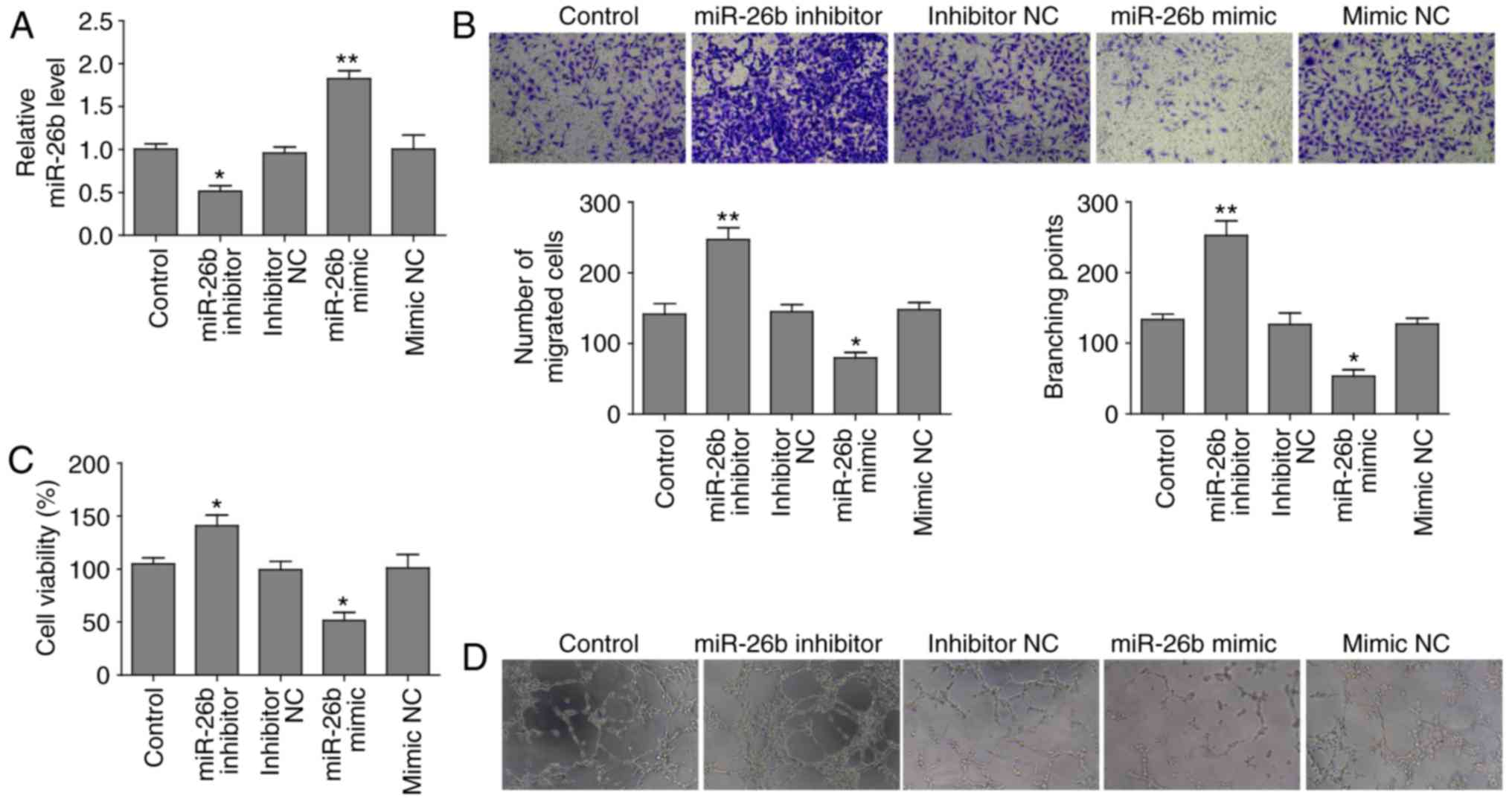
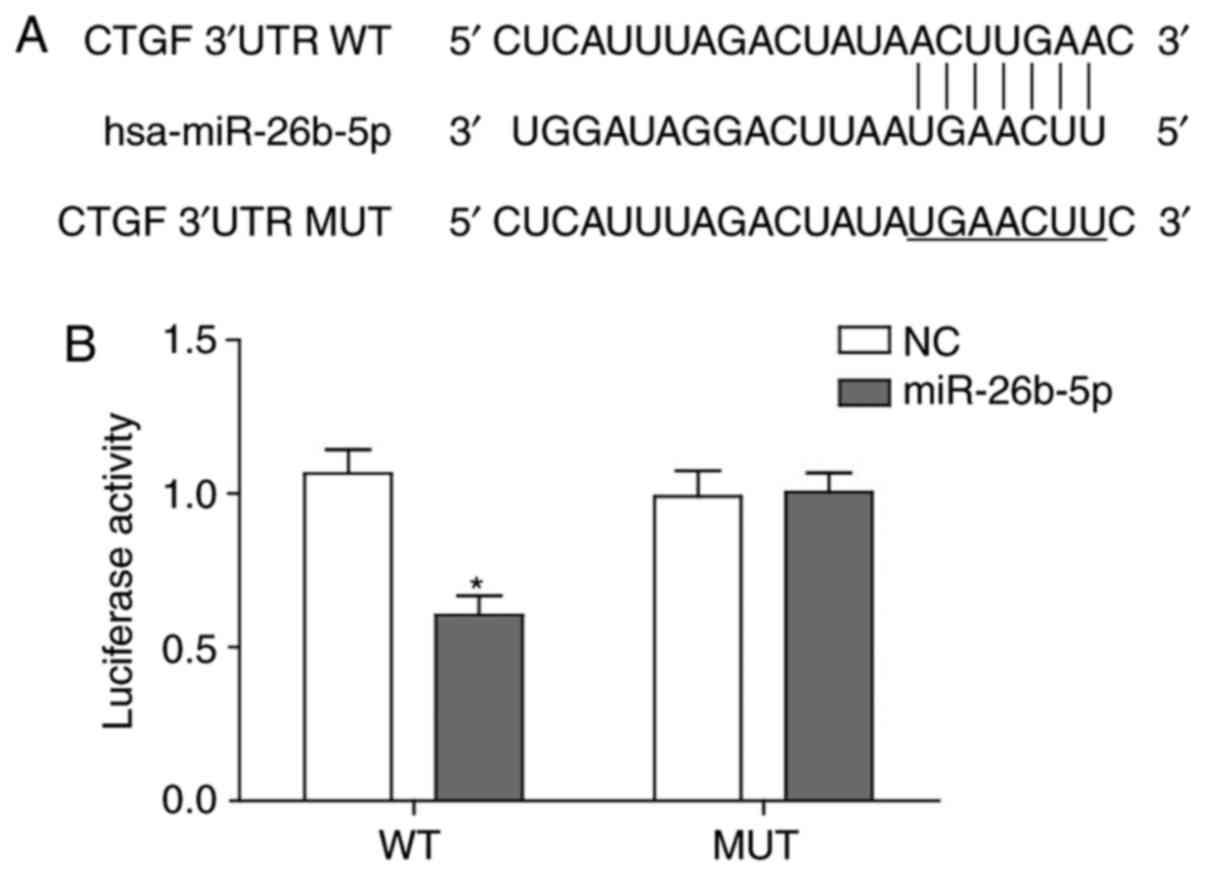
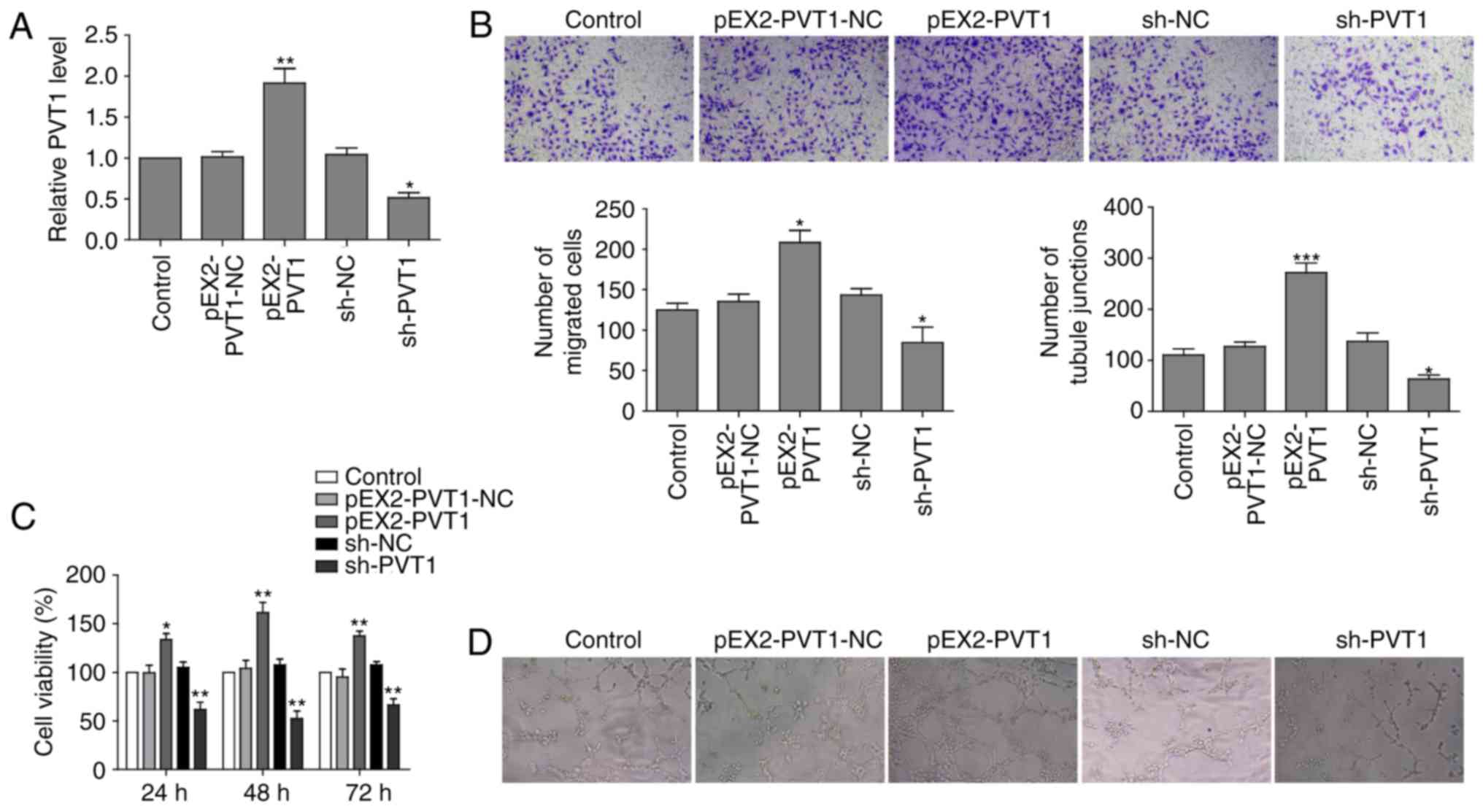
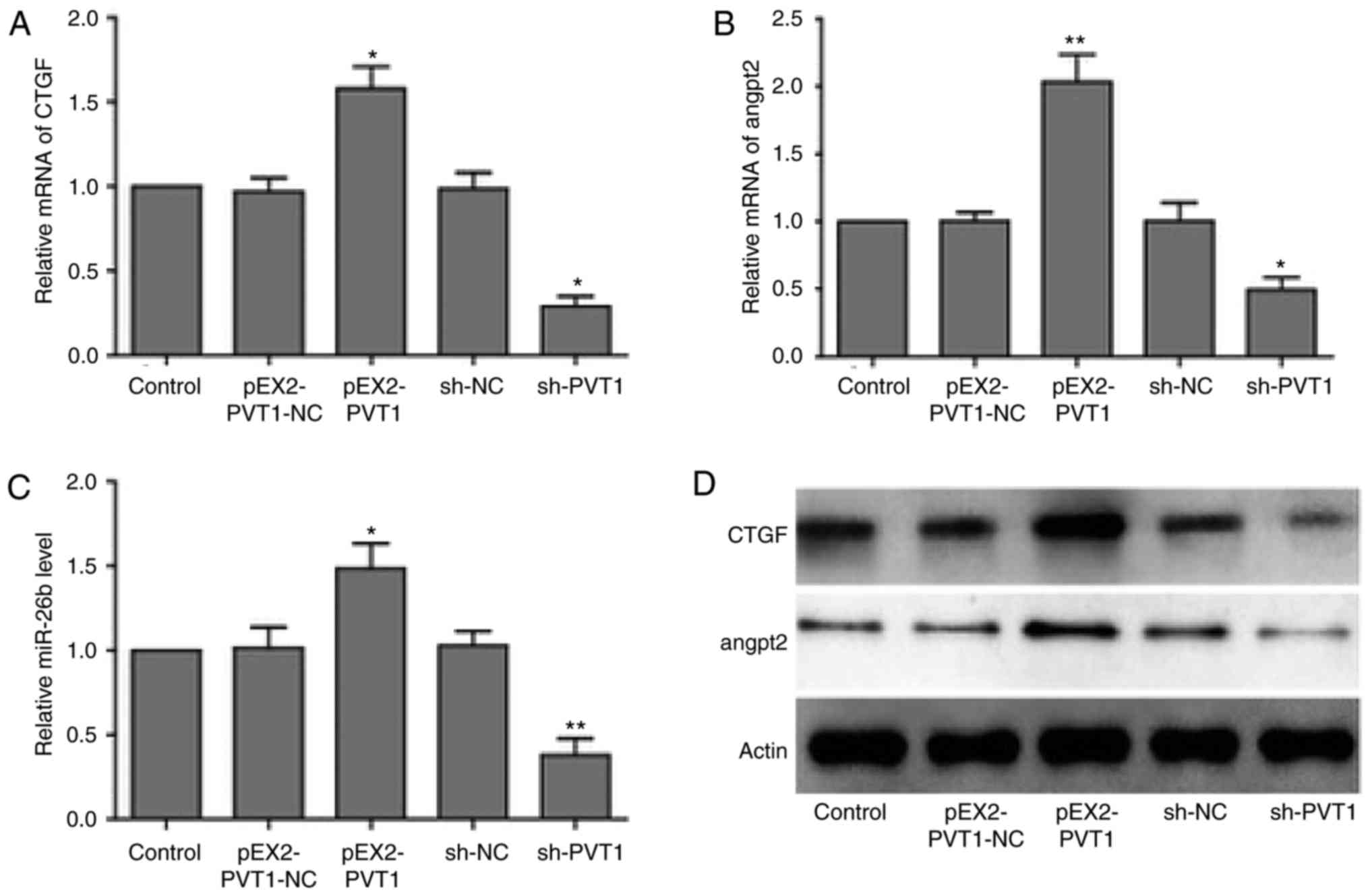
|
1
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Colombo T, Farina L, Macino G and Paci P:
PVT1: A rising star among oncogenic long noncoding RNAs. BioMed Res
Int. 2015:3042082015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yang YR, Zang SZ, Zhong CL, Li YX, Zhao SS
and Feng XJ: Increased expression of the lncRNA PVT1 promotes
tumorigenesis in non-small cell lung cancer. Int J Clin Exp Pathol.
7:6929–6935. 2014.PubMed/NCBI
|
|
4
|
Ding C, Yang Z, Lv Z, Du C, Xiao H, Peng
C, Cheng S, Xie H, Zhou L, Wu J and Zheng S: Long non-coding RNA
PVT1 is associated with tumor progression and predicts recurrence
in hepatocellular carcinoma patients. Oncol Lett. 9:955–963. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ding J, Li D, Gong M, Wang J, Huang X, Wu
T and Wang C: Expression and clinical significance of the long
non-coding RNA PVT1 in human gastric cancer. Onco Targets Ther.
7:1625–1630. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zheng X, Hu H and Li S: High expression of
lncRNA PVT1 promotes invasion by inducing epithelial-to-mesenchymal
transition in esophageal cancer. Oncol Lett. 12:2357–2362. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shen CJ, Cheng YM and Wang CL: LncRNA PVT1
epigenetically silences miR-195 and modulates EMT and
chemoresistance in cervical cancer cells. J Drug Target. 25:1–8.
2017. View Article : Google Scholar
|
|
8
|
Chunharojrith P, Nakayama Y, Jiang X, Kery
RE, Ma J, De La Hoz Ulloa CS, Zhang X, Zhou Y and Klibanski A:
Tumor suppression by MEG3 lncRNA in a human pituitary tumor derived
cell line. Mol Cell Endocrinol. 416:27–35. 2015. View Article : Google Scholar :
|
|
9
|
Gordon FE, Nutt CL, Cheunsuchon P,
Nakayama Y, Provencher KA, Rice KA, Zhou Y, Zhang X and Klibanski
A: Increased expression of angiogenic genes in the brains of mouse
meg3-null embryos. Endocrinology. 151:2443–2452. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fu WM, Lu YF, Hu BG, Liang WC, Zhu X, Yang
HD, Li G and Zhang JF: Long noncoding RNA Hotair mediated
angiogenesis in nasopharyngeal carcinoma by direct and indirect
signaling pathways. Oncotarget. 7:4712–4723. 2016. View Article : Google Scholar :
|
|
11
|
Ma Y, Wang P, Xue Y, Qu C, Zheng J, Liu X,
Ma J and Liu Y: PVT1 affects growth of glioma microvascular
endothelial cells by negatively regulating miR-186. Tumour Biol.
39:1010428317694326. 2017. View Article : Google Scholar
|
|
12
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yu J, Wang F, Yang GH, Wang FL, Ma YN, Du
ZW and Zhang JW: Human microRNA clusters: Genomic organization and
expression profile in leukemia cell lines. Biochem Biophys Res
Commun. 349:59–68. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bentwich I, Avniel A, Karov Y, Aharonov R,
Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, et al:
Identification of hundreds of conserved and nonconserved human
microRNAs. Nature Genet. 37:766–770. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Brennecke J, Hipfner DR, Stark A, Russell
RB and Cohen SM: Bantam encodes a developmentally regulated
microRNA that controls cell proliferation and regulates the
proapoptotic gene hid in Drosophila. Cell. 113:25–36. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Calame K: MicroRNA-155 function in B
Cells. Immunity. 27:825–827. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jopling CL, Yi M, Lancaster AM, Lemon SM
and Sarnow P: Modulation of hepatitis C virus RNA abundance by a
liver-specific MicroRNA. Science. 309:1577–1581. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Icli B, Dorbala P and Feinberg MW: An
emerging role for the miR-26 family in cardiovascular disease.
Trends Cardiovasc Med. 24:241–248. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Icli B, Wara AK, Moslehi J, Sun X, Plovie
E, Cahill M, Marchini JF, Schissler A, Padera RF, Shi J, et al:
MicroRNA-26a regulates pathological and physiological angiogenesis
by targeting BMP/SMAD1 signaling. Circ Res. 113:1231–1241. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu SC, Chuang SM, Hsu CJ, Tsai CH, Wang
SW and Tang CH: CTGF increases vascular endothelial growth
factor-dependent angiogenesis in human synovial fibroblasts by
increasing miR-210 expression. Cell Death Dis. 5:e14852014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Babic AM, Chen CC and Lau LF: Fisp12/mouse
connective tissue growth factor mediates endothelial cell adhesion
and migration through integrin alphavbeta3, promotes endothelial
cell survival, and induces angiogenesis in vivo. Mol Cell Biol.
19:2958–2966. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang BJ, Ding HW and Ma GA: Long noncoding
RNA PVT1 promotes melanoma progression via endogenous sponging
miR-26b. Oncol Res. 2017. View Article : Google Scholar
|
|
23
|
Wang R, Ding X, Zhou S, Li M, Sun L, Xu X
and Fei G: Microrna-26b attenuates monocrotaline-induced pulmonary
vascular remodeling via targeting connective tissue growth factor
(CTGF) and cyclin D1 (CCND1). Oncotarget. 7:72746–72757. 2016.
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
25
|
Kuo CH, Chen PK, Chang BI, Sung MC, Shi
CS, Lee JS, Chang CF, Shi GY and Wu HL: The recombinant lectin-like
domain of thrombomodulin inhibits angiogenesis through interaction
with Lewis Y antigen. Blood. 119:1302–1313. 2012. View Article : Google Scholar
|
|
26
|
Boettger T, Beetz N, Kostin S, Schneider
J, Kruger M, Hein L and Braun T: Acquisition of the contractile
phenotype by murine arterial smooth muscle cells depends on the
Mir143/145 gene cluster. J Clin Invest. 119:2634–2647. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xin M, Small EM, Sutherland LB, Qi X,
McAnally J, Plato CF, Richardson JA, Bassel-Duby R and Olson EN:
MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and
responsiveness of smooth muscle cells to injury. Genes Dev.
23:2166–2178. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dorn GW 2nd, Matkovich SJ, Eschenbacher WH
and Zhang Y: A human 3′ miR-499 mutation alters cardiac mRNA
targeting and function. Circ Res. 110:958–967. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Duan G, Ren C, Zhang Y and Feng S:
MicroRNA-26b inhibits metastasis of osteosarcoma via targeting CTGF
and Smad1. Tumour Biol. 36:6201–6209. 2015. View Article : Google Scholar
|
|
30
|
Guttman M, Amit I, Ga rber M, French C,
Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al:
Chromatin signature reveals over a thousand highly conserved large
non-coding RNAs in mammals. Nature. 458:223–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Khalil AM, Guttman M, Huarte M, Garber M,
Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van
Oudenaarden A, et al: Many human large intergenic noncoding RNAs
associate with chromatin-modifying complexes and affect gene
expression. Proc Natl Acad Sci USA. 106:11667–11672. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Huarte M, Guttman M, Feldser D, Garber M,
Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M,
et al: A large intergenic noncoding RNA induced by p53 mediates
global gene repression in the p53 response. Cell. 142:409–419.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Paci P, Colombo T and Farina L:
Computational analysis identifies a sponge interaction network
between long non-coding RNAs and messenger RNAs in human breast
cancer. BMC Syst Biol. 8:832014. View Article : Google Scholar : PubMed/NCBI
|