42.15, P=0.047) (Table IV). Their methylation levels,
with cut-off β-values of 3.36 (ACSL4 cg15536552) and 3.54 (CPT1C
cg21604803), well differentiated NAFLD patients from normal
controls (ACSL4 cg15536552, AUC: 0.80, 95% CI: 0.62–0.98, P=0.009,
n=33; CPT1C cg21604803, AUC: 0.78, 95% CI: 0.65–0.91, P=0.001,
n=50) (Fig. 6A and B).
 | Table IVACSL4 (cg15536552) and CPT1C
(cg21604803) methylation associated with NAFLD and
borderline/definitive NASH. |
Table IV
ACSL4 (cg15536552) and CPT1C
(cg21604803) methylation associated with NAFLD and
borderline/definitive NASH.
| Methylation
level | NAFLD, n (%) | Control, n (%) | OR (95% CI) | P-value | Adjusted OR (95%
CI) | P-value |
|---|
| ACSL4
(cg15536552) | | | | | | |
| High (≥3.36) | 6 (25) | 7 (77.78) | – | | | |
| Low
(<3.36) | 18 (75) | 2 (22.22) | 10.50
(1.70–64.99) | 0.014 | 11.44
(1.04–125.37)a | 0.046 |
| CPT1C
(cg21604803) | | | | | | |
| High (≥3.54) | 6 (20.69) | 14 (66.67) | – | | – | |
| Low
(<3.54) | 23 (79.31) | 7 (33.33) | 7.67
(2.14–27.49) | 0.001 | 6.57
(1.02–42.15)1 | 0.047 |
| Methylation
level | NAS ≥3, n (%) | NAS <3, n
(%) | OR (95% CI) | – | Adjusted OR (95%
CI) | – |
| ACSL4
(cg15536552) | | | | | | |
| High (≥3.26) | 3 (21.43) | 7 (70) | – | | – | |
| Low
(<3.26) | 11 (78.57) | 3 (30) | 8.56
(1.33–54.95) | 0.024 | 8.72
(1.29–58.78)a | 0.026 |
Hypomethylation of ACSL4 (cg15536552) and
CPT1C (cg21604803) associate with histopathological
classification
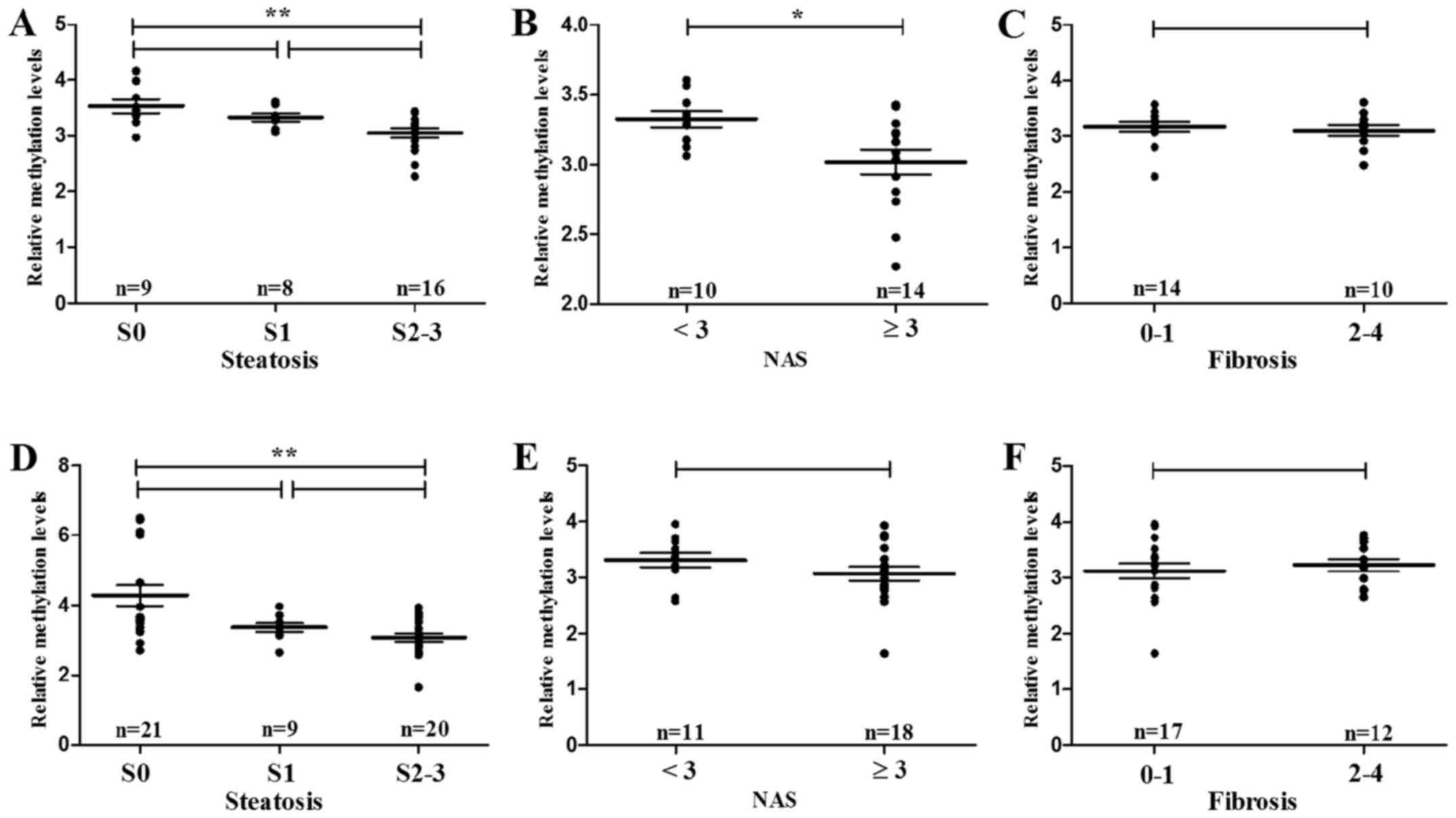
Histopathologically, hypomethylation of ACSL4
(cg15536552) significantly associated with hepatic steatosis (S2–3
vs. S0, P=0.004) and NAS (≥3 vs. <3, P=0.021) (Fig. 7A and B), respectively, in male
subjects. However, no significant correlation was found between
ACSL4 (cg15536552) methylation and liver fibrosis (F2–4 vs. F0–1,
P=0.501) (Fig. 7C). Despite its
noncorrelation with NAS and fibrosis, hypomethylation of CPT1C
(cg21604803) also conferred high risk to hepatic steatosis (S2–3
vs. S0, P=0.007) (Fig. 7D).
Moreover, patients with ACSL4 (cg15536552)
hypomethylation showed increased risk for borderline/definitive
NASH (OR: 8.56, 95% CI: 1.33–54.95, P=0.024). After adjusting for
BMI and HOMA-IR, ACSL4 (cg15536552) hypomethylation qualify itself
for the risk factor of borderline/definitive NASH (OR: 8.72, 95%
CI: 1.29–58.78, P=0.026) (Table
IV). By receiver operating characteristic (ROC) curve,
decreased level of ACSL4 (cg15536552) methylation serve as an
index, with the optimum cut-off β-values of 3.26, for
borderline/definitive NASH in male NAFLD patients (AUC: 0.79, 95%
CI: 0.60–0.97, P=0.019, n=24) (Fig.
6C).
Validation of ACSL4 (cg15536552)
methylation by pyro-sequencing
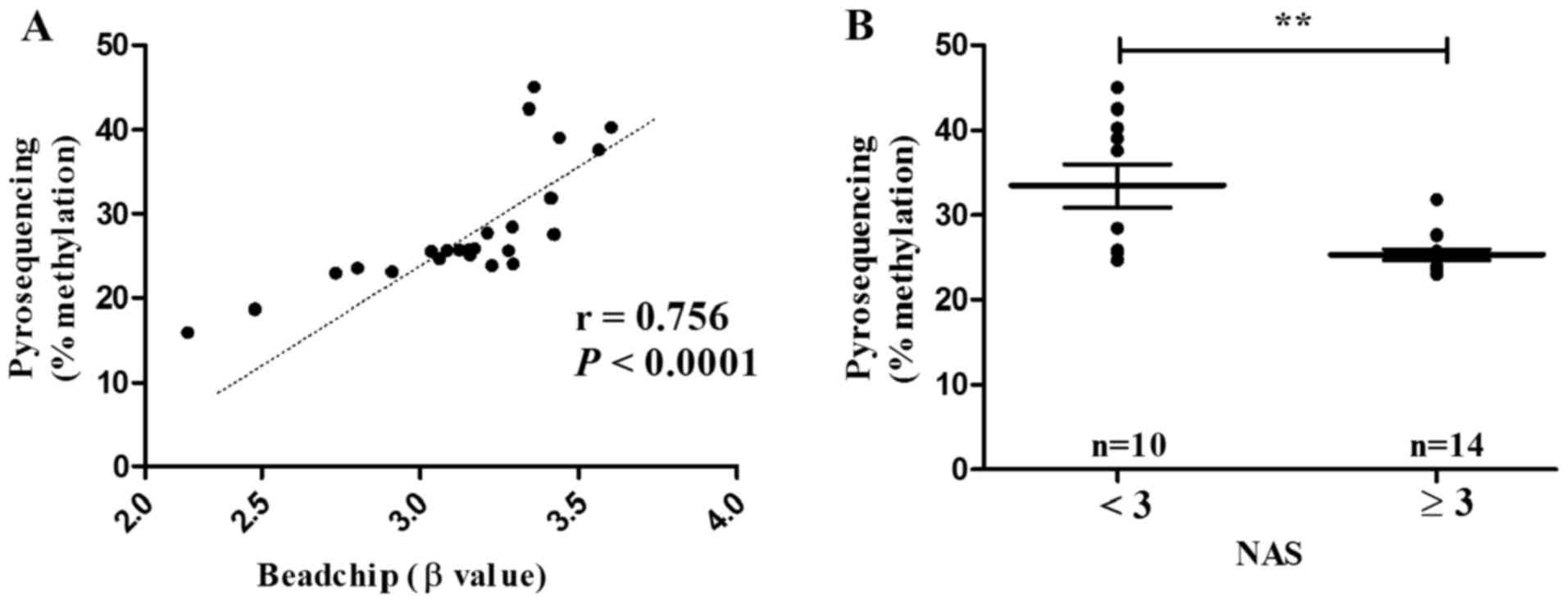
For the sake of its close association with
borderline/definitive NASH, ACSL4 (cg15536552) was subjected to
validation in male NAFLD patients aged ≤50 (n=24). Indeed,
pyrosequencing displayed methylation level of ACSL4 (cg15536552) in
parallel to that of methylation 450K BeadChip (r=0.756,
P<0.0001) (Fig. 8A). The
methylation level of ACSL4 (cg15536552) was also statistically
decreased in patients with borderline/definite NASH (NAS ≥3)
(P=0.004) (Fig. 8B).
Discussion
NAFLD is generally considered to be the result of
environmental, genetic, and epigenetic disorders. DNA methylation
reflects one of the most important patterns of epigenetic
modification, and mediates cross-talk between environmental and
epigenetic factors. To shed light on its role in NAFLD, DNA
methylation was recently subjected to profiling in liver tissue.
First, hepatic DNA methylation was analyzed by genome-wide
profiling in patients with mild to advanced NAFLD (10). Second, global analysis of DNA
methylation was carried out, before and after bariatric surgery, in
morbidly obese patients with NAFLD (25). Strikingly, altered methylation in
CpG sites of genes (i.e., PC, ACLY, PLCG1,
IGF1, IGFBP2, PRKCE, ZNF274,
FGFR2, MAT1A and CASP1), which regulated
glycolipid metabolism, steatohepatitis, fibrosis and
carcinogenesis, demonstrated close association with NAFLD in both
studies (10,26). Hypermethylated promoter of
peroxisome proliferator-activated receptor γ co-activator 1a
(PPARGC1A) significantly correlated to plasma fasting insulin and
HOMA-IR (8). Hypermethylation in
the promoter of glucokinase and L-type pyruvate kinase, which lead
to downregulation of their transcription, is related to both
impaired insulin sensitivity and NAFLD in high-fat diet induced
animal models (9,27). Differences in DNA methylation even
distinguish patients with advanced versus mild NAFLD (10,26). DNA methylation, therefore, is
suggested to be an epigenetic factor deeply involved in NAFLD.
In spite of its critical role in NAFLD, DNA
methylation of liver tissue is prevented from experimental and
clinical application due to the difficulty in sample collection.
Fortunately, peripheral leukocytes instead of organic tissue, has
now been employed in the profiling of DNA methylation related to
various diseases, such as cancers and myelodysplastic syndrome
(13,14,16,25,28). As compared to that of normal
controls, a CpG site in the first intron of FTO shows
significant hypomethylation in the peripheral leukocytes of
patients with type 2 diabetes (13). Increased risk for different types
of cancer (colon, bladder, stomach, breast, head and neck cancer)
has been found in cohorts with the lowest quartile of leukocytic
DNA methylation (15,29–35). Thus DNA methylation of peripheral
leukocytes, serving as alternative for organic tissue to some
extent, may be potential in the identification of NAFLD.
In the present study, genome-wide profiling of DNA
methylation was performed in the peripheral leukocytes obtained
from both NAFLD patients and normal controls. In total, 863 DM CpG
sites characterized the groups of NAFLD patients. In detail, the
percentage of hyper- and hypomethylated loci in the coding region
was much higher than that in other regions of the genome. According
to the non-coding region of genome, hypo- and hypermethylated loci
were abundant in the CGIs of promoter and intergenic region,
respectively. These findings indicate that the DM loci, although
numerically limited, are highly enriched in the biological regions
of the genome.
Dramatically, genomic hypomethylation, representing
78.8% of DM loci, reflected the methylatic characteristics of
peripheral leukocytes in NAFLD patients. Hypo- rather than
hypermethylation, with 76% of DM loci, already reported to dominate
the genome of liver tissues in patients with advanced NAFLD
(10). Hypomethylation also
serves as the epigenetic characteristics of alcohol-related fatty
liver disease (10). It is
further uncovered to underlie the animal model of NASH induced by
methyl-deficient diet (36).
Taken together, hepatocytes and leukocytes of NAFLD patients share
the same pattern of DNA methylation, that of global hypomethylation
(10).
NAFLD-related DM loci of peripheral leukocytes were
then subjected to bioinformatic analysis on the basis of GO and
KEGG algorithm. Interestingly, multiple hypomethylated CpG sites
were focused in critical genes, including ACSL4 (cg15536552,
cg06822229, cg08855111, cg19635884, cg26119746, cg09091181 and
cg10721440), CPT1C (cg21604803), IRS4 (cg06779802) and IKBKG
(cg08417382, cg02869694, cg00813156, cg08560117 and cg08873063), of
adipocytokine signaling pathway. When compared to those in the
control group, NAFLD patients suffered from significantly lowered
β-values in 2 loci of the ACSL4 gene (cg15536552 and cg06822229)
and 1 locus of the CPT1C gene (cg21604803). ACSL4, an important
enzyme regulating the intracellular level of unesterified
arachidonic acid (AA), has been reported overexpressed in
African-American patients with NAFLD or NASH (37). Hepatic fat content and steroid
synthesis are also significantly associated with ACSL4 even after
adjustment for BMI (38).
Functional studies have shown that upregulated ACSL4 accelerates
lipogenesis, whereas downregulated ACSL4 prevents the accumulation
of cellular cholesterol (39). On
the other hand, CPT1C (isoform of CPT1) plays a vital role in the
mitochondrial β-oxidation, energy balance, and hepatic glucose
homeostasis. These results are supposed to reflect the
differentially methylated effect of both peripheral leukocytes and
liver tissue in response to lipogenetic stimuli, and potentiate
leukocytic DNA methylation to be noninvasive biomarker for
NAFLD.
Indeed, statistically hypomethylated ACSL4
(cg15536552) and CPT1C (cg21604803) in peripheral leukocytes
associated with the grade of liver steatosis in this study. Both
loci were verified to be independent variables and risk factors of
NAFLD, even after the adjustment for age, sex and BMI and HOMA-IR.
Their accuracy for the diagnosis of NAFLD, as evaluated by ROC
curves, was 0.80 of ACSL4 cg15536552, and 0.78 of CPT1C cg21604803,
respectively. Moreover, the hypomethylation of ACSL4 (cg15536552)
served as an independent risk factor for borderline/definitive NASH
(NAS ≥3) after the adjustment for BMI and HOMA-IR. Its AUC for the
detection of NAS (≥3) was proved to be 0.79. DNA methylation of
peripheral leukocytes is qualified for the serum biomarker of
NAFLD/NASH with moderate diagnostic efficiency. Noninvasive
biomarker for hepatopathological identification, especially NASH,
has long been the focus of research and clinical interference.
Although multiple indexes [i.e., cytokertin 18 (CK18), procollagen
III, adiponectin, ferritin, tumor necrosis factor-α (TNF-α)] have
been explored (40–42), limited diagnostic efficiency has
made it a difficult task until now. The serum concentration of
CK18, an index for hepatocyte apoptosis, exhibits the most
promising AUC (0.82) for NASH diagnosis (40–42). Dramatically, hypomethylation of
ACSL4 (cg15536552) showed comparable efficacy. Then combination of
these 2 biomarkers may provide us with a better solution for the
noninvasive diagnosis of NAFLD/NASH.
Some limitations of the study should be considered.
First, the DNA methylation was only measured in peripheral
leukocytes. Methylation pattern of whole blood has been reported to
serve as a good proxy for methylation levels from a specific site
of action (43,44). Consistently, both liver tissue and
peripheral leukocytes exhibited global hypomethylation. Despite the
similarity in methylation pattern, there were still different loci
between liver tissue and peripheral leukocytes on the basis of
tissue-specific methylation. Second, sample size of the present
study was relative limited. Clinical trial with large cohorts would
be preferable for the validation of our findings.
In conclusion, NAFLD patients demonstrate global DNA
hypomethylation in peripheral leukocytes when compared to that of
normal controls. Hypomethylated CpG sites of ACSL4 (cg15536552) and
CPT1C (cg21604803), critical genes of adipocytokine signaling
pathway, are associated with the increased risk for NAFLD. Lowered
methylation level of ACSL4 (cg15536552) serves as an index for
borderline/definitive NASH. DNA methylation profiling of peripheral
leukocytes, therefore, may identify noninvasive biomarkers with
potential for the pathological evaluation of NAFLD.
Acknowledgments
The authors would like to thank Professor Ruidan
Zheng and Professor Yuqiang Mi who provided the NAFLD patients,
clinical data and blood samples for this manuscript.
References
|
1
|
Fan JG, Jia JD, Li YM, Wang BY, Lu LG, Shi
JP and Chan LY; Chinese Association for the Study of Liver Disease:
Guidelines for the diagnosis and management of nonalcoholic fatty
liver disease: update 2010: (published in Chinese on Chinese
Journal of Hepatology 2010; 18:163–166). J Dig Dis. 12:38–44. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferreira DM, Simão AL, Rodrigues CM and
Castro RE: Revisiting the metabolic syndrome and paving the way for
microRNAs in non-alcoholic fatty liver disease. FEBS J.
281:2503–2524. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gori M, Arciello M and Balsano C:
MicroRNAs in nonalcoholic fatty liver disease: Novel biomarkers and
prognostic tools during the transition from steatosis to
hepatocarcinoma. Biomed Res Int. 2014:7414652014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lopez-Serra L and Esteller M: Proteins
that bind methylated DNA and human cancer: Reading the wrong words.
Br J Cancer. 98:1881–1885. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bian EB, Zhao B, Huang C, Wang H, Meng XM,
Wu BM, Ma TT, Zhang L, Lv XW and Li J: New advances of DNA
methylation in liver fibrosis, with special emphasis on the
crosstalk between microRNAs and DNA methylation machinery. Cell
Signal. 25:1837–1844. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Robertson KD: DNA methylation and human
disease. Nat Rev Genet. 6:597–610. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Komatsu Y, Waku T, Iwasaki N, Ono W,
Yamaguchi C and Yanagisawa J: Global analysis of DNA methylation in
early-stage liver fibrosis. BMC Med Genomics. 5:52012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sookoian S, Rosselli MS, Gemma C, Burgueño
AL, Fernández Gianotti T, Castaño GO and Pirola CJ: Epigenetic
regulation of insulin resistance in nonalcoholic fatty liver
disease: Impact of liver methylation of the peroxisome
proliferator-activated receptor γ coactivator 1α promoter.
Hepatology. 52:1992–2000. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jiang M, Zhang Y, Liu M, Lan MS, Fei J,
Fan W, Gao X and Lu D: Hypermethylation of hepatic glucokinase and
L-type pyruvate kinase promoters in high-fat diet-induced obese
rats. Endocrinology. 152:1284–1289. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Murphy SK, Yang H, Moylan CA, Pang H,
Dellinger A, Abdelmalek MF, Garrett ME, Ashley-Koch A, Suzuki A,
Tillmann HL, et al: Relationship between methylome and
transcriptome in patients with nonalcoholic fatty liver disease.
Gastroenterology. 145:1076–1087. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Saadeh S, Younossi ZM, Remer EM, Gramlich
T, Ong JP, Hurley M, Mullen KD, Cooper JN and Sheridan MJ: The
utility of radiological imaging in nonalcoholic fatty liver
disease. Gastroenterology. 123:745–750. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dasarathy S, Dasarathy J, Khiyami A,
Joseph R, Lopez R and McCullough AJ: Validity of real time
ultrasound in the diagnosis of hepatic steatosis: A prospective
study. J Hepatol. 51:1061–1067. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Toperoff G, Aran D, Kark JD, Rosenberg M,
Dubnikov T, Nissan B, Wainstein J, Friedlander Y, Levy-Lahad E,
Glaser B, et al: Genome-wide survey reveals predisposing diabetes
type 2-related DNA methylation variations in human peripheral
blood. Hum Mol Genet. 21:371–383. 2012. View Article : Google Scholar :
|
|
14
|
Terry MB, Delgado-Cruzata L, Vin-Raviv N,
Wu HC and Santella RM: DNA methylation in white blood cells:
Association with risk factors in epidemiologic studies.
Epigenetics. 6:828–837. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hou L, Wang H, Sartori S, Gawron A,
Lissowska J, Bollati V, Tarantini L, Zhang FF, Zatonski W, Chow WH,
et al: Blood leukocyte DNA hypomethylation and gastric cancer risk
in a high-risk Polish population. Int J Cancer. 127:1866–1874.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wu HC, Wang Q, Yang HI, Tsai WY, Chen CJ
and Santella RM: Global DNA methylation levels in white blood cells
as a biomarker for hepatocellular carcinoma risk: A nested
case-control study. Carcinogenesis. 33:1340–1345. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sasso M, Tengher-Barna I, Ziol M, Miette
V, Fournier C, Sandrin L, Poupon R, Cardoso AC, Marcellin P, Douvin
C, et al: Novel controlled attenuation parameter for noninvasive
assessment of steatosis using Fibroscan(®): Validation in chronic
hepatitis C. J Viral Hepat. 19:244–253. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wong VW, Vergniol J, Wong GL, Foucher J,
Chan HL, Le Bail B, Choi PC, Kowo M, Chan AW, Merrouche W, et al:
Diagnosis of fibrosis and cirrhosis using liver stiffness
measurement in nonalcoholic fatty liver disease. Hepatology.
51:454–462. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kleiner DE, Brunt EM, Van Natta M, Behling
C, Contos MJ, Cummings OW, Ferrell LD, Liu YC, Torbenson MS,
Unalp-Arida A, et al Nonalcoholic Steatohepatitis Clinical Research
Network: Design and validation of a histological scoring system for
nonalcoholic fatty liver disease. Hepatology. 41:1313–1321. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bedossa P and Poynard T; The METAVIR
Cooperative Study Group: An algorithm for the grading of activity
in chronic hepatitis C. Hepatology. 24:289–293. 1996. View Article : Google Scholar
|
|
21
|
Bibikova M, Barnes B, Tsan C, Ho V,
Klotzle B, Le JM, Delano D, Zhang L, Schroth GP, Gunderson KL, et
al: High density DNA methylation array with single CpG site
resolution. Genomics. 98:288–295. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Downey T: Analysis of a multifactor
microarray study using Partek genomics solution. Methods Enzymol.
411:256–270. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dupuy D, Bertin N, Hidalgo CA, Venkatesan
K, Tu D, Lee D, Rosenberg J, Svrzikapa N, Blanc A, Carnec A, et al:
Genome-scale analysis of in vivo spatiotemporal promoter activity
in Caenorhabditis elegans. Nat Biotechnol. 25:663–668. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Draghici S, Khatri P, Tarca AL, Amin K,
Done A, Voichita C, Georgescu C and Romero R: A systems biology
approach for pathway level analysis. Genome Res. 17:1537–1545.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Steegenga WT, Boekschoten MV, Lute C,
Hooiveld GJ, de Groot PJ, Morris TJ, Teschendorff AE, Butcher LM,
Beck S and Müller M: Genome-wide age-related changes in DNA
methylation and gene expression in human PBMCs. Age (Dordr).
36:96482014. View Article : Google Scholar
|
|
26
|
Ahrens M, Ammerpohl O, von Schönfels W,
Kolarova J, Bens S, Itzel T, Teufel A, Herrmann A, Brosch M,
Hinrichsen H, et al: DNA methylation analysis in nonalcoholic fatty
liver disease suggests distinct disease-specific and remodeling
signatures after bariatric surgery. Cell Metab. 18:296–302. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sookoian S and Pirola CJ: DNA methylation
and hepatic insulin resistance and steatosis. Curr Opin Clin Nutr
Metab Care. 15:350–356. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Aberg KA, McClay JL, Nerella S, Clark S,
Kumar G, Chen W, Khachane AN, Xie L, Hudson A, Gao G, et al:
Methylome-wide association study of schizophrenia: Identifying
blood biomarker signatures of environmental insults. JAMA
Psychiatry. 71:255–264. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lim U, Flood A, Choi SW, Albanes D, Cross
AJ, Schatzkin A, Sinha R, Katki HA, Cash B, Schoenfeld P, et al:
Genomic methylation of leukocyte DNA in relation to colorectal
adenoma among asymptomatic women. Gastroenterology. 134:47–55.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pufulete M, Al-Ghnaniem R, Leather AJ,
Appleby P, Gout S, Terry C, Emery PW and Sanders TA: Folate status,
genomic DNA hypomethylation, and risk of colorectal adenoma and
cancer: A case control study. Gastroenterology. 124:1240–1248.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cash HL, Tao L, Yuan JM, Marsit CJ,
Houseman EA, Xiang YB, Gao YT, Nelson HH and Kelsey KT: LINE-1
hypomethylation is associated with bladder cancer risk among
nonsmoking Chinese. Int J Cancer. 130:1151–1159. 2012. View Article : Google Scholar
|
|
32
|
Wilhelm CS, Kelsey KT, Butler R, Plaza S,
Gagne L, Zens MS, Andrew AS, Morris S, Nelson HH, Schned AR, et al:
Implications of LINE1 methylation for bladder cancer risk in women.
Clin Cancer Res. 16:1682–1689. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Moore LE, Pfeiffer RM, Poscablo C, Real
FX, Kogevinas M, Silverman D, García-Closas R, Chanock S, Tardón A,
Serra C, et al: Genomic DNA hypomethylation as a biomarker for
bladder cancer susceptibility in the Spanish Bladder Cancer Study:
A case-control study. Lancet Oncol. 9:359–366. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Choi JY, James SR, Link PA, McCann SE,
Hong CC, Davis W, Nesline MK, Ambrosone CB and Karpf AR:
Association between global DNA hypomethylation in leukocytes and
risk of breast cancer. Carcinogenesis. 30:1889–1897. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hsiung DT, Marsit CJ, Houseman EA, Eddy K,
Furniss CS, McClean MD and Kelsey KT: Global DNA methylation level
in whole blood as a biomarker in head and neck squamous cell
carcinoma. Cancer Epidemiol Biomarkers Prev. 16:108–114. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tryndyak VP, Han T, Muskhelishvili L,
Fuscoe JC, Ross SA, Beland FA and Pogribny IP: Coupling global
methylation and gene expression profiles reveal key
pathophysiological events in liver injury induced by a
methyl-deficient diet. Mol Nutr Food Res. 55:411–418. 2011.
View Article : Google Scholar
|
|
37
|
Stepanova M, Hossain N, Afendy A, Perry K,
Goodman ZD, Baranova A and Younossi Z: Hepatic gene expression of
Caucasian and African-American patients with obesity-related
non-alcoholic fatty liver disease. Obes Surg. 20:640–650. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Westerbacka J, Kolak M, Kiviluoto T,
Arkkila P, Sirén J, Hamsten A, Fisher RM and Yki-Järvinen H: Genes
involved in fatty acid partitioning and binding, lipolysis,
monocyte/macrophage recruitment, and inflammation are overexpressed
in the human fatty liver of insulin-resistant subjects. Diabetes.
56:2759–2765. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cui M, Xiao Z, Sun B, Wang Y, Zheng M, Ye
L and Zhang X: Involvement of cholesterol in hepatitis B virus X
protein-induced abnormal lipid metabolism of hepatoma cells via
upregulating miR-205-targeted ACSL4. Biochem Biophys Res Commun.
445:651–655. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Purnomo HD, Mundhofir FE, Kasno, Sudijanto
E, Darmono, Daldiyono, Djokomoeljanto R and Faradz SM: Combination
of aspartate aminotranferase and tumor necrosis factor-α as non
invasive diagnostic tools for non alcoholic steatohepatitis (NASH).
Acta Med Indones. 47:16–23. 2015.PubMed/NCBI
|
|
41
|
Tamimi TI, Elgouhari HM, Alkhouri N,
Yerian LM, Berk MP, Lopez R, Schauer PR, Zein NN and Feldstein AE:
An apoptosis panel for nonalcoholic steatohepatitis diagnosis. J
Hepatol. 54:1224–1229. 2011. View Article : Google Scholar :
|
|
42
|
Arulanandan A and Loomba R: Non-invasive
testing for NASH and NASH with advanced fibrosis: Are we there yet?
Curr Hepatol Rep. 14:109–118. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Heyn H and Esteller M: DNA methylation
profiling in the clinic: Applications and challenges. Nat Rev
Genet. 13:679–692. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lowe R, Slodkowicz G, Goldman N and Rakyan
VK: The human blood DNA methylome displays a highly distinctive
profile compared with other somatic tissues. Epigenetics.
10:274–281. 2015. View Article : Google Scholar : PubMed/NCBI
|