Introduction
Previous studies have reported that protein-coding
genes account for only a small proportion of the genome, while the
larger remaining part includes genes that are either not
transcribed or are transcribed as non-coding RNA (1,2).
Currently, the function of non-coding RNA merits further
investigation. Generally, long non-coding RNAs (lncRNAs) do not
encode proteins and have a length of at least 200 nucleotides
(3). Research has confirmed the
involvement of lncRNAs in the process of transcription and
post-transcription in tumor cells (4–6);
however, the mechanisms of lncRNA action in myocytes have received
relatively limited attention.
In 2006, myocardial infarction-associated transcript
(MIAT) was identified as an lncRNAs that confers a risk of
myocardial infraction (7). MIAT
is also known as retinal non-coding RNA 2, AK028326 or Gomafu
(8). Subsequent studies have
reported that MIAT participates in a variety of diseases and
cellular processes, including myocardial infarction, microvascular
dysfunction (9), paranoid
schizophrenia (10), neurogenic
commitment (11) and the
formation of nuclear bodies (12). However, only a limited number of
studies have examined MIAT in myocyte hypertrophy (13,14). MIAT was reported to suppress
microRNA (miR)-150 expression in cardiomyocytes (14). Furthermore, previous studies have
demonstrated that cardiomyocyte hypertrophy is modulated by miR-150
via its regulatory effect on the expression of the transcriptional
co-activator P300 (15). However,
it remains unclear whether MIAT is involved in the pathway of
miR-150-5p/P300. Therefore, the present study investigated the
involvement of MIAT in hypertrophy in vitro and in
vivo through cell and animal models.
Materials and methods
Animal care and use
This study conformed to the Guide for the Care and
Use of Laboratory Animals as published by the National Institutes
of Health (NIH publication no. 85-23; revised 1996). All procedures
were performed in accordance with the Ethical Standards of the
Committee on Animal Experimentation of China Medical University
(project identification code, SCXK-2013-0001; Shenyang, China), and
were approved by the Ethical Standards of the Committee on Animal
Experimentation of the First Hospital of China Medical University
(no. 2017010). Healthy male specific-pathogen-free C57BL/6J mice
(age, 8 weeks; weight, 19–21 g) were purchased from the Laboratory
Animal Center of China Medical University, and cared for as
reported previously (16). Mice
were kept in a 12 h light-dark cycle, temperature-controlled
(22±2°C) and humidity-controlled (55±5%) environment, and fed a
standard chow diet for at least 1 week before experimentation.
Animal procedures and
echocardiography
Mice were randomized into two groups, and
intraperitoneally injected with 20 µg/kg of isoproterenol
(ISO; diluted with 0.9% NaCl; n=4) or vehicle (0.9% NaCl; n=4)
twice a day for 7 days. Transthoracic echocardiography was
performed in conscious mice under isofluorane anesthesia (1%
isoflurane; 0.6-liter flow of O2) using a Vevo 2100
imaging system (FUJIFILM VisualSonics Inc., Toronto, Canada). B and
M-mode images were obtained, and other data were collected from
each mouse group prior to and following the injection week. For
obtaining the left ventricular images, the cursor was positioned at
the level just posterior to the chordae tendinae, perpendicular to
the interventricular septum and left ventricular posterior wall.
Left ventricular posterior wall dimensions at diastole (LVPWd) and
interventricular septal thickness at diastole (IVSd) were measured
from the left ventricle-left ventricular internal dimension at
diastole.
The mice were then sacrificed and weighed,
subsequently the hearts were harvested, rinsed and weighted, and
the tibias were measured. Ventricles were either stored in TRIzol
reagent (Thermo Fisher Scientific, Inc., Waltham, MA, USA) for RNA
extraction or radio immunoprecipitation assay lysis buffer (RIPA)
for protein extraction.
Histological analyses
Excised mice hearts were immediately fixed in 10%
neutral buffered formalin for 24 h at room temperature after a
brief rinse with cold PBS, followed by dehydration and embedding in
paraffin wax. Left ventricular sections (4 µm) were cut and
stained with hematoxylin and eosin (H&E) to measure
cardiomyocyte cross-sectional area. Briefly, 4-µm microtome
sections were deparaffinized with xylene and rehydrated through
graded concentrations of ethanol (100, 95 and 80%), followed by
staining with hematoxylin for 4 min at room temperature.
Subsequently, slides were rinsed in deionized water and dipped in
0.25% acidic ethanol for 2 sec. Following washing with deionized
water, the slides were immersed in eosin for 2 min at room
temperature. Slides were then rinsed thoroughly in three changes of
absolute alcohol. Finally, the slides were washed with xylene and
then mounted in Permount.
Histology images were obtained with a Leica DM6000
microscope and a DFC420 C camera (Leica Microsystems GmbH, Wetzlar,
Germany; magnifications, ×10 and 40). The cardiomyocyte
cross-sectional area per nucleus was measured. The selection of
cardiomyocytes for measurement was based on clearly defined
sarcolemma and associated round nuclei. Cross-sectional areas in
over 200 randomly chosen cardiomyocytes from at least 16 fields per
group were evaluated using ImageJ software (version 1.48, National
Institutes of Health, Bethesda, MD, USA).
Primary culture of neonatal rat
ventricular myocytes (NRVMs) and treatment
NRVMs were isolated as previously described
(17). Briefly, newborn
Sprague-Dawley rats (1–3-days-old) were sacrificed, their hearts
were removed and the atria were immediately separated. Cold Hank’s
equilibrium salt solution with no Ca2+ and
Mg2+ (Gibco; Thermo Fisher Scientific, Inc.) was used to
store the ventricle tissues. Subsequent to transferring the
ventricles to trypsin-0.125% EDTA (Invitrogen; Thermo Fisher
Scientific, Inc.) and storing overnight at 4°C (16–20 h),
collagenase II (Worthington Biochemical Corporation, Lakewood, NJ,
USA) was used for further digestion at 37°C for 30 min in a shaking
water bath. Isolated cells were prepared in an uncoated flask at
37°C to remove cardiac fibroblasts, and cells were harvested from
the supernatants after 1.5 h. NRVMs were seeded in plating medium
containing 45% Dulbecco’s modified Eagle’s medium (DMEM), 45% M199
medium, 10% fetal bovine serum, 100 U/ml penicillin and 100
µg/ml streptomycin (all obtained from Thermo Fisher
Scientific, Inc.) for 24 h at 37°C. The following day, the plating
medium was replaced by maintenance medium containing 50% DMEM, 50%
M199 without serum, 100 U/ml penicillin and 100 µg/ml
streptomycin, and incubated for an additional 24 h at 37°C prior to
being used for experimentation. NRVMs were incubated with or
without 10 µM ISO for 24 h.
Cell transfection
MIAT siRNA (si-MIAT) and the negative control siRNA
(si-NC) were purchased from GenePharma Co., Ltd. (Shanghai, China).
Their sequences were as follows: Negative control siRNA,
5′-UUCUCCGAACGUGUCACGUTT-3′ (sense) and 5′-ACGUGACACGUUCGGAGAATT-3′
(antisense); MIAT siRNA, 5′-GGUGUUAAGACUUGGUUUCTT-3′ (sense) and
5′-GAAACCAAGUCUUAACACCTT-3′ (antisense). NRVMs were transfected
with 100 nM MIAT siRNA or negative control for 6 h using
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to manufacturer instructions. Briefly,
NRVMs were seeded into a 6-well plate at 6×105 cells per
well in 2 ml plating medium for 24 h at 37°C, and then incubated in
serum-free maintenance medium for at least 24 h. When 60–70%
confluence was reached, transfected NRVMs were then treated with or
without ISO (10 µm) for 48 h at 37°C. Cells were collected
after 48 h for quantification of mRNA and miRNA expression.
Immunofluorescence staining and
measurement of NRVM cell area
NRVMs were stained with phalloidin conjugated with
Alexa Fluor 594, and nuclei were stained with DAPI (all from
Invitrogen; Thermo Fisher Scientific, Inc.). Slides were visualized
using an Olympus AX70 Provis fluorescence microscope at a ×40
magnification (Olympus Corporation, Tokyo, Japan). In each group,
30 cells were counted, and the fields-of-view of cells were
randomly selected under each condition. The surface area was
estimated by the ratio of control cells. Each experiment was
repeated in triplicate.
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA from the tissue and NRVMs was extracted
using TRIzol reagent (Thermo Fisher Scientific, Inc.) according to
manufacturer’s protocol. RNAs with an OD260/OD280 ratio in the
range of 1.80–2.20 were used for downstream assays, and 1 µg
RNA from each sample was reverse transcribed into cDNA using a
high-capacity cDNA RT kit (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The RT conditions were conducted as per the
manufacturer’s protocols. Subsequently, qPCR was performed to
quantify the mRNA expression levels using Real-Time
SYBR® Green PCR Master Mix (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA) on the CFX384 Touch Real-Time PCR Detection
system (Bio-Rad Laboratories, Inc.), with GAPDH serving as an
internal control. Expression levels were analyzed using the
2−ΔΔCq method (18).
Primers sequences used in the project are listed in Table I.
 | Table ISequences of polymerase chain
reaction primers. |
Table I
Sequences of polymerase chain
reaction primers.
| Target ID | Sequence |
|---|
| GAPDH | Forward:
5′-AGTGCCAGCCTCGTCTCATA-3′ |
| Reverse:
5′-GGTAACCAGGCGTCCGATAC-3′ |
| MIAT | Forward:
5′-TCTCTGGTGCTTCCCTCCTT-3′ |
| Reverse:
5′-GATCTAAGCTTGAGCCCCCA-3′ |
| ANP | Forward:
5′-CTCCCAGGCCATATTGGAG-3′ |
| Reverse:
5′-TCCAGGTGGTCTAGCAGGTT-3′ |
| BNP | Forward:
5′-GAGTCCTTCGGTCTCAAGGC-3′ |
| Reverse:
5′-CAACTTCAGTGCGTTACAGCC-3′ |
| P300 | Forward:
5′-GACCCTCAGCTTTTAGGAATCC-3′ |
| Reverse:
5′-TGCCGTAGCAACACAGTGTCT-3′ |
For miRNA analysis, cDNA was synthesized using the
TaqMan™ MicroRNA Reverse Transcription kit (Applied Biosystems;
Thermo Fisher Scientific, Inc.). The miR-150-5p expression were
determined by the TaqMan™ MicroRNA assay (cat. no. A25576, Applied
Biosystems; Thermo Fisher Scientific, Inc.) and Taqman 2X universal
PCR master mix (cat. no. 4304437, Applied Biosystems; Thermo Fisher
Scientific, Inc.) on the CFX384 Touch Real-Time PCR Detection
System (Bio-Rad Laboratories, Inc.), with U6 small nuclear RNA (U6)
assay (cat. no. 4427975, Applied Biosystems; Thermo Fisher
Scientific, Inc.) serving as an internal control. Expression levels
were analyzed using the 2−ΔΔCq method (18).
Western blot analysis
Samples from mice and NRVMs were lysed in radio
immunoprecipitation assay lysis buffer (RIPA, cat. no. 9806, Cell
Signaling Technology, Inc., Danvers, MA, USA) with protease
inhibitors. Protein concentrations were then quantified using a
bicinchoninic acid assay kit (Thermo Fisher Scientific, Inc.). Cell
lysate (20 µg protein) was isolated with 10% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis, and the protein was
transferred to polyvinylidene difluoride membranes. The membranes
were subsequently blocked using 5% BSA for 1 h at room temperature,
and incubated with atrial natriuretic peptide (ANP) primary
antibody (cat. no. ab189921; 1:2,000; Abcam, Cambridge, MA, USA)
overnight at 4°C. The membrane was also incubated with GAPDH (cat.
no. 5174; 1:20,000; Cell Signaling Technology, Inc.) for 1 h at
room temperature. Subsequently, a horseradish peroxidase-linked
anti-rabbit IgG secondary antibody (cat. no. 7074; 1:20,000; Cell
Signaling Technology, Inc.) was then used incubated for 1 h at room
temperature. An enhanced chemiluminescence reagent (SuperSignal™
West Femto Maximum Sensitivity Substrate; Thermo Fisher Scientific,
Inc.) was used to visualize the bands, and semi-quantification of
western blot bands was conducted by comparison against the GAPDH
bands using ImageJ software (version 1.48, National Institutes of
Health, Bethesda, MD, USA).
Statistical analysis
Data are reported as the mean ± standard error.
Comparisons between groups were analyzed by double-sided Student’s
t-test, or with analysis of variance and Turkey’s test for
experiments with more than two subgroups. P<0.05 was considered
to indicate a statistically significant difference. Statistical
analysis was performed using IBM SPSS software (version 24; IBM
Corp., Armonk, NY, USA).
Results
MIAT, miR-150-5p and P300 expression
levels are associated with cardiomyocyte hypertrophy induced by ISO
in vivo
ISO was injected in mice in order to build a cardiac
hypertrophy model. Mice were injected 20 mg ISO/kg twice per day
over 7 days, and cardiac hypertrophy in the ISO and control group
mice was analyzed. Echocardiography, histopathology and
morphometric analysis were conducted to confirm that ISO induced
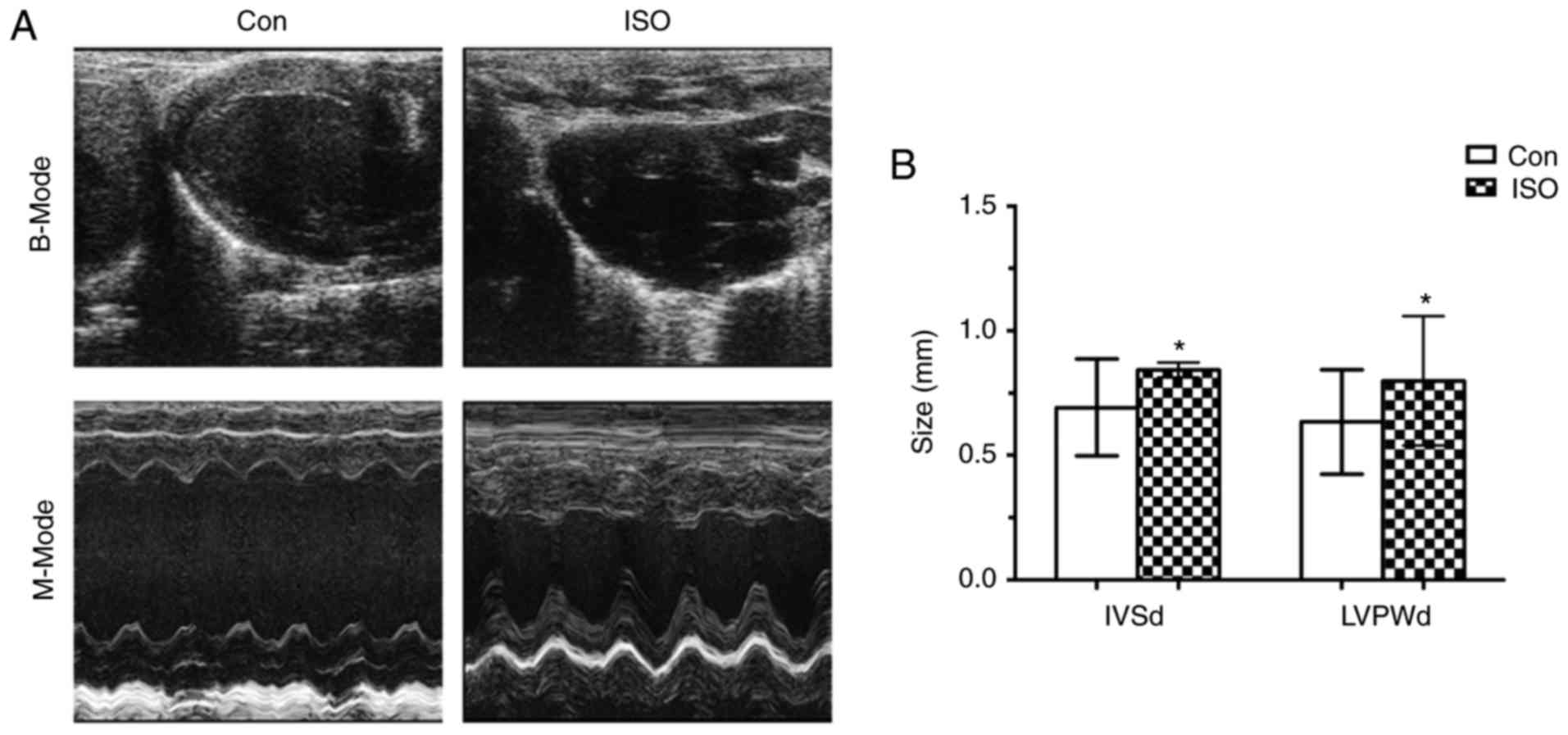
cardiac hypertrophy. Firstly, we performed echocardiography to
measure the cardiac parameters in B and M modes after 7 day’s ISO
injection. The IVSd and LVPWd in ISO mice were significantly higher
than in control mice (0.843±0.03 vs. 0.69±0.19, P<0.05;
0.80±0.26 vs. 0.63±0.21, P<0.05; respectively; Fig. 1A and B). In addition, heart
anatomy of the ISO and control mice was conducted. As presented in
Table II, after the heart weight
(HW) was normalized to tibia length (TL), a higher HW/TL ratio was
observed in ISO mice than that of control mice (0.083±0.007 vs.
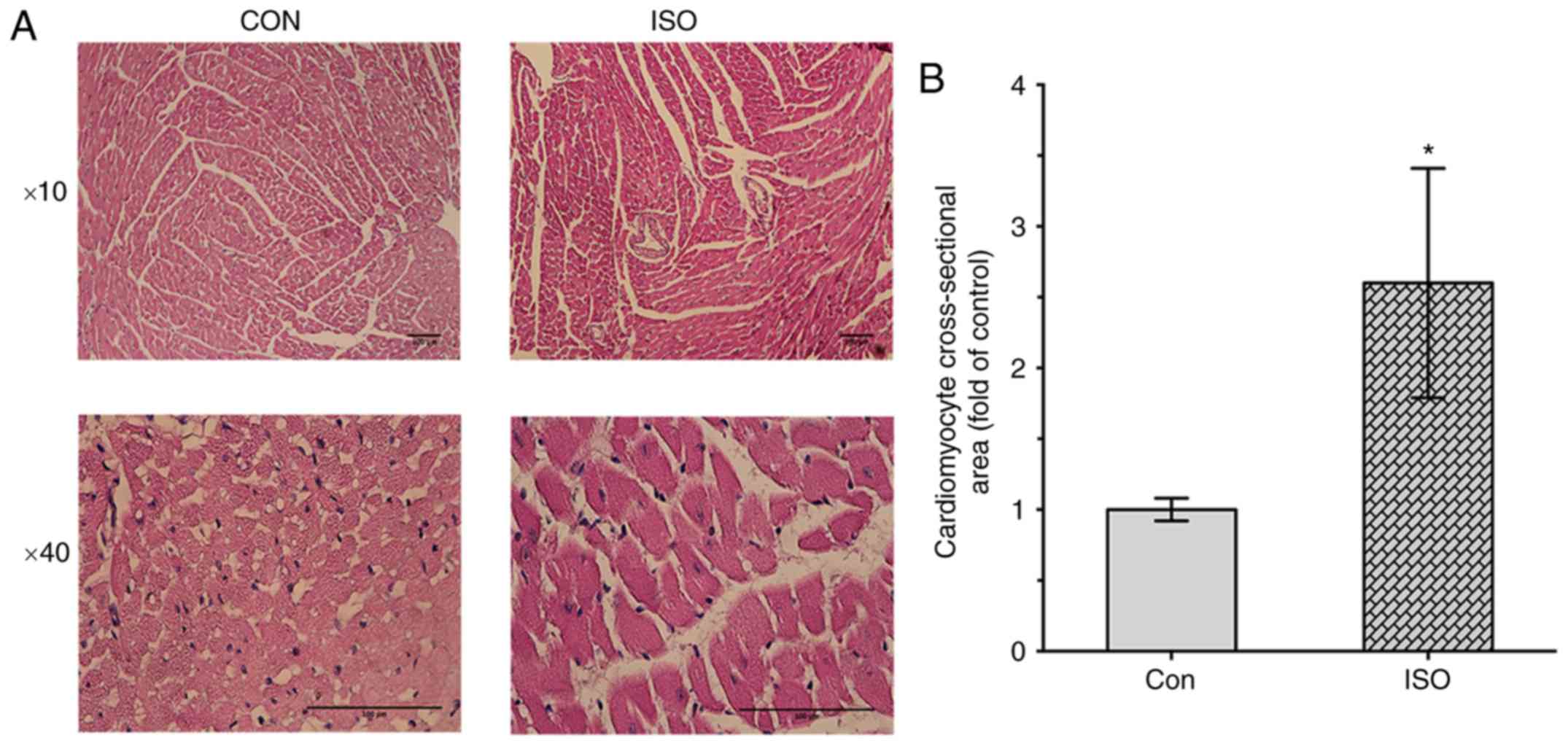
0.108±0.0014, P<0.05). Furthermore, histological analysis
revealed that cardiomyocyte cross-sectional area was signifi-cantly
increased in ISO-induced group compared with the control group
(1±0.08 vs. 2.6±0.81, P<0.05, respectively; Fig. 2A and B).
 | Table IIMorphometric analysis. |
Table II
Morphometric analysis.
| Parameter | Con | ISO | P-value |
|---|
| BW (g) |
27.75000±0.89000 |
26.75000±0.61000 | 0.78000 |
| HW (mg) |
149.00000±1.30000 |
192.00000±2.10000 | 0.03800 |
| HW/BW |
0.00530±0.00043 |
0.00729±0.00010 | 0.01900 |
| TL (mm) |
17.80000±0.20000 |
17.80000±0.15000 | 0.95000 |
| HW/TL |
0.08300±0.00700 |
0.10800±0.00140 | 0.03260 |
To investigate whether MIAT and miR-150-5p were
involved in the development of cardiac hypertrophy in vivo,
MIAT and miR-150-5p expression levels in heart tissue were assessed
by RT-qPCR. As shown in Fig. 3,
the expression levels of ANP and BNP were higher in the ISO group
(Fig. 3A). The protein expression
levels of ANP were also analyzed, which revealed a significant
increase in the ISO group compared with in the control group
(Fig. 3B). Additionally, MIAT
expression levels were significantly increased in the ISO group
compared with control groups (Fig.
3C). miR-150 expression levels were significantly decreased in
ISO group compared with control group (Fig. 3D). Conversely, the expression
levels of P300 in the ISO group were ~5-fold higher in comparison
with that of the control group (Fig.
3E). The results of the present study suggested that MIAT,
miR-150-5p and P300 may be involved in the hypertrophic process of
myocytes and affect the cardiac function in vivo.
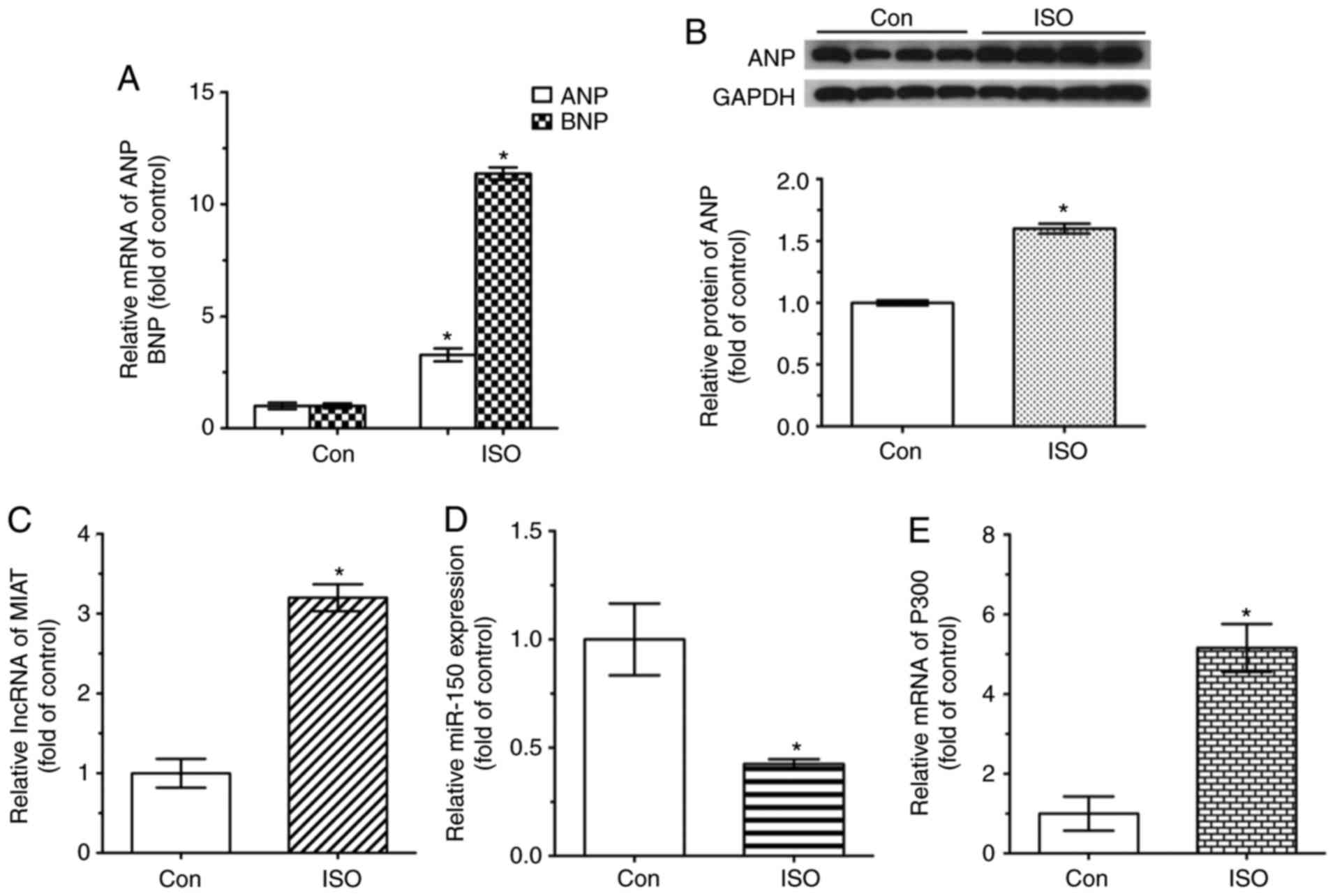
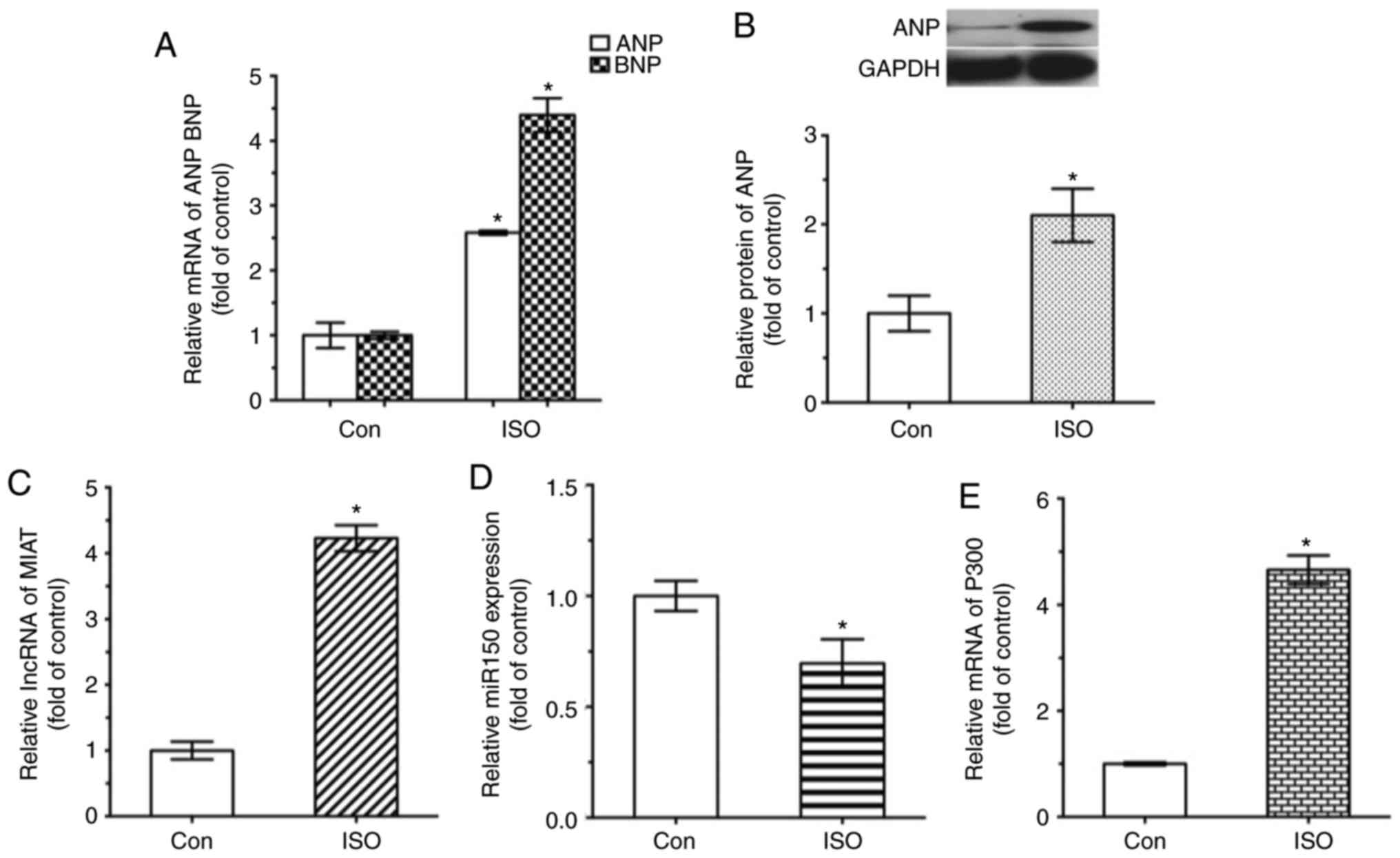 | Figure 3Expression levels of MIAT, miR-150-5p
and P300 in ISO-induced hypertrophy mouse model. mRNA and protein
levels were analyzed by reverse transcription-quantitative
polymerase chain reaction and western blot analysis, respectively.
(A) ANP and BNP mRNA, (B) ANP protein, (C) MIAT mRNA, (D)
miR-150-5p and (E) P300 mRNA expression levels are presented.
*P<0.05 vs. control group (n=4). ANP, atrial
natriuretic peptide; BNP, brain natriuretic peptide; Con, control;
ISO, isoproterenol; MIAT, myocardial infarction-associated
transcript; miR, microRNA. |
MIAT, miR-150-5p, and P300 expression is
associated with cardiomyocyte hypertrophy induced by ISO in
vitro
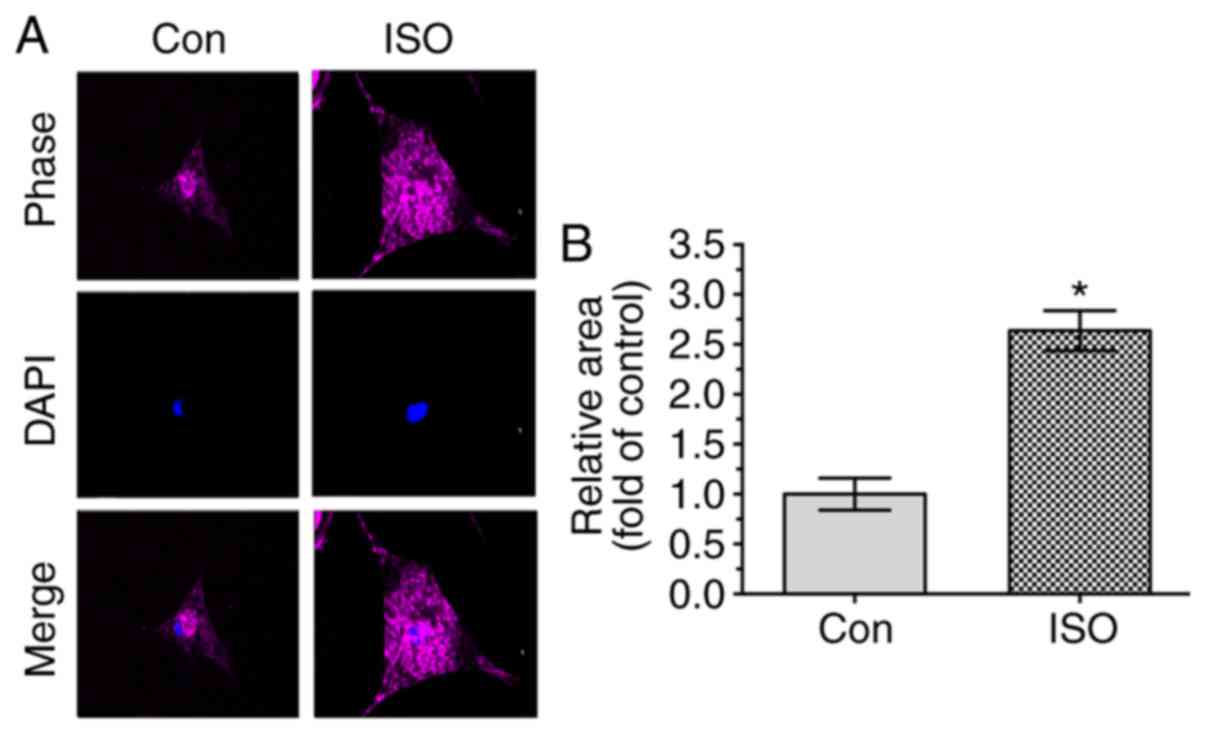
Since ISO was observed to lead to myocyte
hypertrophy in vivo, the effect of ISO in NRVMs was also
examined in vitro. NRVMs were incubated for 24 h in the
presence or absence of 10 µM ISO, and their surface areas
were then measured. As shown in Fig.
4, the myocyte size was increased in the group treated with ISO
compared with that in the control group. Furthermore, the
expression levels of ANP and BNP, biomarkers of cardiomyocyte
hypertrophy, were observed to be significantly upregulate in the
ISO group by using RT-qPCR and western blotting (Fig. 5A and B).
 | Figure 5Expression levels of MIAT, miR-150-5p
and P300 in NRVMs induced by ISO. mRNA and protein levels were
analyzed by reverse transcription-quantitative polymerase chain
reaction and western blot analysis, respectively. (A) ANP and BNP
mRNA, (B) ANP protein, (C) MIAT mRNA, (D) miR-150-5p and (E) P300
mRNA expression levels are shown. *P<0.05 vs. Con
group (n=3). ANP, atrial natriuretic peptide; BNP, brain
natriuretic peptide; ISO, isoproterenol; Con, control; MIAT,
myocardial infarction-associated transcript; miR, microRNA. |
Subsequently, MIAT expression was assessed in
ISO-treated NRVMs. As shown in Fig.
5, it was determined that the level of MIAT was significantly
increased in cardiomyocytes with ISO-induced hypertrophy,
suggesting that MIAT was associated with hypertrophy (Fig. 5C). Furthermore, miR-150-5p
expression was also assessed, and it was observed that miR-150-5p
was markedly decreased in ISO-induced NRVMs (Fig. 5D). Additionally, compared with in
the control group, P300 expression levels were significantly
increased in NRVMs treated with ISO (Fig. 5E). These results suggested that
MIAT, miR-150-5p, and P300 expression may be associated with
cardiomyocyte hypertrophy induced by ISO in vitro.
MIAT may upregulate P300 expression by
suppression of miR-150-5p
Since MIAT and miR-150-5p were observed to be
connected with hypertrophy in the aforementioned results, it was
then hypothesized that the pathological effect of MIAT in cardiac
hypertrophy may be associated with miR-150-5p. To test this
hypothesis, we measured ANP, BNP, P300 and miR-150 expression in
NRVMs by RT-qPCR after transfecting NRVMs with si-MIAT (Fig. 6). The results revealed that MIAT
expression levels were downregulated in NRVMs following
transfection with si-MIAT for 48 h (Fig. 6A). Furthermore, the NRVMs were
treated with or without ISO following transfection with si-NC or
si-MIAT, which demonstrated a significant decrease in the
expression levels of ANP and BNP within si-MIAT cells than that of
si-NC cells (Fig. 6B). In
addition, miR-150-5p expression was significantly increased in
si-MIAT cells compared with in si-NC cells, (Fig. 6C). Similar results with the
expression levels of P300 expression (Fig. 6D). These results indicated that
MIAT may upregulate the expression of ANP, BNP and P300 expression
by suppressing miR-150-5p in NRVMs.
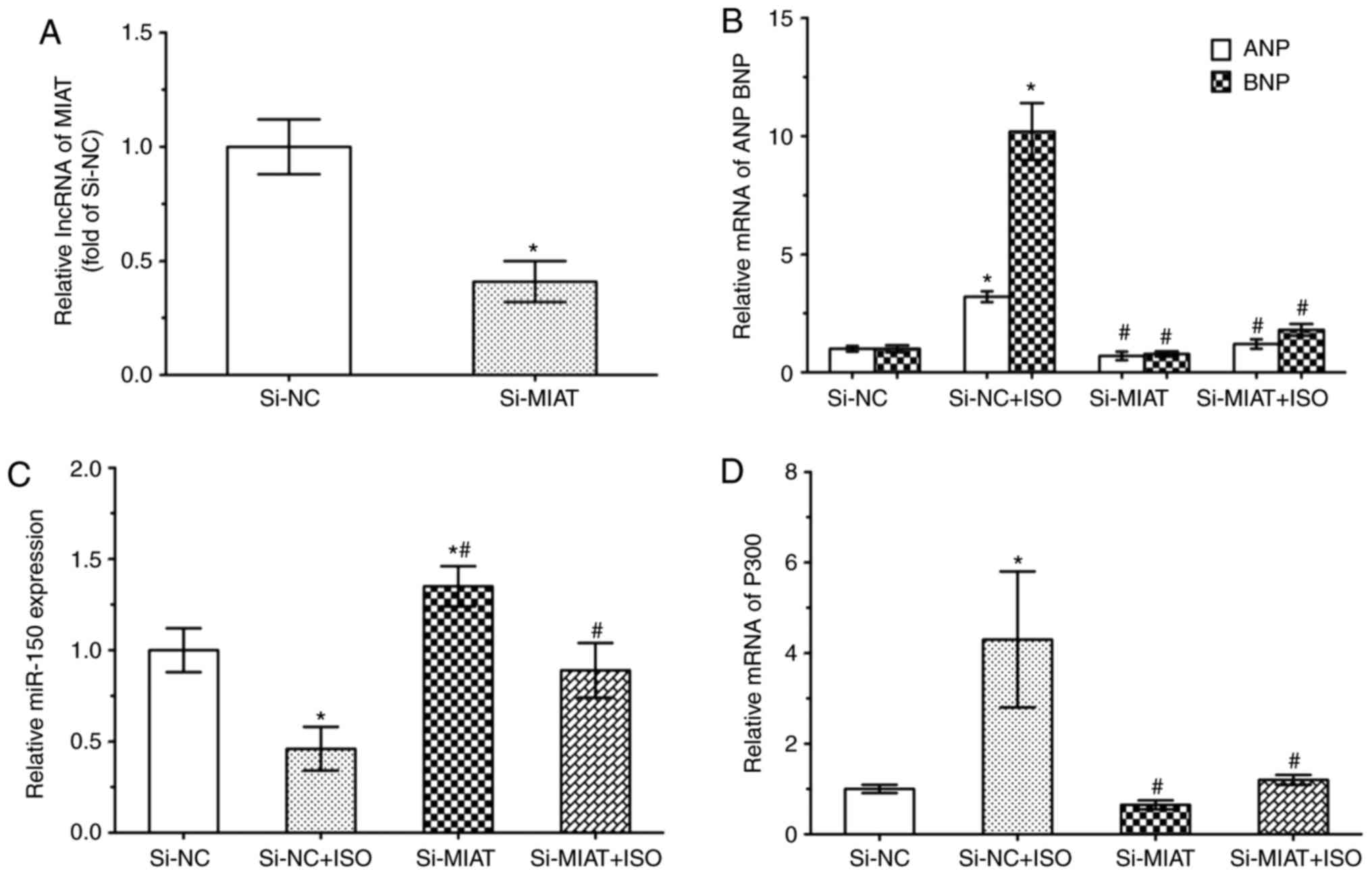 | Figure 6MIAT upregulates P300 expression by
suppressing miR-150-5p. NRVMs were transfected with si-MIAT (100
nM) or si-NC (100 nM) for 6 h, and then treated with or without ISO
(10 µm) for 48 h. Reverse transcription-quantitative
polymerase chain reaction revealed the expression levels of (A)
MIAT, (B) ANP and BNP expression, (C) miR-150-5p and (D) P300.
*P<0.05 vs. si-NC group (n=3); #P<0.05
vs. si-NC + ISO group (n=3). ANP, atrial natriuretic peptide; BNP,
brain natriuretic peptide; si-, small interfering RNA; miR,
microRNA; NC, negative control; ISO, isoproterenol; MIAT,
myocardial infarction-associated transcript; miR, microRNA. |
Discussion
Previous studies have demonstrated that lncRNAs are
correlated with cardiac diseases (19–21); however, few studies have focused
on the effect of lncRNAs in cardiac hypertrophy. MIAT is an lncRNA
isolated as a candidate gene for myocardial infarction and is
expressed predominantly in the heart and brain tissues (8). It has been reported that the
abnormal expression of MIAT is involved in cell proliferation,
apoptosis and migration in numerous diseases, including in
myocardial infarction, microvascular dysfunction and diabetes
(7,22,23). However, whether the lncRNA MIAT
has a regulatory effect on hypertrophy remains known.
The present study attempted to investigate the
effect of MIAT in cardiac hypertrophy and explore the possible
underlying mechanisms. The study findings were as follows: i) MIAT
was upregulated by ISO, as demonstrated in vivo and in
vitro; ii) subsequent to the silencing of MIAT, the expression
levels of ANP and BNP decreased in ISO-treated NRVM cardiomyocytes,
demonstrating the possible association between MIAT and
hypertrophy; and iii) MIAT silencing was associated with increased
miR-150-5p and decreased P300 expression levels in hypertrophic
NRVMs.
It has been indicated that miR-150 mediates the
function of MIAT, and that MIAT may function as a regulator of
miR-150 (24). Thus, as expected,
MIAT silencing in the current study increased the expression of
miR-150-5p in myocytes and decreased P300 expression. Conversely,
ISO treatment suppressed miR-150-5p and enhanced P300 expression in
NRVMs. These results suggested the involvement of the
MIAT/miR-150-5p/P300 network in cardiac hypertrophy; P300 may also
be a downstream target of MIAT.
The importance of P300 in the development of cell
proliferation and differentiation is well-known (25–27). By binding specifically to
phosphorylated CREB protein, P300 mediates the regulation of the
cAMP gene (28,29). Previous studies have reported that
P300 contains a bromodomain and is involved in interleukin-6
signaling (30,31). This gene has been identified as
potentially serving an important role in the regulation of
hypoxia-induced genes (32,33). Furthermore, the P300 protein may
activate transcription by binding to transcription factors; thus,
P300 is also known as a transcriptional coacti-vator (34–36). Previous studies have demonstrated
that miR-150 regulates the expression of the transcriptional
co-activator P300 (37,38). Accordingly, the present study
identified that P300 was a downstream target of
MIAT/miR-150-5p.
In conclusion, the results of the present study
confirmed that the lncRNA MIAT was associated with the
miR-150-5p/P300 signaling pathway in cardiac hypertrophy, providing
novel insights for understanding the function of lncRNAs in the
pathogenesis of cardiac hypertrophy.
Acknowledgments
Not applicable.
Funding
This study was supported by grants from the National
Key R&D Program of China (grant no. 2017YFC1307600) and the
Natural Science Foundation of Liaoning Province (grant no.
2013021090).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors’ contributions
ZL and YL conducted the experiments and wrote the
study. XG, GS and QM made substantial contributions to the design
of the present study. YD performed data analysis, and produced the
figures and tables. GZ conducted animal experiments and wrote the
corresponding results of the paper. YS performed cell experiments
and wrote the corresponding results of the paper. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
All procedures were performed in accordance with the
Ethical Standards of the Committee on Animal Experimentation of
China Medical University (project identification code,
SCXK-2013-0001; Shenyang, China), and were approved by the Ethical
Standards of the Committee on Animal Experimentation of the First
Hospital of China Medical University (no. 2017010).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bertels F, Gokhale CS and Traulsen A:
Discovering complete quasispecies in bacterial genomes. Genetics.
206:2149–2157. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wu DD, Irwin DM and Zhang YP: De novo
origin of human protein-coding genes. PLoS Genet. 7:e10023792011.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang D, Xiong M, Xu C, Xiang P and Zhong
X: Long noncoding RNAs: An overview. Methods Mol Biol.
1402:287–295. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Meller VH, Joshi SS and Deshpande N:
Modulation of chromatin by noncoding RNA. Annu Rev Genet.
49:673–695. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tan JY, Vance KW, Varela MA, Sirey T,
Watson LM, Curtis HJ, Marinello M, Alves S, Steinkraus BR, Cooper
S, et al: Cross-talking noncoding RNAs contribute to cell-specific
neurodegeneration in SCA7. Nat Struct Mol Biol. 21:955–961. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Beck ZT, Xing Z and Tran EJ: LncRNAs:
Bridging environmental sensing and gene expression. RNA Biol.
13:1189–1196. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ishii N, Ozaki K, Sato H, Mizuno H, Saito
S, Takahashi A, Miyamoto Y, Ikegawa S, Kamatani N, Hori M, et al:
Identification of a novel non-coding RNA, MIAT, that confers risk
of myocardial infarction. J Hum Genet. 51:1087–1099. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liao J, He Q, Li M, Chen Y, Liu Y and Wang
J: LncRNA MIAT: Myocardial infarction associated and more. Gene.
578:158–161. 2016. View Article : Google Scholar
|
|
9
|
Jiang Q, Shan K, Qun-Wang X, Zhou RM, Yang
H, Liu C, Li YJ, Yao J, Li XM, Shen Y, et al: Long non-coding
RNA-MIAT promotes neurovascular remodeling in the eye and brain.
Oncotarget. 7:49688–49698. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rao SQ, Hu HL, Ye N, Shen Y and Xu Q:
Genetic variants in long non-coding RNA MIAT contribute to risk of
paranoid schizophrenia in a Chinese Han population. Schizophr Res.
166:125–130. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Aprea J, Prenninger S, Dori M, Ghosh T,
Monasor LS, Wessendorf E, Zocher S, Massalini S, Alexopoulou D,
Lesche M, et al: Transcriptome sequencing during mouse brain
development identifies long non-coding RNAs functionally involved
in neurogenic commitment. EMBO J. 32:3145–3160. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ishizuka A, Hasegawa Y, Ishida K, Yanaka K
and Nakagawa S: Formation of nuclear bodies by the lncRNA
Gomafu-associating proteins Celf3 and SF1. Genes Cells. 19:704–721.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li Y, Wang J, Sun L and Zhu S: LncRNA
myocardial infarction-associated transcript (MIAT) contributed to
cardiac hypertrophy by regulating TLR4 via miR-93. Eur J Pharmacol.
818:508–517. 2018. View Article : Google Scholar
|
|
14
|
Zhu XH, Yuan YX, Rao SL and Wang P: LncRNA
MIAT enhances cardiac hypertrophy partly through sponging miR-150.
Eur Rev Med Pharmacol Sci. 20:3653–3660. 2016.PubMed/NCBI
|
|
15
|
Li J, Liu K, Gao X, Yao B, Huo K, Cheng Y,
Cheng X, Chen D, Wang B, Sun W, et al: Oxygen- and
nitrogen-enriched 3D porous carbon for supercapacitors of high
volumetric capacity. ACS Appl Mater Interfaces. 7:24622–24628.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Qu Q, Liu Y, Yan X, Fan X, Liu N and Wu G:
A Novel pentapeptide targeting integrin β3-subunit inhibits
platelet aggregation and its application in rat for thrombosis
prevention. Front Pharmacol. 7:492016. View Article : Google Scholar
|
|
17
|
McCommis KS, Douglas DL, Krenz M and
Baines CP: Cardiac-specific hexokinase 2 overexpression attenuates
hypertrophy by increasing pentose phosphate pathway flux. J Am
Heart Assoc. 2:e0003552013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-delta delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
19
|
Ounzain S, Micheletti R, Beckmann T,
Schroen B, Alexanian M, Pezzuto I, Crippa S, Nemir M, Sarre A,
Johnson R, et al: Genome-wide profiling of the cardiac
transcriptome after myocardial infarction identifies novel
heart-specific long non-coding RNAs. Eur Heart J. 36:353a–368a.
2015. View Article : Google Scholar
|
|
20
|
Guo X, Chang Q, Pei H, Sun X, Qian X, Tian
C and Lin H: Long non-coding RNA-mRNA correlation analysis reveals
the potential role of HOTAIR in pathogenesis of sporadic thoracic
aortic aneurysm. Eur J Vasc Endovasc Surg. 54:303–314. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kitow J, Derda AA, Beermann J, Kumarswarmy
R, Pfanne A, Fendrich J, Lorenzen JM, Xiao K, Bavendiek U,
Bauersachs J and Thum T: Mitochondrial long noncoding RNAs as blood
based biomarkers for cardiac remodeling in patients with
hyper-trophic cardiomyopathy. Am J Physiol Heart Circ Physiol.
311:H707–H712. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yan B, Yao J, Liu JY, Li XM, Wang XQ, Li
YJ, Tao ZF, Song YC, Chen Q and Jiang Q: lncRNA-MIAT regulates
microvascular dysfunction by functioning as a competing endogenous
RNA. Circ Res. 116:1143–1156. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang J, Chen M, Chen J, Lin S, Cai D,
Chen C and Chen Z: Long non-coding RNA MIAT acts as a biomarker in
diabetic retinopathy by absorbing miR-29b and regulating cell
apoptosis. Biosci Rep. 37:BSR201700362017. View Article : Google Scholar :
|
|
24
|
Zhang HY, Zheng FS, Yang W and Lu JB: The
long non-coding RNA MIAT regulates zinc finger E-box binding
homeobox 1 expression by sponging miR-150 and promoteing cell
invasion in non-small-cell lung cancer. Gene. 633:61–65. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Otero M, Peng H, Hachem KE, Culley KL,
Wondimu EB, Quinn J, Asahara H, Tsuchimochi K, Hashimoto K and
Goldring MB: ELF3 modulates type II collagen gene (COL2A1)
transcription in chondrocytes by inhibiting SOX9-CBP/p300-driven
histone acetyltransferase activity. Connect Tissue Res. 58:15–26.
2017. View Article : Google Scholar :
|
|
26
|
Tikhanovich I, Zhao J, Bridges B, Kumer S,
Roberts B and Weinman SA: Arginine methylation regulates
c-Myc-dependent transcription by altering promoter recruitment of
the acetyltransferase p300. J Biol Chem. 292:13333–13344. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Clark MD, Kumar GS, Marcum R, Luo Q, Zhang
Y and Radhakrishnan I: Molecular basis for the mechanism of
constitutive CBP/p300 coactivator recruitment by CRTC1-MAML2 and
Its implications in cAMP signaling. Biochemistry. 54:5439–5446.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Naqvi S, Martin KJ and Arthur JS: CREB
phosphorylation at Ser133 regulates transcription via distinct
mechanisms downstream of cAMP and MAPK signalling. Biochem J.
458:469–479. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hou T, Ray S, Lee C and Brasier AR: The
STAT3 NH2-terminal domain stabilizes enhanceosome assembly by
interacting with the p300 bromodomain. J Biol Chem.
283:30725–30734. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zgheib C, Zouein FA, Chidiac R, Kurdi M
and Booz GW: Calyculin A reveals serine/threonine phosphatase
protein phosphatase 1 as a regulatory nodal point in canonical
signal transducer and activator of transcription 3 signaling of
human microvascular endothelial cells. J Interferon Cytokine Res.
32:87–94. 2012. View Article : Google Scholar :
|
|
31
|
Zhou CH, Zhang XP, Liu F and Wang W:
Modeling the interplay between the HIF-1 and p53 pathways in
hypoxia. Sci Rep. 5:138342015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kato H, Tamamizu-Kato S and Shibasaki F:
Histone deacetylase 7 associates with hypoxia-inducible factor
1alpha and increases transcriptional activity. J Biol Chem.
279:41966–41974. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kasper LH, Fukuyama T, Lerach S, Chang Y,
Xu W, Wu S, Boyd KL and Brindle PK: Genetic interaction between
mutations in c-Myb and the KIX domains of CBP and p300 affects
multiple blood cell lineages and influences both gene activation
and repression. PloS One. 8:e826842013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang F, Marshall CB, Yamamoto K, Li GY,
Gasmi-Seabrook GM, Okada H, Mak TW and Ikura M: Structures of KIX
domain of CBP in complex with two FOXO3a transactivation domains
reveal promiscuity and plasticity in coactivator recruitment. Proc
Natl Acad Sci USA. 109:6078–6083. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bedford DC, Kasper LH, Wang R, Chang Y,
Green DR and Brindle PK: Disrupting the CH1 domain structure in the
acetyltransferases CBP and p300 results in lean mice with increased
metabolic control. Cell Metab. 14:219–230. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Teufel DP, Freund SM, Bycroft M and Fersht
AR: Four domains of p300 each bind tightly to a sequence spanning
both trans-activation subdomains of p53. Proc Natl Acad Sci USA.
104:7009–7014. 2007. View Article : Google Scholar
|
|
37
|
Duan Y, Zhou B, Su H, Liu Y and Du C:
miR-150-5p regulates high glucose-induced cardiomyocyte hypertrophy
by targeting the transcriptional co-activator p300. Exp Cell Res.
319:173–184. 2013. View Article : Google Scholar
|
|
38
|
Prakhar P, Holla S, Ghorpade DS, Gilleron
M, Puzo G, Udupa V and Balaji KN: Ac2PIM-responsivmiR-150-5p and
miR-143 target receptor-interacting protein kinase 2 and
transforming growth factor beta-activated kinase 1 to suppress
NOD2-induced immunomodulators. J Biol Chem. 290:26576–26586. 2015.
View Article : Google Scholar : PubMed/NCBI
|