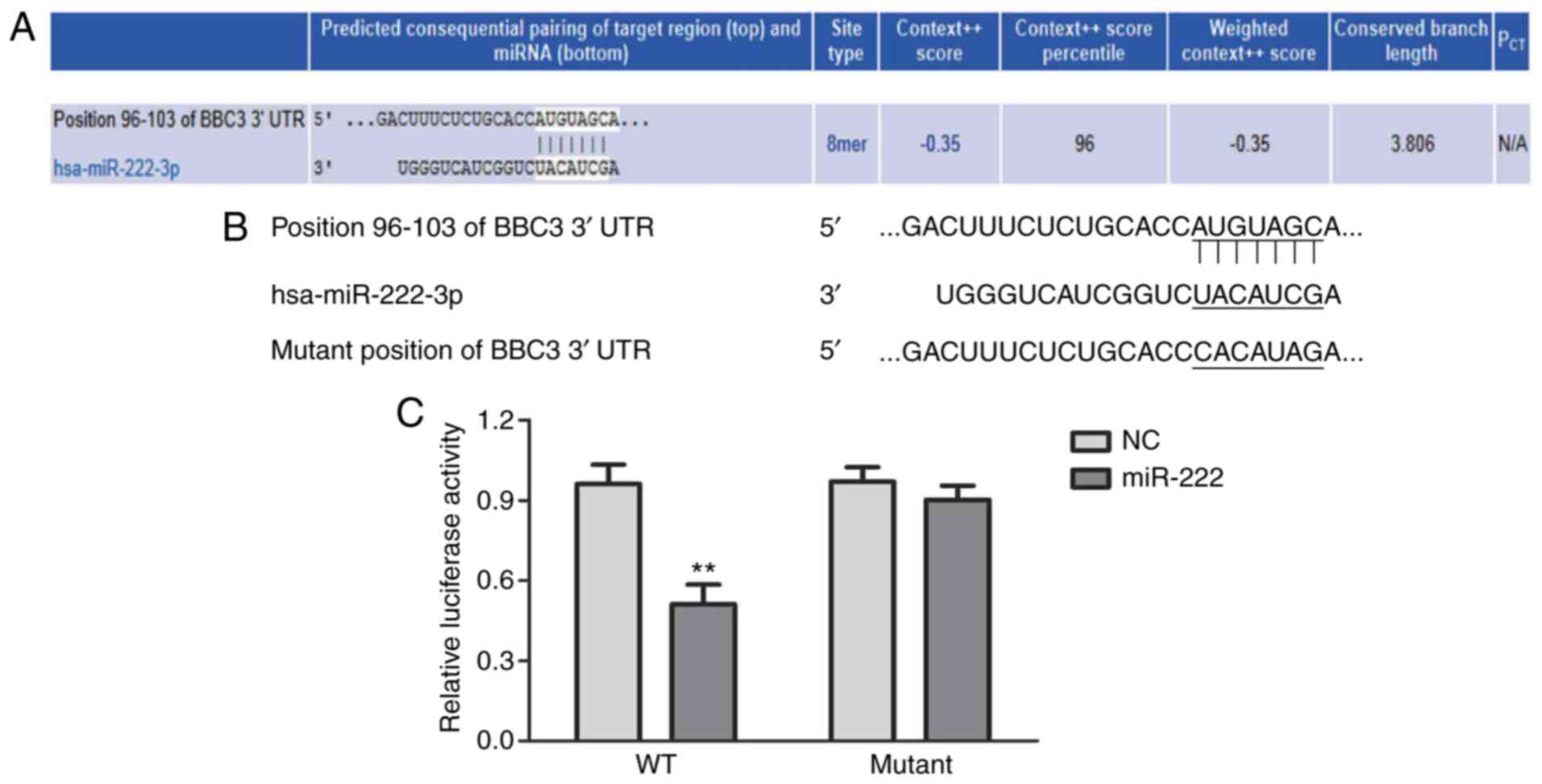
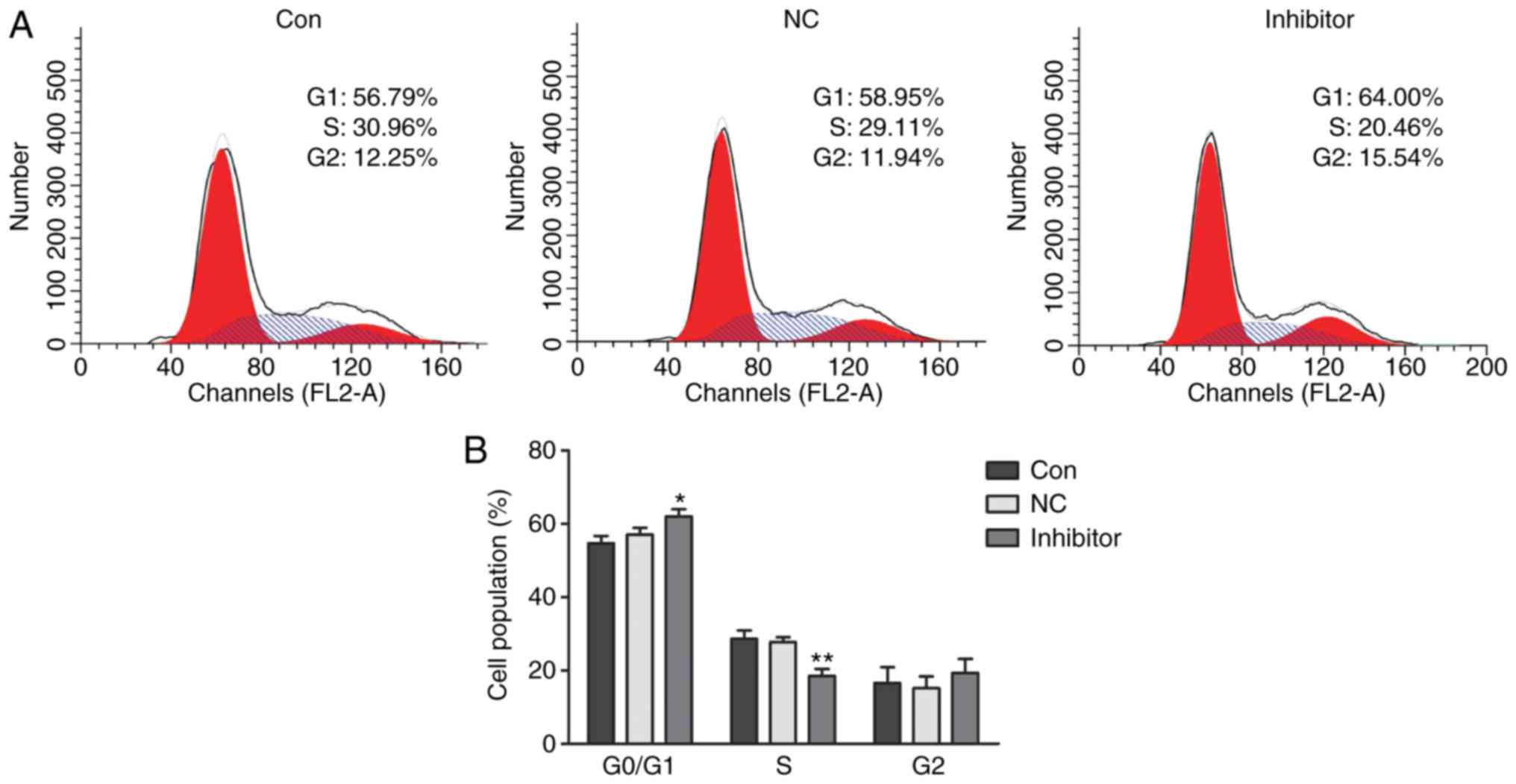
|
1
|
Adult Primary Liver Cancer Treatment
(PDQ®)-Patient Version. NCI. 6 July 2016. Archived from the
original on October 2016. Retrieved 29 September 2016.
|
|
2
|
World Cancer Report: World Health
Organization. 20145(6)2014. ISBN: 9283204298
|
|
3
|
Tsujimoto Y, Finger LR, Yunis J, Nowell PC
and Croce CM: Cloning of the chromosome breakpoint of neoplastic B
cells with the t (14;18) chromosome translocation. Science.
226:1097–1099. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cleary ML, Smith SD and Sklar J: Cloning
and structural analysis of cDNAs for bcl-2 and a hybrid
bcl-2/immunoglobulin transcript resulting from the t (14;18)
translocation. Cell. 47:19–28. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yu J, Zhang L, Hwang PM, Kinzler KW and
Vogelstein B: PUMA induces the rapid apoptosis of colorectal cancer
cells. Mol Cell. 7:673–682. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nakano K and Vousden KH: PUMA, a novel
proapoptotic gene, is induced by p53. Mol Cell. 7:683–694. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chipuk JE and Green DR: PUMA cooperates
with direct activator proteins to promote mitochondrial outer
membrane permeabilization and apoptosis. Cell Cycle. 8:2692–2696.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Thakur VS, Ruhul Amin AR, Paul RK, Gupta
K, Hastak K, Agarwal MK, Jackson MW, Wald DN, Mukhtar H and Agarwal
ML: p53-Dependent p21-mediated growth arrest pre-empts and protects
HCT116 cells from PUMA-mediated apoptosis induced by EGCG. Cancer
Lett. 296:225–232. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yu J and Zhang L: PUMA, a potent killer
with or without p53. Oncogene. 27(Suppl 1): S71–S83. 2008.
View Article : Google Scholar
|
|
10
|
Lee RC, Feinbaum RL, Ambros V and Feinbaum
A: The C. elegans heterochronic gene lin-4 encodes small RNAs with
antisense complementarity to lin-14. Cell. 75:843–854. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wightman B, Ha I and Ruvkun G:
Posttranscriptional regulation of the heterochronic gene lin-14 by
lin-4 mediates temporal pattern formation in C. elegans. Cell.
75:855–862. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cel. 116:281–297. 2004.
View Article : Google Scholar
|
|
14
|
Brennecke J, Hipfner DR, Stark A, Russell
RB and Cohen SM: Bantam encodes a developmentally regulated
microRNA that controls cell proliferation and regulates the
proapoptotic gene hid in Drosophila. Cell. 113:25–36. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Poy MN, Eliasson L, Krutzfeldt J, Kuwajima
S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P
and Stoffel M: A pancreatic islet-specific microRNA regulates
insulin secretion. Nature. 432:226–230. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen CZ, Li L, Lodish HF and Bartel DP:
MicroRNAs modulate hematopoietic lineage differentiation. Science.
303:83–86. 2004. View Article : Google Scholar
|
|
17
|
Bentwich I, Avniel A, Karov Y, Aharonov R,
Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, et al:
Identification of hundreds of conserved and nonconserved human
microRNAs. Nat Genet. 37:766–770. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Trang P, Weidhaas JB and Slack FJ:
MicroRNAs as potential cancer therapeutics. Oncogene. 27(Suppl 2):
S52–S57. 2008. View Article : Google Scholar
|
|
19
|
Li C, Feng Y, Coukos G and Zhang L:
Therapeutic microRNA strategies in human cancer. AAPS J.
11:747–757. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fasanaro P, Greco S, Ivan M, Capogrossi MC
and Martelli F: microRNA: Emerging therapeutic targets in acute
ischemic diseases. Pharmacol Ther. 125:92–104. 2010. View Article : Google Scholar
|
|
21
|
Gyugos M, Lendvai G, Kenessey I,
Schlachter K, Halász J, Nagy P, Garami M, Jakab Z, Schaff Z and
Kiss A: MicroRNA expression might predict prognosis of epithelial
hepatoblastoma. Virchows Arch. 464:419–427. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
von Frowein J, Pagel P, Kappler R, von
Schweinitz D, Roscher A and Schmid I: MicroRNA-492 is processed
from the keratin 19 gene and up-regulated in metastatic
hepatoblastoma. Hepatology. 53:833–842. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
López-Terrada D, Cheung SW, Finegold MJ
and Knowles BB: HepG2 is a hepatoblastoma-derived cell line. Hum
Pathol. 40:1512–1515. 2009. View Article : Google Scholar
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
25
|
Yang YF, Wang F, Xiao JJ, Song Y, Zhao YY,
Cao Y, Bei YH and Yang CQ: MiR-222 overexpression promotes
proliferation of human hepatocellular carcinoma HepG2 cells by
downregulating p27. Int J Clin Exp Med. 7:893–902. 2014.PubMed/NCBI
|
|
26
|
Yang X, Yang YM, Gan R, Zhao LX, Li W,
Zhou HB, Wang XJ, Lu JX and Meng QH: Down-regulation of miR-221 and
miR-222 restrain prostate cancer cell proliferation and migration
that is partly mediated by activation of SIRT1. Plos One.
9:e988332014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pandhare-Dash J, Mantri CK, Gong Y, Chen Z
and Dash C: XMRV accelerates cellular proliferation,
transformational activity, and invasiveness of prostate cancer
cells by downregulating p27 (Kip1). Prostate. 72:886–897. 2012.
View Article : Google Scholar
|
|
28
|
Salvesen GS: Caspases: Opening the boxes
and interpreting the arrows. Cell Death Differ. 9:3–5. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ghavami S, Hashemi M, Ande SR, Yeganeh B,
Xiao W, Eshraghi M, Bus CJ, Kadkhoda K, Wiechec E, Halayko AJ and
Los M: Apoptosis and cancer: Mutations within caspase genes. J Med
Genet. 46:497–510. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Stennicke HR and Salvesen GS: Biochemical
characteristics of caspases-3, -6, -7, and -8. J Biol Chem.
272:25719–25723. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Motokura T, Bloom T, Kim HG, Jüppner H,
Ruderman JV, Kronenberg HM and Arnold A: A novel cyclin encoded by
a bcl1-linked candidate oncogene. Nature. 350:512–515. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lew DJ, Dulić V and Reed SI: Isolation of
three novel human cyclins by rescue of G1 cyclin (Cln) function in
yeast. Cell. 66:1197–1206. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Withers DA, Harvey RC, Faust JB, Melnyk O,
Carey K and Meeker TC: Characterization of a candidate bcl-1 gene.
Mol Cell Biol. 11:4846–4853. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Inaba T, Matsushime H, Valentine M,
Roussel MF, Sherr CJ and Look AT: Genomic organization, chromosomal
localization, and independent expression of human cyclin D genes.
Genomics. 13:565–574. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Diehl JA: Cycling to cancer with cyclin
D1. Cancer Biol Ther. 1:226–231. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jares P, Colomer D and Campo E: Genetic
and molecular pathogenesis of mantle cell lymphoma: Perspectives
for new targeted therapeutics. Nat Rev Cancer. 7:750–762. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Thomas GR, Nadiminti H and Regalado J:
Molecular predictors of clinical outcome in patients with head and
neck squamous cell carcinoma. Int J Exp Pathol. 86:347–363. 2005.
View Article : Google Scholar : PubMed/NCBI
|