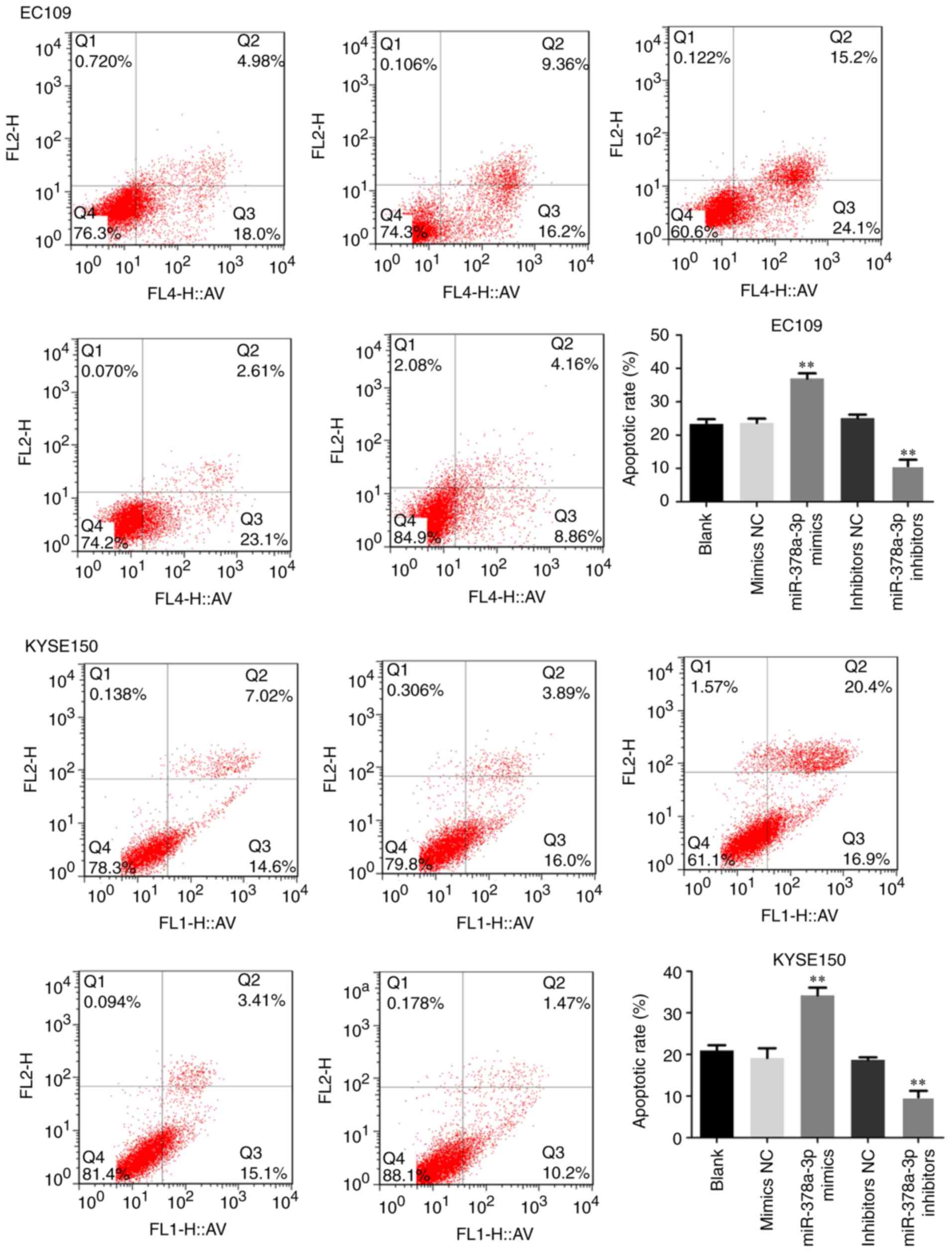
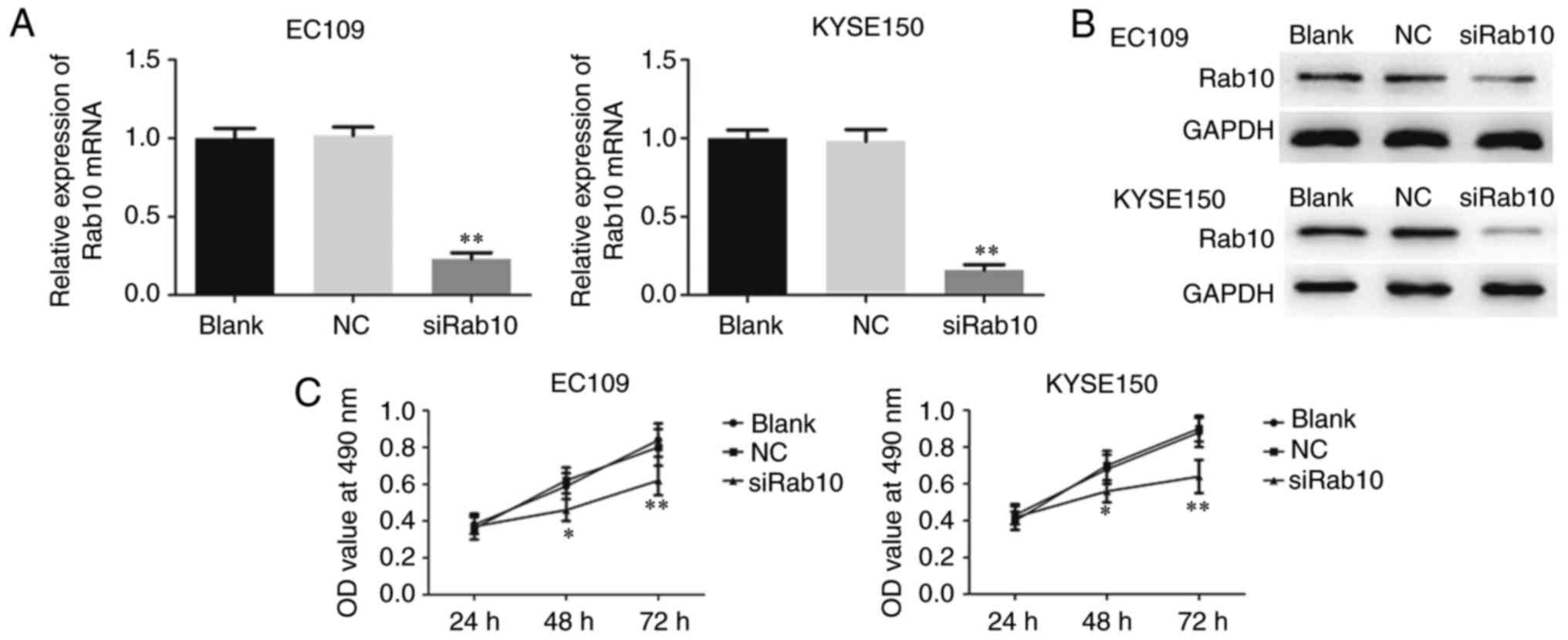
|
1
|
Arnold M, Soerjomataram I, Ferlay J and
Forman D: Global incidence of esophageal cancer by histological
subtype in 2012. Gut. 64:381–387. 2015. View Article : Google Scholar
|
|
2
|
Lagergren J: Oesophageal cancer in 2014:
Advance in curatively intended treatment. Nat Rev Gastroenterol
Hepatol. 12:74–75. 2015. View Article : Google Scholar
|
|
3
|
Stoner GD and Gupta A: Etiology and
chemoprevention of esophageal squamous cell carcinoma.
Carcinogenesis. 22:1737–1746. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Enzinger PC and Mayer RJ: Esophageal
cancer. N Engl J Med. 349:2241–2252. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Calin GA and Croce CM: MicroRNAs
signatures in human cancers. Nat Rev Cancer. 6:857–866. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kloosterman WP and Plasterk RH: The
diverse functions of microRNAs in animal development and diseases.
Dev Cell. 11:441–450. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang Y, Sui J, Shen X, Li C, Yao W, Hong
W, Peng H, Pu Y, Yin L and Liang G: Differential expression
profiles of microRNAs as potential biomarkers for the early
diagnosis of lung cancer. Oncol Rep. 37:3543–3553. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li Z, Jiang J, Tian L, Li X, Chen J, Li S,
Li C and Yang Z: A plasma mir-125a-5p as a novel biomarker for
Kawasaki disease and induces apoptosis in HUVECs. Plos One.
12:e01754072017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen Y, Luo D, Tian W, Li Z and Zhang X:
Demethylation of miR-495 inhibits cell proliferation, migration and
promotes apoptosis by targeting STAT-3 in breast cancer. Oncol Rep.
37:3581–3589. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Arias Sosa LA, Cuspoca Orduz AF and Bernal
Gomez BM: Deregulation of microRNAs in gastric cancer: Up
regulation by miR-21 and miR-106. Rev Gastroenterol Peru. 37:65–70.
2017.In Spanish. PubMed/NCBI
|
|
12
|
Sharma P, Saini N and Sharma R: MiR-107
functions as a tumor suppressor in human esophageal squamous cell
carcinoma and targets Cdc42. Oncol Rep. 37:3116–3127. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Eichner LJ, Perry MC, Dufour CR, Bertos N,
Park M, St-Pierre J and Giguère V: MiR-378(*) mediates metabolic
shift in breast cancer cells via the PGC-1β/ERRγ transcriptional
pathway. Cell Metab. 12:352–361. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ikeda K, Horie-Inoue K, Ueno T, Suzuki T,
Sato W, Shigekawa T, Osaki A, Saeki T, Berezikov E, Mano H and
Inoue S: MiR-378a-3p modulates tamoxifen sensitivity in breast
cancer MCF-7 cells through targeting GOLT1A. Sci Rep. 5:131702015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jeon TI, Park JW, Ahn J, Jung CH and Ha
TY: Fisetin protects against hepatosteatosis in mice by inhibiting
miR-378. Mol Nutr Food Res. 57:1931–1937. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Krist B, Florczyk U,
Pietraszek-Gremplewicz K, Józkowicz A and Dulak J: The Role of
miR-378a in metabolism, angionesis, and muscle biology. Int J
Endocrinol. 2015:2817562015. View Article : Google Scholar
|
|
17
|
Babbey CM, Bacallao RL and Dunn KW: Dunn.
Rab10 associates with primary cilia and the exocyst complex in
renal epithelial cells. Am J Physiol Renal Physiol. 299:F495–F506.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sano H, Peck GR, Kettenbach AN, Gerber SA
and Lienhard GE: Insulin-stimulated GLUT4 protein translocation in
adipocytes requires the Rab10 guanine nucleotide exchange factor
Dennd4C. J Biol Chem. 286:16541–16545. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Barbosa MD, Johnson SA, Achey K, Gutierrez
MJ, Wakeland EK, Zerial M and Kingsmore SF: The Rab protein family:
Genetic mapping of six Rab genes in the mouse. Genomics.
30:439–444. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
He H, Dai F, Yu L, She X, Zhao Y, Jiang J,
Chen X and Zhao S: Identification and characterization of nine
novel human small GTPases showing variable expressions in liver
cancer tissues. Gene Expr. 10:231–242. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hummel R, Sie C, Waston DI, Wang T, Ansar
A, Michael MZ, Van der Hoek M, Haier J and Hussey DJ: MicroRNAs
signatures in chemotherapy resistant esophageal cancer cell lines.
World J Gastroenterol. 20:14904–14912. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2-ΔΔC T method. Methods. 25:402–408.
2001. View Article : Google Scholar
|
|
23
|
Croce CM: Causes and consequences of
microRNAs dysregulation in cancer. Nat Rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Prieto-Garcia E, Diaz-Garcia CV,
Garcia-Ruiz I and Agulló-Ortuño MT: Epithelial-to-mesenchymal
transition in tumor progression. Med Oncol. 34:1222017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Megiorni F, Cialfi S, McDowell HP, Felsanj
A, Camero S, Guffanti A, Pizer B, Clerico A, De Grazia A, Pizzuti
A, et al: Deep Sequencing the microRNA profile in rhabdomysarcoma
reveals down-regulation of miR-378 family members. BMC Cancer.
14:8802014. View Article : Google Scholar
|
|
26
|
Nagalingam RS, Sundaresan NR, Noor M,
Gupta MP, Solaro RJ and Gupta M: Deficiency of
cardiomyocyte-specific microRNA-378 contributes to the development
of cardiac fibrosis involving a transforming growth factor β (TGF
β1)-dependent paracrine mechanism. J Biol Chem. 289:27199–27214.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hyun J, Wang S, Kim J, Rao KM, Park SY,
Chung I, Ha CS, Kim SW, Yun YH and Jung Y: MicroRNA-378 limits
activation of hepatic stellate cells and liver fibrosis by
suppressing Gli3. Nat Commun. 7:109932016. View Article : Google Scholar
|
|
28
|
Stenmark H: Rab GTPase as coordinates of
vesicle traffic. Nat Rev Mol Cell Biol. 10:513–525. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Grosshans BL, Ortiz D and Novick P: Rabs
and their effectors: Achieving specificity in membrane traffic.
Proc Natl Acad Sci USA. 103:11821–11827. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang P, Liu H, Wang Y, Liu O, Zhang J,
Gleason A, Yang Z, Wang H, Shi A and Grant BD: RAB-10 promotes
EHBP-1 bridging of filamentous actin and tubular recycling
endosomes. PLoS Genet. 12:e10060932016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ito G, Katsemonova K, Tonelli F, Lis P,
Baptista MA, Shpiro N, Duddy G, Wilson S, Ho PW, Ho SL, et al:
Phos-tag analysis of Rab10 phosphorylation by LRRK2: A powerful
assay for assessing kinase function and inhibitors. Biochem J.
473:2671–2685. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lee H, Shin N, Song M, Kang UB, Yeom J,
Lee C, Ahn YH, Yoo JS, Paik YK and Kim H: Analysis of nuclear high
mobility group box 1 (HMGB1)-binding proteins in colon cancer
cells: Clustering with proteins involved in secretion and
extracellular function. J Proteome Res. 9:4661–4670. 2010.
View Article : Google Scholar : PubMed/NCBI
|