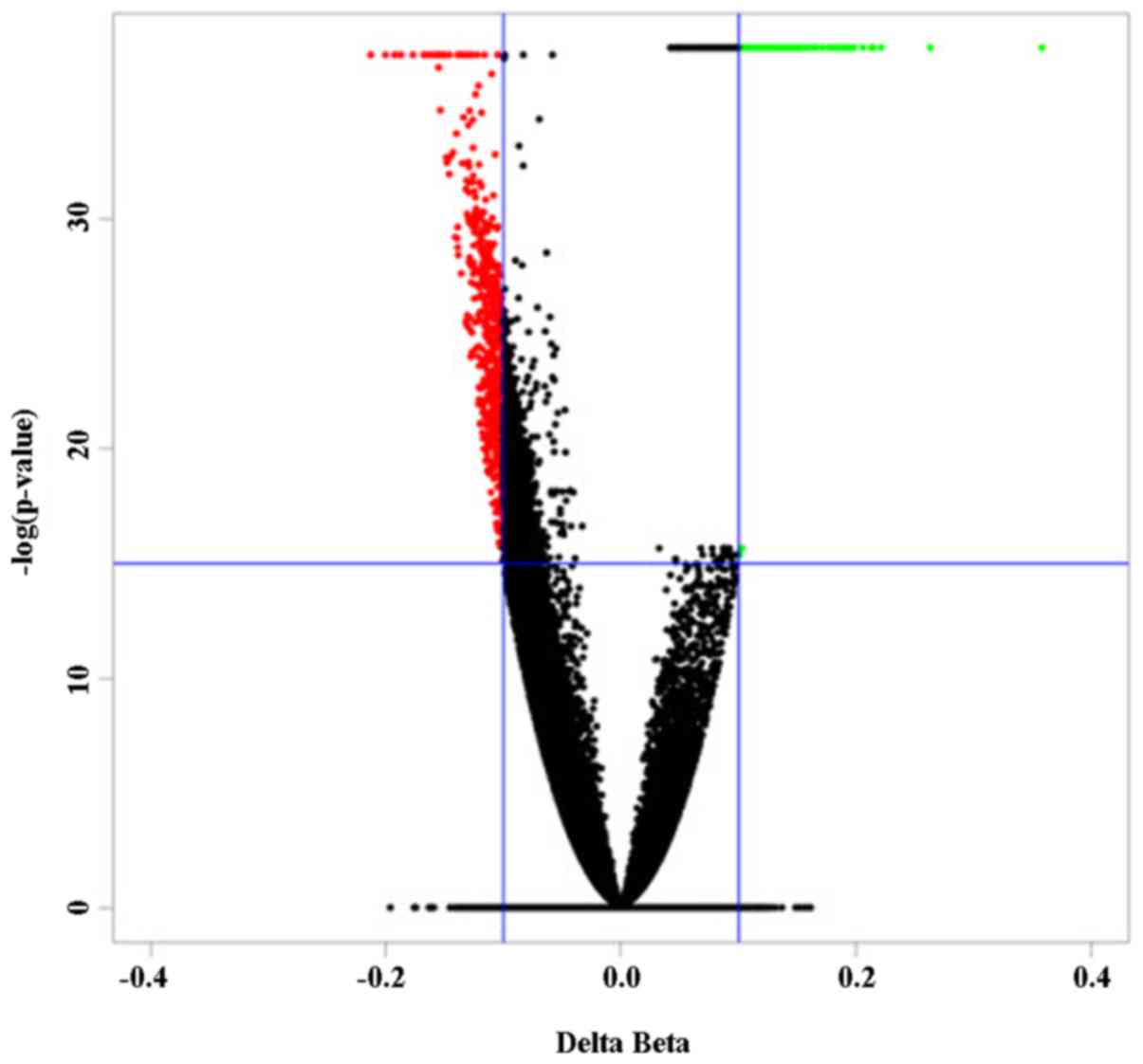
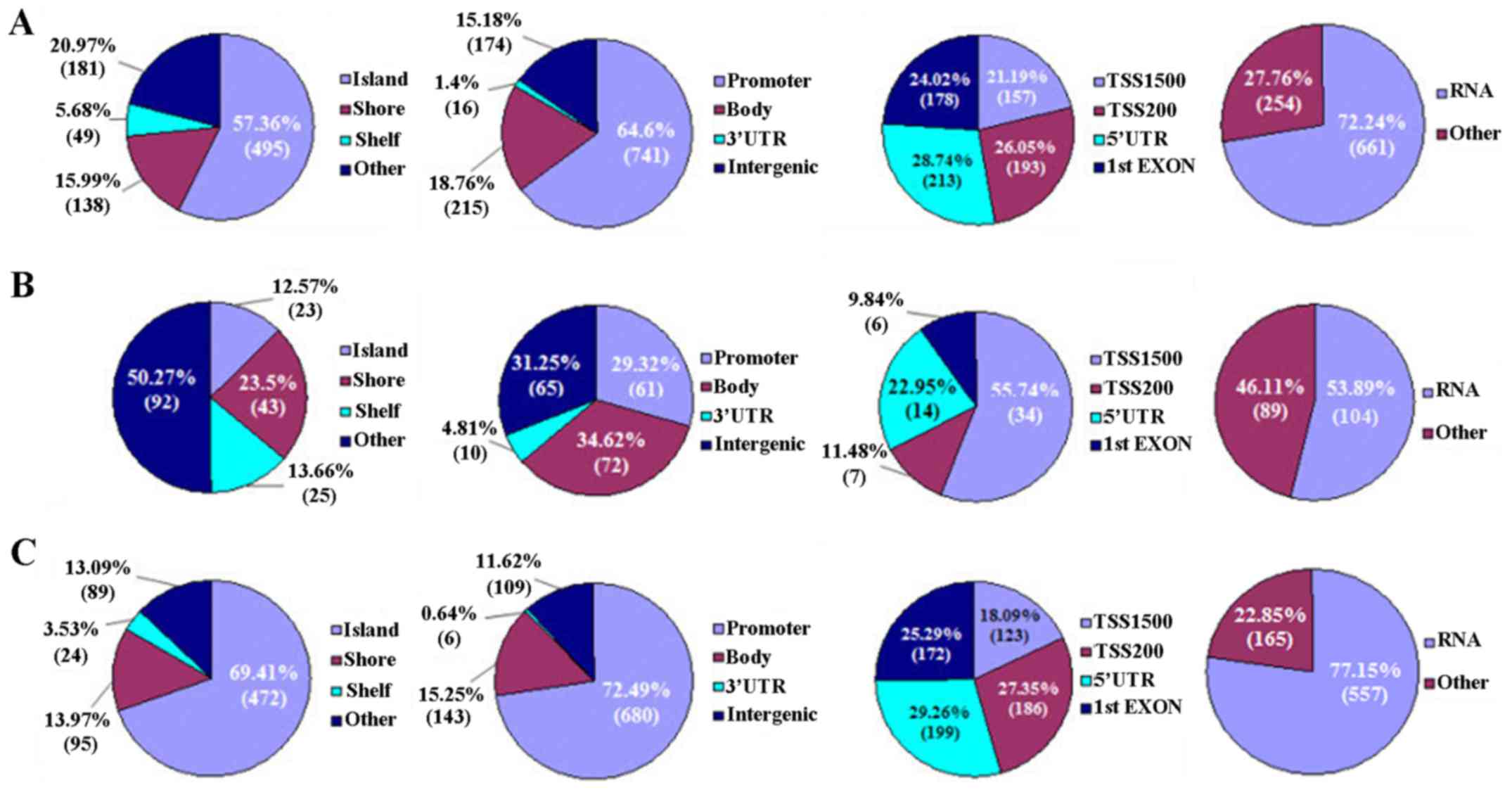
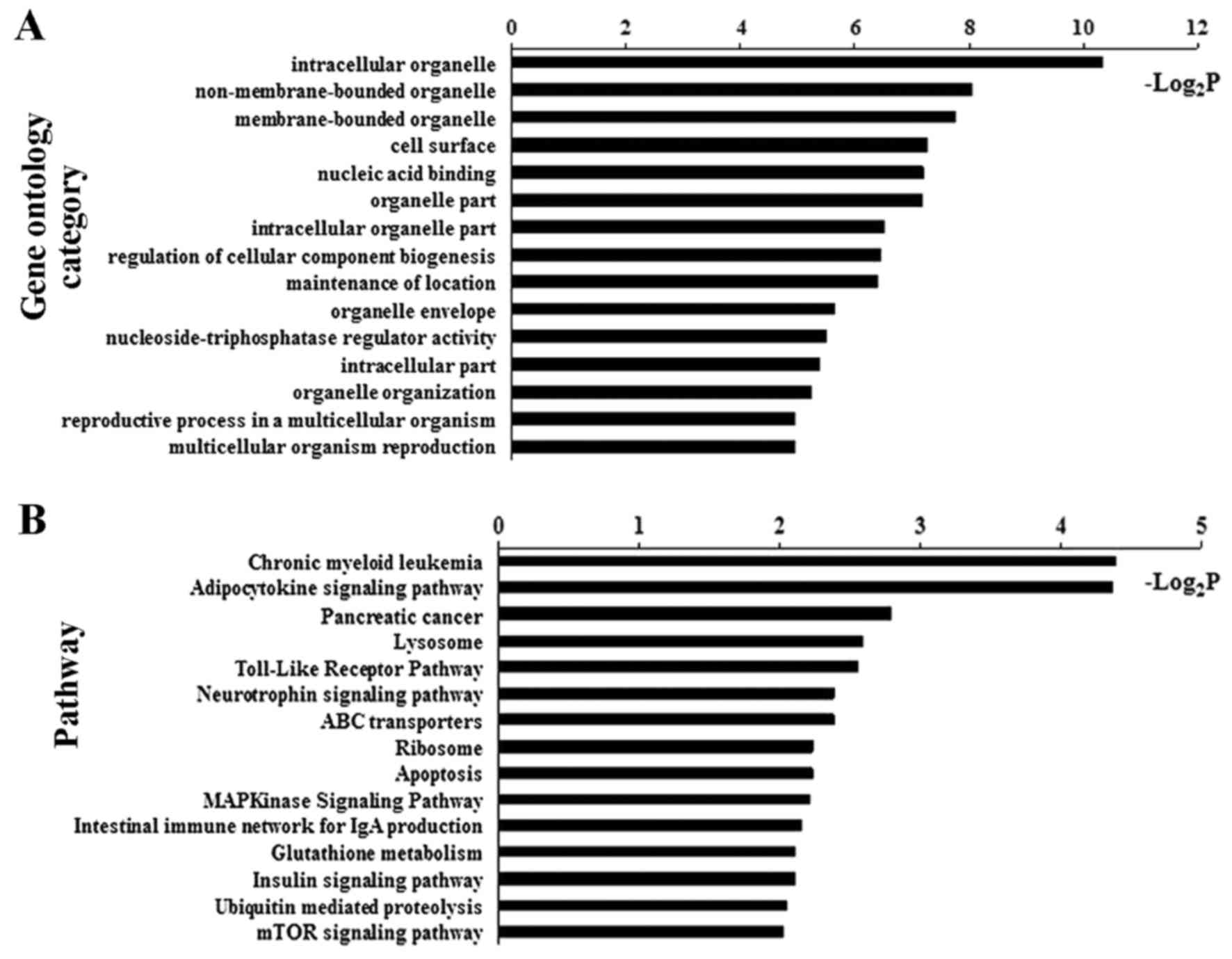
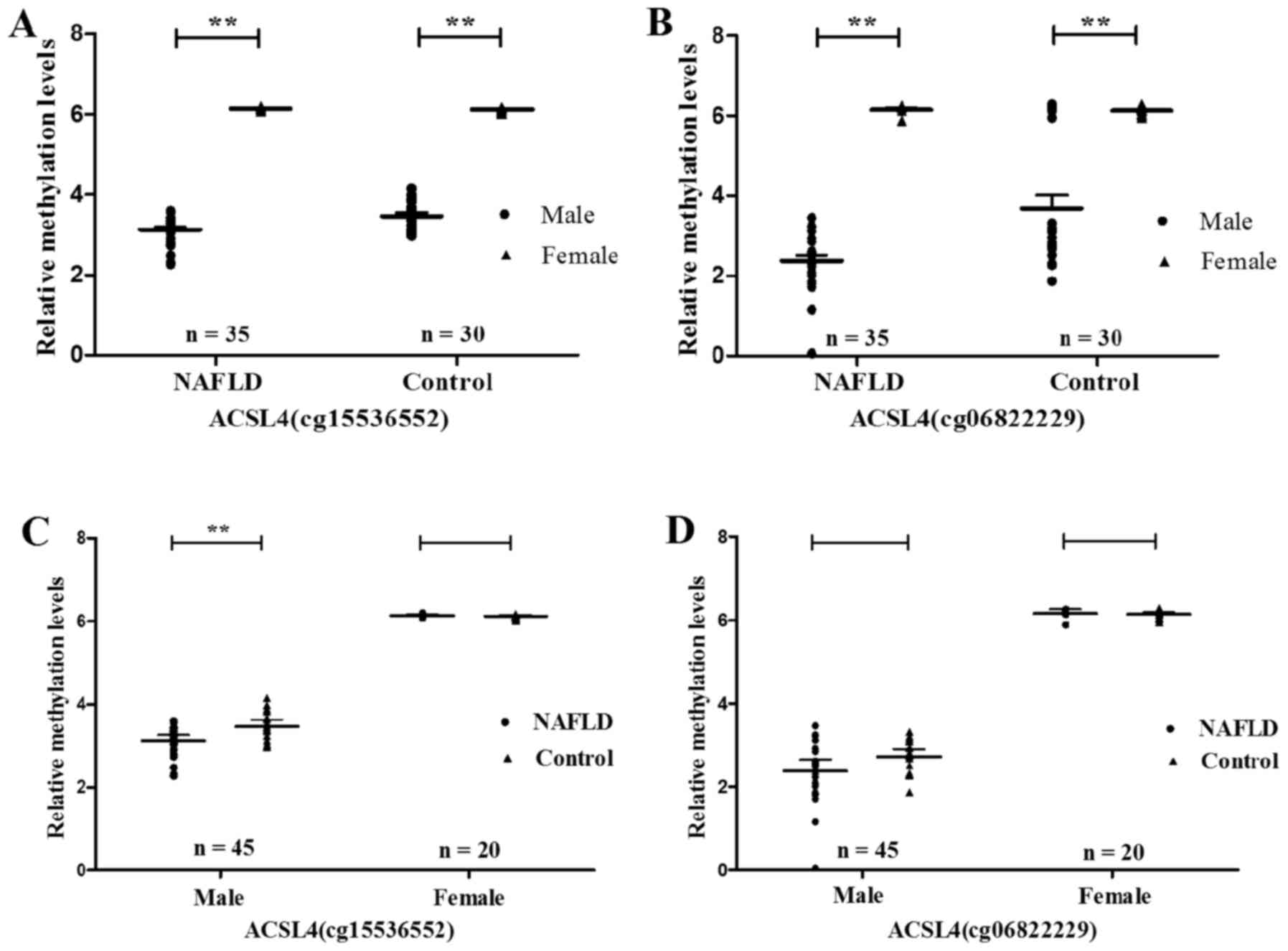
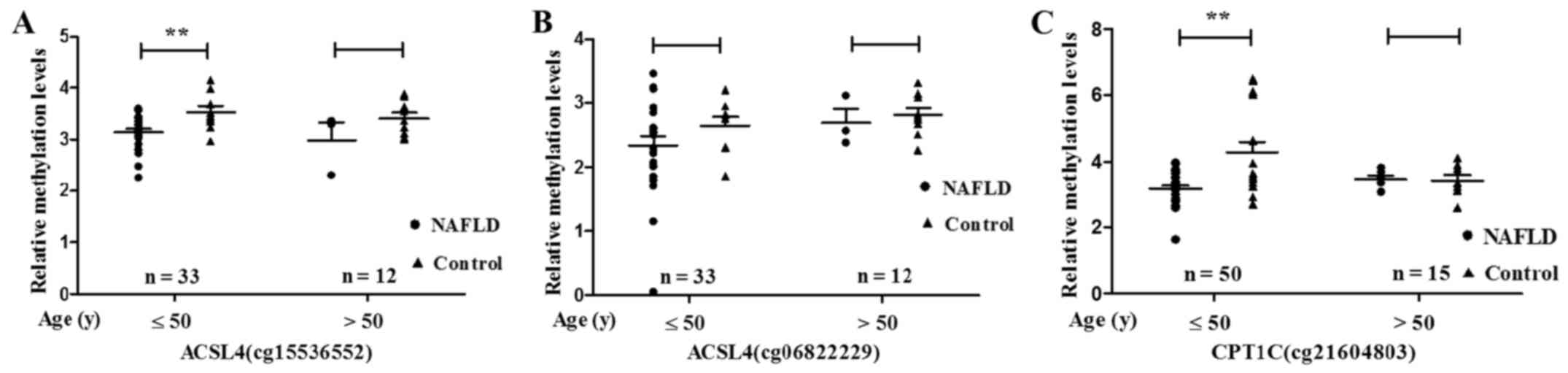
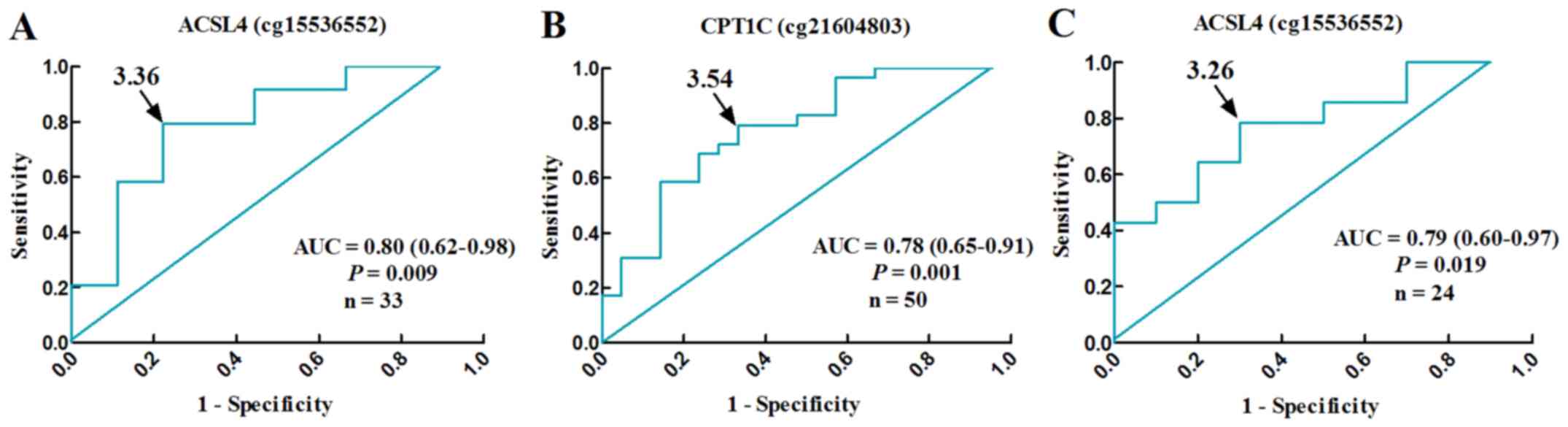
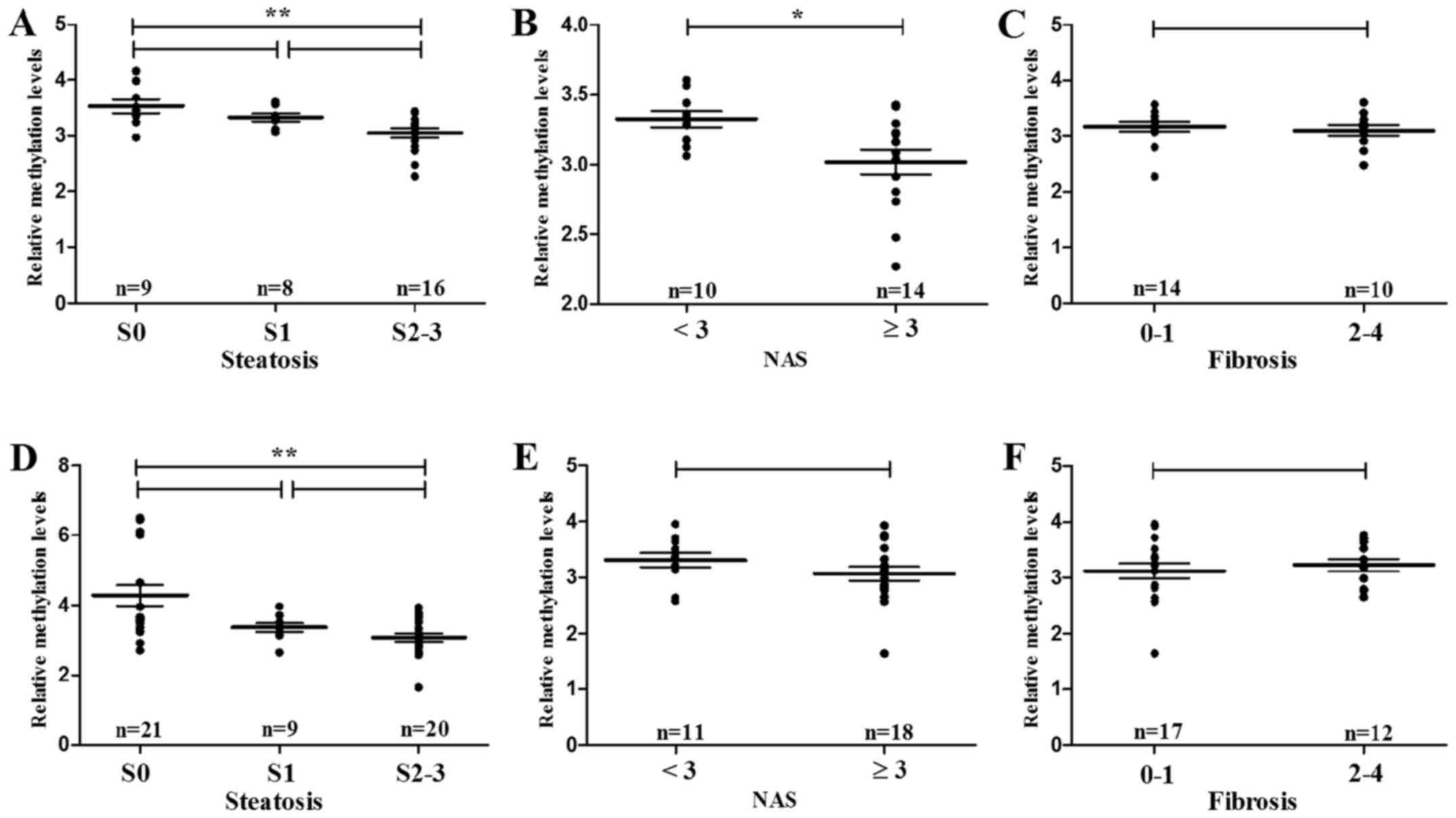
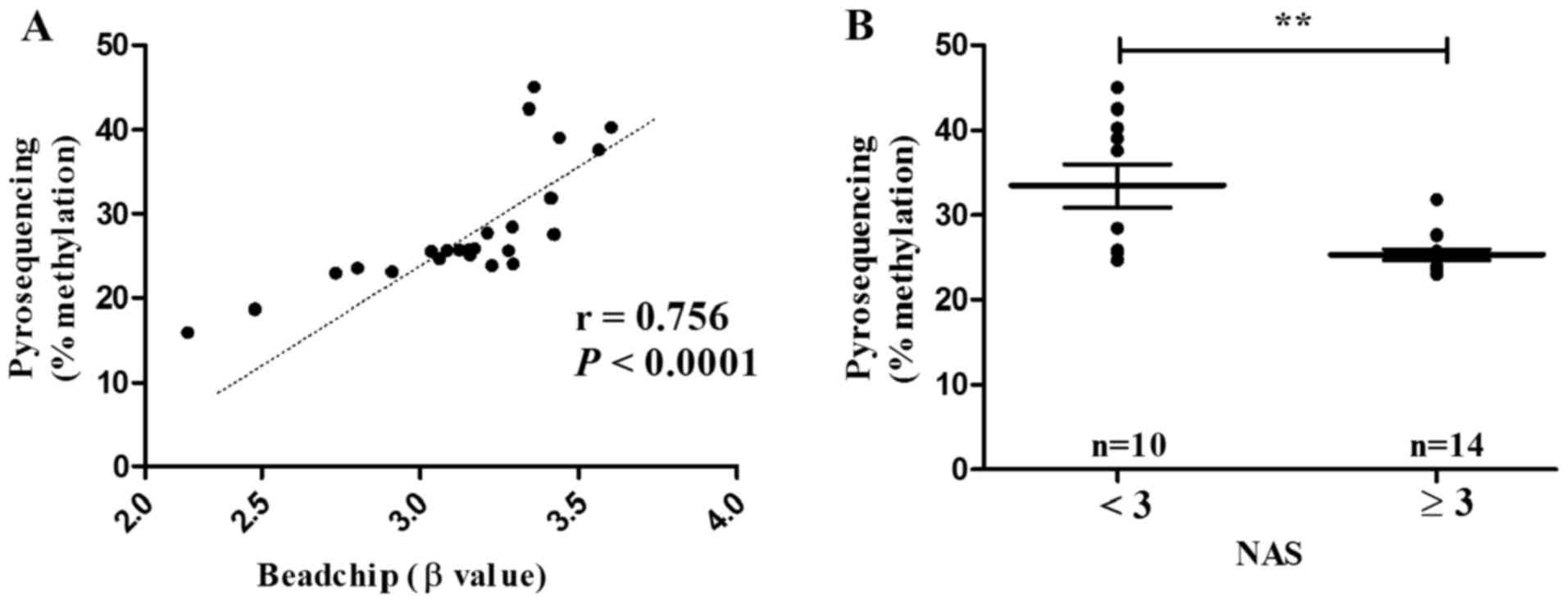
|
1
|
Fan JG, Jia JD, Li YM, Wang BY, Lu LG, Shi
JP and Chan LY; Chinese Association for the Study of Liver Disease:
Guidelines for the diagnosis and management of nonalcoholic fatty
liver disease: update 2010: (published in Chinese on Chinese
Journal of Hepatology 2010; 18:163–166). J Dig Dis. 12:38–44. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferreira DM, Simão AL, Rodrigues CM and
Castro RE: Revisiting the metabolic syndrome and paving the way for
microRNAs in non-alcoholic fatty liver disease. FEBS J.
281:2503–2524. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gori M, Arciello M and Balsano C:
MicroRNAs in nonalcoholic fatty liver disease: Novel biomarkers and
prognostic tools during the transition from steatosis to
hepatocarcinoma. Biomed Res Int. 2014:7414652014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lopez-Serra L and Esteller M: Proteins
that bind methylated DNA and human cancer: Reading the wrong words.
Br J Cancer. 98:1881–1885. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bian EB, Zhao B, Huang C, Wang H, Meng XM,
Wu BM, Ma TT, Zhang L, Lv XW and Li J: New advances of DNA
methylation in liver fibrosis, with special emphasis on the
crosstalk between microRNAs and DNA methylation machinery. Cell
Signal. 25:1837–1844. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Robertson KD: DNA methylation and human
disease. Nat Rev Genet. 6:597–610. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Komatsu Y, Waku T, Iwasaki N, Ono W,
Yamaguchi C and Yanagisawa J: Global analysis of DNA methylation in
early-stage liver fibrosis. BMC Med Genomics. 5:52012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sookoian S, Rosselli MS, Gemma C, Burgueño
AL, Fernández Gianotti T, Castaño GO and Pirola CJ: Epigenetic
regulation of insulin resistance in nonalcoholic fatty liver
disease: Impact of liver methylation of the peroxisome
proliferator-activated receptor γ coactivator 1α promoter.
Hepatology. 52:1992–2000. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jiang M, Zhang Y, Liu M, Lan MS, Fei J,
Fan W, Gao X and Lu D: Hypermethylation of hepatic glucokinase and
L-type pyruvate kinase promoters in high-fat diet-induced obese
rats. Endocrinology. 152:1284–1289. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Murphy SK, Yang H, Moylan CA, Pang H,
Dellinger A, Abdelmalek MF, Garrett ME, Ashley-Koch A, Suzuki A,
Tillmann HL, et al: Relationship between methylome and
transcriptome in patients with nonalcoholic fatty liver disease.
Gastroenterology. 145:1076–1087. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Saadeh S, Younossi ZM, Remer EM, Gramlich
T, Ong JP, Hurley M, Mullen KD, Cooper JN and Sheridan MJ: The
utility of radiological imaging in nonalcoholic fatty liver
disease. Gastroenterology. 123:745–750. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dasarathy S, Dasarathy J, Khiyami A,
Joseph R, Lopez R and McCullough AJ: Validity of real time
ultrasound in the diagnosis of hepatic steatosis: A prospective
study. J Hepatol. 51:1061–1067. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Toperoff G, Aran D, Kark JD, Rosenberg M,
Dubnikov T, Nissan B, Wainstein J, Friedlander Y, Levy-Lahad E,
Glaser B, et al: Genome-wide survey reveals predisposing diabetes
type 2-related DNA methylation variations in human peripheral
blood. Hum Mol Genet. 21:371–383. 2012. View Article : Google Scholar :
|
|
14
|
Terry MB, Delgado-Cruzata L, Vin-Raviv N,
Wu HC and Santella RM: DNA methylation in white blood cells:
Association with risk factors in epidemiologic studies.
Epigenetics. 6:828–837. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hou L, Wang H, Sartori S, Gawron A,
Lissowska J, Bollati V, Tarantini L, Zhang FF, Zatonski W, Chow WH,
et al: Blood leukocyte DNA hypomethylation and gastric cancer risk
in a high-risk Polish population. Int J Cancer. 127:1866–1874.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wu HC, Wang Q, Yang HI, Tsai WY, Chen CJ
and Santella RM: Global DNA methylation levels in white blood cells
as a biomarker for hepatocellular carcinoma risk: A nested
case-control study. Carcinogenesis. 33:1340–1345. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sasso M, Tengher-Barna I, Ziol M, Miette
V, Fournier C, Sandrin L, Poupon R, Cardoso AC, Marcellin P, Douvin
C, et al: Novel controlled attenuation parameter for noninvasive
assessment of steatosis using Fibroscan(®): Validation in chronic
hepatitis C. J Viral Hepat. 19:244–253. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wong VW, Vergniol J, Wong GL, Foucher J,
Chan HL, Le Bail B, Choi PC, Kowo M, Chan AW, Merrouche W, et al:
Diagnosis of fibrosis and cirrhosis using liver stiffness
measurement in nonalcoholic fatty liver disease. Hepatology.
51:454–462. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kleiner DE, Brunt EM, Van Natta M, Behling
C, Contos MJ, Cummings OW, Ferrell LD, Liu YC, Torbenson MS,
Unalp-Arida A, et al Nonalcoholic Steatohepatitis Clinical Research
Network: Design and validation of a histological scoring system for
nonalcoholic fatty liver disease. Hepatology. 41:1313–1321. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bedossa P and Poynard T; The METAVIR
Cooperative Study Group: An algorithm for the grading of activity
in chronic hepatitis C. Hepatology. 24:289–293. 1996. View Article : Google Scholar
|
|
21
|
Bibikova M, Barnes B, Tsan C, Ho V,
Klotzle B, Le JM, Delano D, Zhang L, Schroth GP, Gunderson KL, et
al: High density DNA methylation array with single CpG site
resolution. Genomics. 98:288–295. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Downey T: Analysis of a multifactor
microarray study using Partek genomics solution. Methods Enzymol.
411:256–270. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dupuy D, Bertin N, Hidalgo CA, Venkatesan
K, Tu D, Lee D, Rosenberg J, Svrzikapa N, Blanc A, Carnec A, et al:
Genome-scale analysis of in vivo spatiotemporal promoter activity
in Caenorhabditis elegans. Nat Biotechnol. 25:663–668. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Draghici S, Khatri P, Tarca AL, Amin K,
Done A, Voichita C, Georgescu C and Romero R: A systems biology
approach for pathway level analysis. Genome Res. 17:1537–1545.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Steegenga WT, Boekschoten MV, Lute C,
Hooiveld GJ, de Groot PJ, Morris TJ, Teschendorff AE, Butcher LM,
Beck S and Müller M: Genome-wide age-related changes in DNA
methylation and gene expression in human PBMCs. Age (Dordr).
36:96482014. View Article : Google Scholar
|
|
26
|
Ahrens M, Ammerpohl O, von Schönfels W,
Kolarova J, Bens S, Itzel T, Teufel A, Herrmann A, Brosch M,
Hinrichsen H, et al: DNA methylation analysis in nonalcoholic fatty
liver disease suggests distinct disease-specific and remodeling
signatures after bariatric surgery. Cell Metab. 18:296–302. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sookoian S and Pirola CJ: DNA methylation
and hepatic insulin resistance and steatosis. Curr Opin Clin Nutr
Metab Care. 15:350–356. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Aberg KA, McClay JL, Nerella S, Clark S,
Kumar G, Chen W, Khachane AN, Xie L, Hudson A, Gao G, et al:
Methylome-wide association study of schizophrenia: Identifying
blood biomarker signatures of environmental insults. JAMA
Psychiatry. 71:255–264. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lim U, Flood A, Choi SW, Albanes D, Cross
AJ, Schatzkin A, Sinha R, Katki HA, Cash B, Schoenfeld P, et al:
Genomic methylation of leukocyte DNA in relation to colorectal
adenoma among asymptomatic women. Gastroenterology. 134:47–55.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pufulete M, Al-Ghnaniem R, Leather AJ,
Appleby P, Gout S, Terry C, Emery PW and Sanders TA: Folate status,
genomic DNA hypomethylation, and risk of colorectal adenoma and
cancer: A case control study. Gastroenterology. 124:1240–1248.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cash HL, Tao L, Yuan JM, Marsit CJ,
Houseman EA, Xiang YB, Gao YT, Nelson HH and Kelsey KT: LINE-1
hypomethylation is associated with bladder cancer risk among
nonsmoking Chinese. Int J Cancer. 130:1151–1159. 2012. View Article : Google Scholar
|
|
32
|
Wilhelm CS, Kelsey KT, Butler R, Plaza S,
Gagne L, Zens MS, Andrew AS, Morris S, Nelson HH, Schned AR, et al:
Implications of LINE1 methylation for bladder cancer risk in women.
Clin Cancer Res. 16:1682–1689. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Moore LE, Pfeiffer RM, Poscablo C, Real
FX, Kogevinas M, Silverman D, García-Closas R, Chanock S, Tardón A,
Serra C, et al: Genomic DNA hypomethylation as a biomarker for
bladder cancer susceptibility in the Spanish Bladder Cancer Study:
A case-control study. Lancet Oncol. 9:359–366. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Choi JY, James SR, Link PA, McCann SE,
Hong CC, Davis W, Nesline MK, Ambrosone CB and Karpf AR:
Association between global DNA hypomethylation in leukocytes and
risk of breast cancer. Carcinogenesis. 30:1889–1897. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hsiung DT, Marsit CJ, Houseman EA, Eddy K,
Furniss CS, McClean MD and Kelsey KT: Global DNA methylation level
in whole blood as a biomarker in head and neck squamous cell
carcinoma. Cancer Epidemiol Biomarkers Prev. 16:108–114. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tryndyak VP, Han T, Muskhelishvili L,
Fuscoe JC, Ross SA, Beland FA and Pogribny IP: Coupling global
methylation and gene expression profiles reveal key
pathophysiological events in liver injury induced by a
methyl-deficient diet. Mol Nutr Food Res. 55:411–418. 2011.
View Article : Google Scholar
|
|
37
|
Stepanova M, Hossain N, Afendy A, Perry K,
Goodman ZD, Baranova A and Younossi Z: Hepatic gene expression of
Caucasian and African-American patients with obesity-related
non-alcoholic fatty liver disease. Obes Surg. 20:640–650. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Westerbacka J, Kolak M, Kiviluoto T,
Arkkila P, Sirén J, Hamsten A, Fisher RM and Yki-Järvinen H: Genes
involved in fatty acid partitioning and binding, lipolysis,
monocyte/macrophage recruitment, and inflammation are overexpressed
in the human fatty liver of insulin-resistant subjects. Diabetes.
56:2759–2765. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cui M, Xiao Z, Sun B, Wang Y, Zheng M, Ye
L and Zhang X: Involvement of cholesterol in hepatitis B virus X
protein-induced abnormal lipid metabolism of hepatoma cells via
upregulating miR-205-targeted ACSL4. Biochem Biophys Res Commun.
445:651–655. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Purnomo HD, Mundhofir FE, Kasno, Sudijanto
E, Darmono, Daldiyono, Djokomoeljanto R and Faradz SM: Combination
of aspartate aminotranferase and tumor necrosis factor-α as non
invasive diagnostic tools for non alcoholic steatohepatitis (NASH).
Acta Med Indones. 47:16–23. 2015.PubMed/NCBI
|
|
41
|
Tamimi TI, Elgouhari HM, Alkhouri N,
Yerian LM, Berk MP, Lopez R, Schauer PR, Zein NN and Feldstein AE:
An apoptosis panel for nonalcoholic steatohepatitis diagnosis. J
Hepatol. 54:1224–1229. 2011. View Article : Google Scholar :
|
|
42
|
Arulanandan A and Loomba R: Non-invasive
testing for NASH and NASH with advanced fibrosis: Are we there yet?
Curr Hepatol Rep. 14:109–118. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Heyn H and Esteller M: DNA methylation
profiling in the clinic: Applications and challenges. Nat Rev
Genet. 13:679–692. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lowe R, Slodkowicz G, Goldman N and Rakyan
VK: The human blood DNA methylome displays a highly distinctive
profile compared with other somatic tissues. Epigenetics.
10:274–281. 2015. View Article : Google Scholar : PubMed/NCBI
|