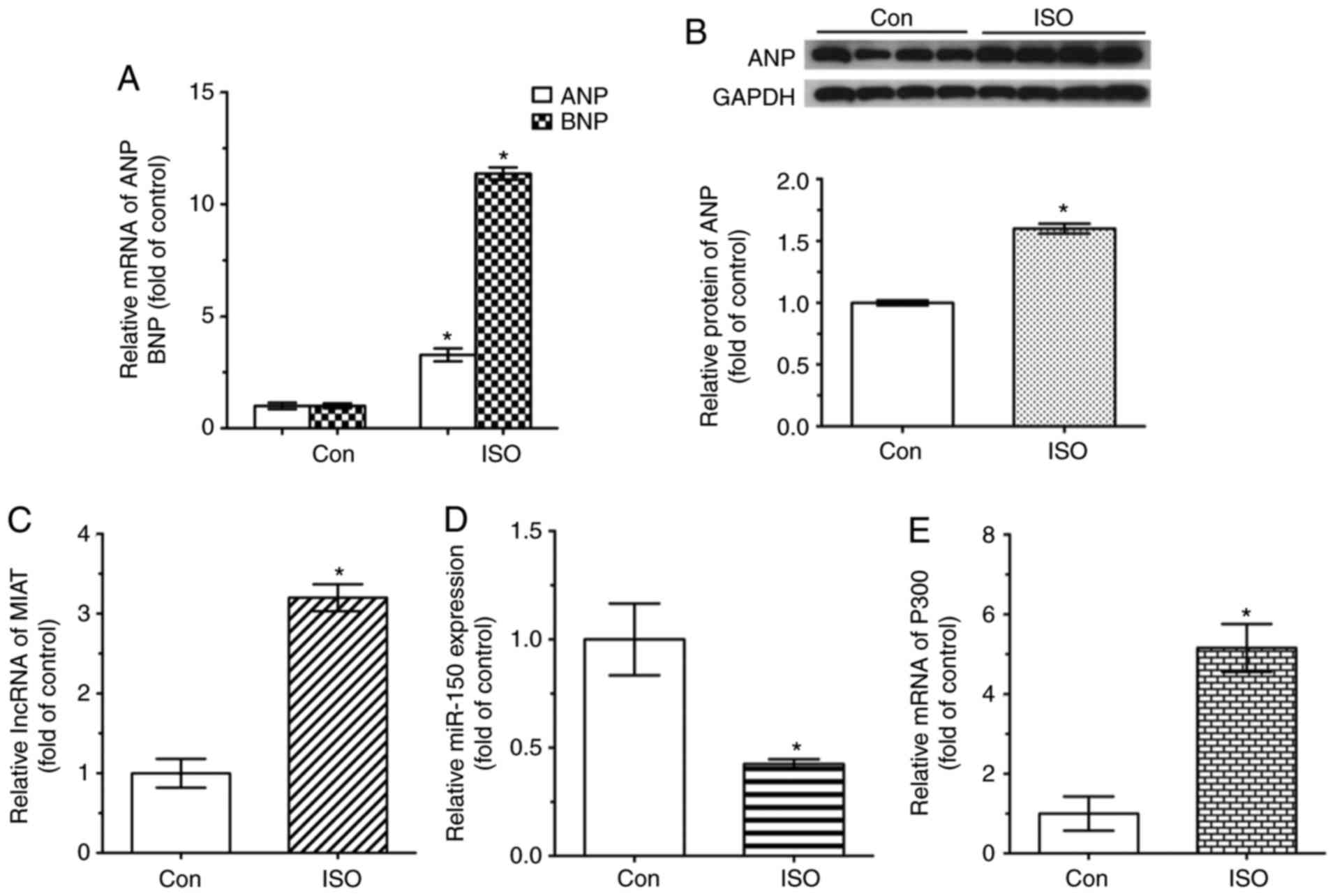
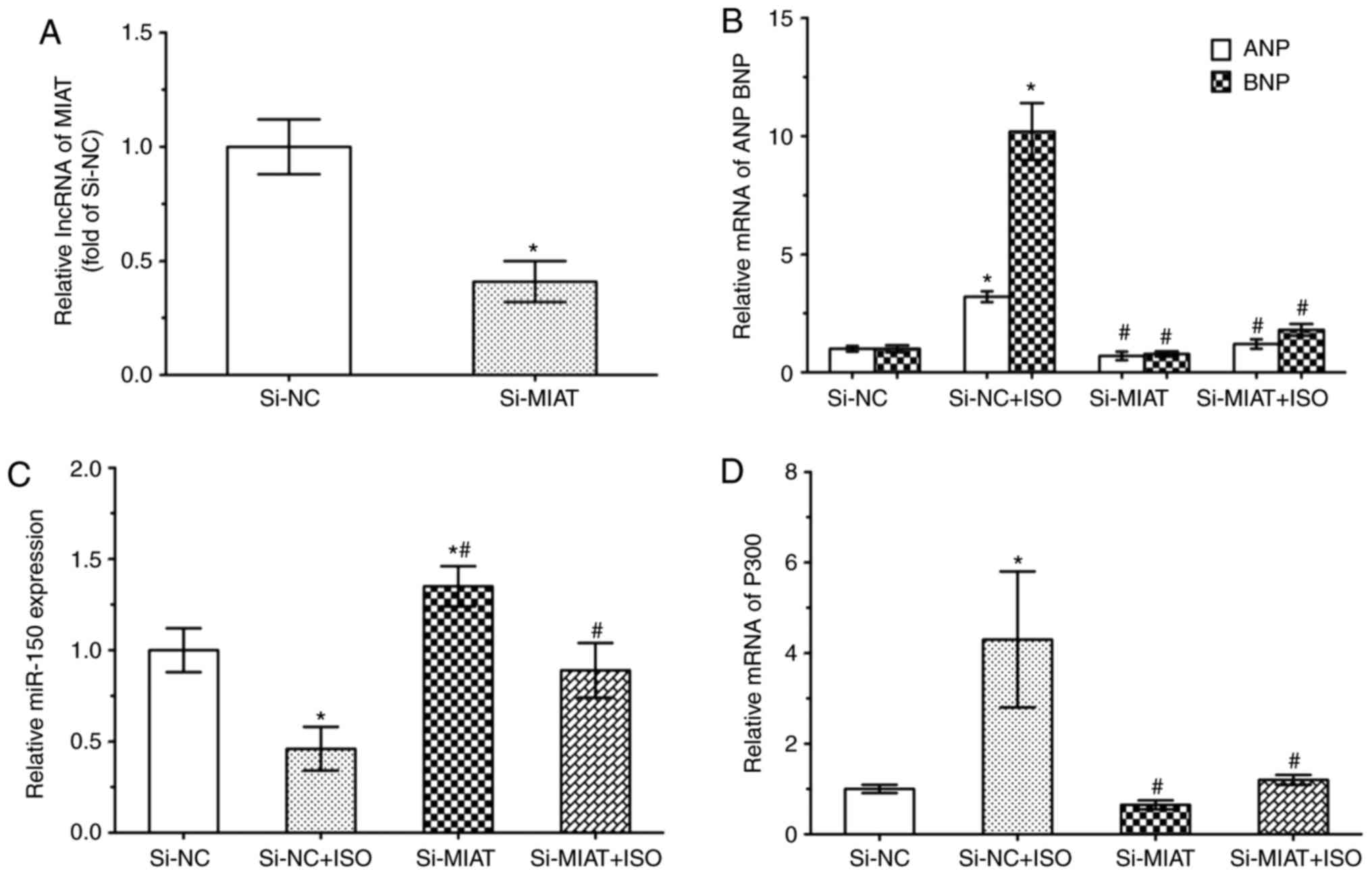
|
1
|
Bertels F, Gokhale CS and Traulsen A:
Discovering complete quasispecies in bacterial genomes. Genetics.
206:2149–2157. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wu DD, Irwin DM and Zhang YP: De novo
origin of human protein-coding genes. PLoS Genet. 7:e10023792011.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang D, Xiong M, Xu C, Xiang P and Zhong
X: Long noncoding RNAs: An overview. Methods Mol Biol.
1402:287–295. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Meller VH, Joshi SS and Deshpande N:
Modulation of chromatin by noncoding RNA. Annu Rev Genet.
49:673–695. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tan JY, Vance KW, Varela MA, Sirey T,
Watson LM, Curtis HJ, Marinello M, Alves S, Steinkraus BR, Cooper
S, et al: Cross-talking noncoding RNAs contribute to cell-specific
neurodegeneration in SCA7. Nat Struct Mol Biol. 21:955–961. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Beck ZT, Xing Z and Tran EJ: LncRNAs:
Bridging environmental sensing and gene expression. RNA Biol.
13:1189–1196. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ishii N, Ozaki K, Sato H, Mizuno H, Saito
S, Takahashi A, Miyamoto Y, Ikegawa S, Kamatani N, Hori M, et al:
Identification of a novel non-coding RNA, MIAT, that confers risk
of myocardial infarction. J Hum Genet. 51:1087–1099. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liao J, He Q, Li M, Chen Y, Liu Y and Wang
J: LncRNA MIAT: Myocardial infarction associated and more. Gene.
578:158–161. 2016. View Article : Google Scholar
|
|
9
|
Jiang Q, Shan K, Qun-Wang X, Zhou RM, Yang
H, Liu C, Li YJ, Yao J, Li XM, Shen Y, et al: Long non-coding
RNA-MIAT promotes neurovascular remodeling in the eye and brain.
Oncotarget. 7:49688–49698. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rao SQ, Hu HL, Ye N, Shen Y and Xu Q:
Genetic variants in long non-coding RNA MIAT contribute to risk of
paranoid schizophrenia in a Chinese Han population. Schizophr Res.
166:125–130. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Aprea J, Prenninger S, Dori M, Ghosh T,
Monasor LS, Wessendorf E, Zocher S, Massalini S, Alexopoulou D,
Lesche M, et al: Transcriptome sequencing during mouse brain
development identifies long non-coding RNAs functionally involved
in neurogenic commitment. EMBO J. 32:3145–3160. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ishizuka A, Hasegawa Y, Ishida K, Yanaka K
and Nakagawa S: Formation of nuclear bodies by the lncRNA
Gomafu-associating proteins Celf3 and SF1. Genes Cells. 19:704–721.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li Y, Wang J, Sun L and Zhu S: LncRNA
myocardial infarction-associated transcript (MIAT) contributed to
cardiac hypertrophy by regulating TLR4 via miR-93. Eur J Pharmacol.
818:508–517. 2018. View Article : Google Scholar
|
|
14
|
Zhu XH, Yuan YX, Rao SL and Wang P: LncRNA
MIAT enhances cardiac hypertrophy partly through sponging miR-150.
Eur Rev Med Pharmacol Sci. 20:3653–3660. 2016.PubMed/NCBI
|
|
15
|
Li J, Liu K, Gao X, Yao B, Huo K, Cheng Y,
Cheng X, Chen D, Wang B, Sun W, et al: Oxygen- and
nitrogen-enriched 3D porous carbon for supercapacitors of high
volumetric capacity. ACS Appl Mater Interfaces. 7:24622–24628.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Qu Q, Liu Y, Yan X, Fan X, Liu N and Wu G:
A Novel pentapeptide targeting integrin β3-subunit inhibits
platelet aggregation and its application in rat for thrombosis
prevention. Front Pharmacol. 7:492016. View Article : Google Scholar
|
|
17
|
McCommis KS, Douglas DL, Krenz M and
Baines CP: Cardiac-specific hexokinase 2 overexpression attenuates
hypertrophy by increasing pentose phosphate pathway flux. J Am
Heart Assoc. 2:e0003552013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-delta delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
19
|
Ounzain S, Micheletti R, Beckmann T,
Schroen B, Alexanian M, Pezzuto I, Crippa S, Nemir M, Sarre A,
Johnson R, et al: Genome-wide profiling of the cardiac
transcriptome after myocardial infarction identifies novel
heart-specific long non-coding RNAs. Eur Heart J. 36:353a–368a.
2015. View Article : Google Scholar
|
|
20
|
Guo X, Chang Q, Pei H, Sun X, Qian X, Tian
C and Lin H: Long non-coding RNA-mRNA correlation analysis reveals
the potential role of HOTAIR in pathogenesis of sporadic thoracic
aortic aneurysm. Eur J Vasc Endovasc Surg. 54:303–314. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kitow J, Derda AA, Beermann J, Kumarswarmy
R, Pfanne A, Fendrich J, Lorenzen JM, Xiao K, Bavendiek U,
Bauersachs J and Thum T: Mitochondrial long noncoding RNAs as blood
based biomarkers for cardiac remodeling in patients with
hyper-trophic cardiomyopathy. Am J Physiol Heart Circ Physiol.
311:H707–H712. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yan B, Yao J, Liu JY, Li XM, Wang XQ, Li
YJ, Tao ZF, Song YC, Chen Q and Jiang Q: lncRNA-MIAT regulates
microvascular dysfunction by functioning as a competing endogenous
RNA. Circ Res. 116:1143–1156. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang J, Chen M, Chen J, Lin S, Cai D,
Chen C and Chen Z: Long non-coding RNA MIAT acts as a biomarker in
diabetic retinopathy by absorbing miR-29b and regulating cell
apoptosis. Biosci Rep. 37:BSR201700362017. View Article : Google Scholar :
|
|
24
|
Zhang HY, Zheng FS, Yang W and Lu JB: The
long non-coding RNA MIAT regulates zinc finger E-box binding
homeobox 1 expression by sponging miR-150 and promoteing cell
invasion in non-small-cell lung cancer. Gene. 633:61–65. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Otero M, Peng H, Hachem KE, Culley KL,
Wondimu EB, Quinn J, Asahara H, Tsuchimochi K, Hashimoto K and
Goldring MB: ELF3 modulates type II collagen gene (COL2A1)
transcription in chondrocytes by inhibiting SOX9-CBP/p300-driven
histone acetyltransferase activity. Connect Tissue Res. 58:15–26.
2017. View Article : Google Scholar :
|
|
26
|
Tikhanovich I, Zhao J, Bridges B, Kumer S,
Roberts B and Weinman SA: Arginine methylation regulates
c-Myc-dependent transcription by altering promoter recruitment of
the acetyltransferase p300. J Biol Chem. 292:13333–13344. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Clark MD, Kumar GS, Marcum R, Luo Q, Zhang
Y and Radhakrishnan I: Molecular basis for the mechanism of
constitutive CBP/p300 coactivator recruitment by CRTC1-MAML2 and
Its implications in cAMP signaling. Biochemistry. 54:5439–5446.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Naqvi S, Martin KJ and Arthur JS: CREB
phosphorylation at Ser133 regulates transcription via distinct
mechanisms downstream of cAMP and MAPK signalling. Biochem J.
458:469–479. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hou T, Ray S, Lee C and Brasier AR: The
STAT3 NH2-terminal domain stabilizes enhanceosome assembly by
interacting with the p300 bromodomain. J Biol Chem.
283:30725–30734. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zgheib C, Zouein FA, Chidiac R, Kurdi M
and Booz GW: Calyculin A reveals serine/threonine phosphatase
protein phosphatase 1 as a regulatory nodal point in canonical
signal transducer and activator of transcription 3 signaling of
human microvascular endothelial cells. J Interferon Cytokine Res.
32:87–94. 2012. View Article : Google Scholar :
|
|
31
|
Zhou CH, Zhang XP, Liu F and Wang W:
Modeling the interplay between the HIF-1 and p53 pathways in
hypoxia. Sci Rep. 5:138342015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kato H, Tamamizu-Kato S and Shibasaki F:
Histone deacetylase 7 associates with hypoxia-inducible factor
1alpha and increases transcriptional activity. J Biol Chem.
279:41966–41974. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kasper LH, Fukuyama T, Lerach S, Chang Y,
Xu W, Wu S, Boyd KL and Brindle PK: Genetic interaction between
mutations in c-Myb and the KIX domains of CBP and p300 affects
multiple blood cell lineages and influences both gene activation
and repression. PloS One. 8:e826842013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang F, Marshall CB, Yamamoto K, Li GY,
Gasmi-Seabrook GM, Okada H, Mak TW and Ikura M: Structures of KIX
domain of CBP in complex with two FOXO3a transactivation domains
reveal promiscuity and plasticity in coactivator recruitment. Proc
Natl Acad Sci USA. 109:6078–6083. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bedford DC, Kasper LH, Wang R, Chang Y,
Green DR and Brindle PK: Disrupting the CH1 domain structure in the
acetyltransferases CBP and p300 results in lean mice with increased
metabolic control. Cell Metab. 14:219–230. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Teufel DP, Freund SM, Bycroft M and Fersht
AR: Four domains of p300 each bind tightly to a sequence spanning
both trans-activation subdomains of p53. Proc Natl Acad Sci USA.
104:7009–7014. 2007. View Article : Google Scholar
|
|
37
|
Duan Y, Zhou B, Su H, Liu Y and Du C:
miR-150-5p regulates high glucose-induced cardiomyocyte hypertrophy
by targeting the transcriptional co-activator p300. Exp Cell Res.
319:173–184. 2013. View Article : Google Scholar
|
|
38
|
Prakhar P, Holla S, Ghorpade DS, Gilleron
M, Puzo G, Udupa V and Balaji KN: Ac2PIM-responsivmiR-150-5p and
miR-143 target receptor-interacting protein kinase 2 and
transforming growth factor beta-activated kinase 1 to suppress
NOD2-induced immunomodulators. J Biol Chem. 290:26576–26586. 2015.
View Article : Google Scholar : PubMed/NCBI
|