Introduction
Non-alcoholic fatty liver disease (NAFLD) is a
manifestation of metabolic syndrome in the liver and is
characterized by ectopic lipid accumulation in hepatocytes without
excess alcohol intake, which encompasses a spectrum from simple
steatosis to steatohepatitis, cirrhosis and hepatic carcinoma
(1). Over the last decade, NAFLD
has emerged as one of the most common chronic liver diseases, with
a prevalence of 20–30% in the general population in China (2), however, its pathogenesis remains to
be elucidated. NAFLD is closely associated with diabetes. Abnormal
glucolipid metabolism can result in fatty degeneration in
hepatocytes (3,4). Consequently, examination of the
mechanism underlying abnormal glucolipid metabolism is particularly
important for elucidating the pathogenesis of NAFLD and identifying
novel therapeutic approaches for its treatment.
Carbohydrate responsive element binding protein
(ChREBP) has been recognized as a key transcription factor of lipid
synthesis, which is activated by high glucose, and its target genes
involve glycolytic enzymes, gluconeogenic enzymes and lipid
synthases (5–9). In response to an excessive glucose
influx in hepatocytes, the expression and transcriptional activity
of ChREBP are increased via liver X receptor (LXR) and
phosphopentose pathways, respectively, which induce the
transcription and expression of the downstream target gene fatty
acid synthase (FASN), the key enzyme of lipid de novo
synthesis, promoting the conversion of glucose to lipid (10–12). Therefore, ChREBP is considered a
hub for glucolipid metabolism and a crucial regulator of de
novo lipogenesis (11,13–15).
In hepatocytes, the overexpression of FASN caused by excessive
activation of ChREBP can result in hepatic steatosis and further
progression of NAFLD (16–18).
Eukaryotic gene transcription is a highly regulated
process, wherein the transformation of the chromatin structure is
essential. All types of biological molecular signals ultimately
exert their effect on the chromatin structure through various
signaling pathways. However, chromatin is intrinsically positioned
in a condensed and closed state, which limits various biological
processes that require DNA as a template; therefore, the binding of
any signaling molecules, including transcription factors, to the
transcriptional regulatory elements of a target gene require
accompanied changes in the local chromatin structure to activate or
inhibit gene transcription. Due to the high stability of genes,
signaling molecules generally affect gene alternative expression by
altering the structure of chromatin without altering the DNA
sequence. Epigenetics describes the reversible changes in gene
activity, which can be inherited through generations and cell
divisions, and eventually affects the phenotype through alterations
to the chromatin structure and DNA methylation to enable the fine
tuning of gene transcription without variation in the DNA sequence;
this is vital in the differentiation of cells and the development
of specific diseases (19,20).
Previous advances in the understanding of epigenetics and its
prevalence as a contributor to the development of metabolic
syndrome, including obesity and diabetes, have resulted in a marked
increase in interest (21–24).
As an important component of epigenetics, histone modification
requires the cooperation of various enzymes, co-activators and
transcriptional factors, including histone acetyltransferase (HAT),
histone deacetylase (HDAC) and p300 (25,26). Through histone modification, the
interaction between histones and DNA, and between the promoter
region of target genes and transcription factors, can be affected
such that the condensation status of the chromatin can be altered
to facilitate or inhibit gene transcription. ChREBP regulates the
selective transcription of FASN by directly binding to the
carbohydrate response element (ChoRE) found in the FASN promoter.
However, no histone modifications of FASN modulated by ChREBP have
been reported; therefore, their investigation is important in
understanding glucolipid metabolism disorders and abnormal lipid
deposition in the liver.
In the present study, HepG2 and L02 cell lines were
selected as model cell lines. First, the association between
histone modifications and transcription of FASN upon stimulation
with high glucose, or following its withdrawal, was examined.
Second, the effect of ChREBP on histone modifications in FASN
promoter regions was determined using RNA interference and
overexpression approaches. Finally, the effect of the histone
acetylation pattern of FASN on hepatocellular lipogenesis was
examined using the HAT inhibitor. The present study aimed to
provide further insight into the pathophysiology of hepatic
steatosis induced by high glucose and to identify the patterns of
FASN histone modifications as a novel strategy for NAFLD therapy
based on epigenetic mechanisms.
Materials and methods
Cell culture and reagents
The human normal liver cell line (L02 cells) and the
hepatocellular carcinoma cell line (HepG2 cells) were obtained from
the Cell Bank of the Institute of Biochemistry and Cell Biology
(Shanghai, China). The HepG2 cells were cultured in Dulbecco’s
modified Eagle’s medium (Hyclone; GE Healthcare, Life Sciences,
Logan, UT, USA) containing 10% fetal bovine serum (FBS; Hyclone; GE
Healthcare Life Sciences). The L02 cells were cultured in RPMI-1640
medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
containing 10% FBS (Hyclone; GE Healthcare Life Sciences). Both of
the cell lines were cultured at 37°C with 5% CO2 in a
humidified incubator. The optimal glucose concentrations for HepG2
or L02 were analyzed using CCK8 assay kit (Applygen Technologies,
Inc., Beijing, China), which revealed that 50 and 30 mM were the
optimal glucose concentrations for the HepG2 and L02 cells,
respectively. Garcinol, an HAT inhibitor, was obtained from Cayman
Chemical Company (Ann Arbor, MI, USA) and the treatment was 5
µM in 5 ml cell culture medium for 24 h as previously
described (27).
Hepatic triglyceride (TG) assay
To determine the content of TG in the hepatocytes,
the level of TG was assayed using a TG assay kit (Applygen
Technologies, Inc.) according to the manufacturer’s recommended
protocol.
Oil Red O staining
The cultured cells were rinsed with PBS three times
for 5 min each and fixed with 4% paraformaldehyde for 30 min at
37°C. The fixed cells were washed with PBS and stained with Oil Red
O (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) for 10–15 min at
room temperature. Subsequently, the stained cells were washed with
PBS, counterstained with hematoxylin for 6–10 sec, and then rinsed
with distilled water. Representative photomicrographs were captured
at higher magnifications using a system incorporated into an
optical microscope.
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
analysis
Total RNA from the hepatocytes was extracted using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and was
reverse transcribed using the RevertAid First Strand cDNA Synthesis
kit (Fermentas; Thermo Fisher Scientific, Inc.) according to the
manufacturer’s protocol. The qPCR analysis was performed using the
KAPA SYBR® FAST qPCR kit Mater Mix ABI Prism™ (Kapa
Biosystems, Boston, MA, USA) with the amplifying primer pairs:
FASN, forward 5′-TTCTACGGCTCCACGCTCTTCC-3′ and reverse
5′-GAAGAGTCTTCGTCAGCCAGGA-3′; and ChREBP, forward
5′-GTCGGAGAACTTGCAGGAGT-3′ and reverse 5′-CCTTCCAGCGACGGTCATC-3′.
β-actin, forward 5′-TGACGGTCAGGTCATCACTATCGG-3′ and reverse
5′-TTGATCTTCATGGTGATAGGAGCGA-3′ served as an internal control. qPCR
master mix was as follows: 10 µl 2× Master Mix, 0.4
µl 10 µm forward primer, 0.4 µl 10 µm
reverse primer, 20 ng template cDNA, and moderate PCR-grade buffer,
the total volume was 20 µl. The thermocycling steps were as
follows: i) Enzyme activation: 95°C for 3 min, followed by 40
cycles of ii) Denaturation: 95°C for 3 sec and iii) Annealing: 60°C
for 20 sec. All samples were run in triplicate. Gene expression was
normalized against β-actin, and the relative mRNA expression was
calculated using the 2−ΔΔCq method (28).
Western blot analysis
Total protein of hepatocytes was extracted using
Protein Extraction kit (Sigma-Aldrich; Merck KGaA) according to the
manufacturer’s protocol. Briefly, 80 µg protein lysates were
separated by 5–15% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis and subsequently transferred onto polyvinylidene
difluoride membranes. Following blocking with PBST containing 5%
fatty acid-free milk, the membranes were incubated overnight at 4°C
with specific primary antibodies against ChREBP (1:1,000; cat.no.
ab157153; Abcam, Cambridge, MA, USA), FASN (1:200; cat. no.
sc-20140; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) or
β-actin (1:1,000; cat. no. SP124; EMD Millipore; Billerica, MA,
USA) and the corresponding HRP-conjugated secondary antibodies
(1:2,000; cat. no. ab6721; Abcam) at room temperature (25°C) for 2
h. The protein bands were visualized using ECL western blotting
reagents. The band intensities were quantified using Quantity
One-4.6.2 software (Bio-Rad Laboratories, Inc., Hercules, CA,
USA).
Plasmids, small interfering RNA (siRNA)
and transient transfection
ChREBP-specific siRNA (siRNA-ChREBP) and
GV102-scramble (negative control) plasmids were purchased from
Shanghai GenePharma Biotechnology (Shanghai, China). The target
sequences of the siRNA-ChREBP and GV102-scramble were
5′-GCAGAAGACAGCTGAGTACAT-3′ and 5′-GTTCTCCGAACGTGTCACGT-3′,
respectively. Cells at 85–90% confluence were transfected with
either siRNA-ChREBP or siRNA-scramble using PolyJet in vitro
DNA transfection reagent (SignaGen Laboratories, Rockville, MD,
USA). The transfection efficiency, which was measured by
quantifying the number of EGFP-positive cells using a fluorescence
microscope, was ~70–80%. For the overexpression experiments, a
plasmid expressing human wild-type ChREBP (GenBank accession no.
NG_009307) was also purchased from GenePharma Biotechnology. The
cells were transfected with the ChREBP plasmid or pEX3-scramble
(negative control) using PolyJet, as described above.
Chromatin immunoprecipitation (ChIP)
assay
The cells were cross-linked with 1% formaldehyde for
10 min at 37°C, and crude nuclei were purified using a previously
described protocol (27). The
crude nuclei were sonicated to produce chromatin fragments of ~500
bp, as determined by agarose gel electrophoresis. For
immunoprecipitation, the antibodies used in the ChIP assay were as
follows: Anti-acetyl-histone H3 (cat. no. 06-599; EMD Millipore),
anti-acetyl-histone H4 (cat. no. 06-866; EMD Millipore),
anti-phospho-histone H3 serine 10 (H3S10, cat. no. 04-817; EMD
Millipore), and anti-trimethyl-histone H4 lysine 20 (cat. no.
07-463; EMD Millipore), in addition to anti-trimethyl-histone H3
lysine 4 (H3K4, cat. no. ab12209; Abcam), anti-trimethyl-histone H3
lysine 9 (H3K9, cat. no. ab8898; Abcam) and anti-ChREBP (cat. no.
ab157153; Abcam). For each ChIP assay, 2.5 µg of antibody
was added and the samples were incubated overnight at 4°C. The
antibody-chromatin complexes were precipitated by overnight
incubation with Protein A Agarose/Salmon Sperm DNA at 4°C. The
DNA-protein crosslinks were reversed by incubation at 65°C for 4 h,
followed by proteinase K treatment. DNA was recovered using a
purification column (Qiagen GmbH, Hilden, Germany). The ChIP and
input DNA samples were quantified by RT-qPCR analysis, and each
assay was performed in triplicate. For the FASN regulatory regions,
the following were selected: ChoRE (forward
5′-CTTTGTCCGCACCACACCAGG-3′ and reverse
5′-AGACCCGAGACGGACGTCCAC-3′), −2 kb (forward
5′-TGGGAACACGATGGGAGAAC-3′ and reverse 5′-ATCGTGGGACCTTTGCAGC-3′)
upstream from the transcription start site (TSS), promoter (forward
5′-GATGGCCGCGGTTTAAATAGC-3′ and reverse 5′-AGAGACGGCAGCGGC-3′), and
exon2 (forward 5′-CGGGAAGCTGCCAGAGT-3′ and reverse
5′-TTCTGGGACAACCTCATCGG-3′).
Statistical analysis
SPSS software (version 17.0; SPSS, Inc., Chicago,
IL, USA) was used to analyze the data which are expressed as the
mean ± standard deviation. All experiments were performed in
triplicate. Differences among groups were determined using the
Student Newman Keuls test. P<0.05 was considered to indicate a
statistically significant difference.
Results
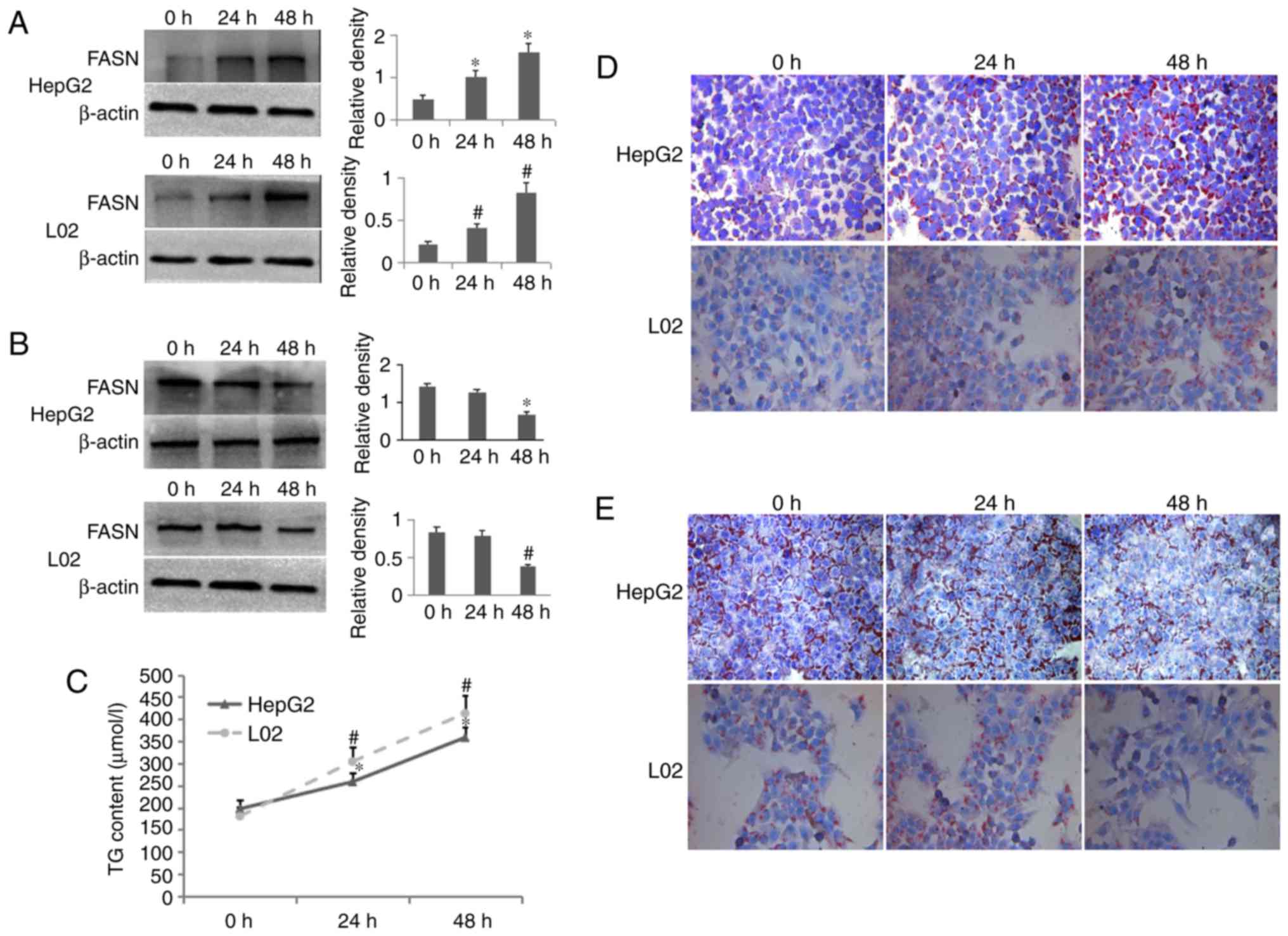
Enhancement of lipogenesis in hepatocytes
induced by high glucose
The HepG2 and L02 cells were incubated with high
glucose for 0, 24 and 48 h. Western blot analysis revealed that
high glucose significantly increased the protein expression of FASN
in the HepG2 and L02 cells in a time-dependent manner (Fig. 1A). Following stimulation of the
HepG2 and L02 cells with high glucose for 48 h in, the culture
medium containing high glucose was replaced with normal medium,
termed ‘glucose removal’, which was followed by the measurement of
FASN protein levels using western blot analyses at 0, 24 and 48 h
later. As shown in Fig. 1B, when
high glucose was removed for 48 h in the HepG2 and L02 cells, the
protein level of FASN was markedly decreased, compared with the
level at 0 h glucose removal, although no significant difference
was found between 24 and 0 h glucose removal. As shown in Fig. 1C, the TG contents of the HepG2 and
L02 cells were increased over time. To further substantiate this
data, the levels of fat droplets were examined in HepG2 and L02
cells using Oil Red O staining analysis. The high glucose treatment
induced fat droplets in the HepG2 and L02 cells, with the same
pattern observed using western blot analyses and the TG content
assay (Fig. 1D). However, in the
two cell lines at 48 h glucose removal, the fat droplets were
decreased compared with those at 0 h glucose removal, although no
significant difference was found between 24 and 0 h glucose removal
(Fig. 1E). Taken together, these
results suggested that high glucose induced the enhancement of
lipogenesis in hepatocytes.
Association between histone modifications
and transcription of FASN in the presence of high glucose and
following its removal
Following the observation of the increased
lipogenesis potential of hepatocyte lines treated with high
glucose, the time spectrum of the mRNA expression of FASN was
investigated in the presence of high glucose or following its
removal using RT-qPCR analysis. The mRNA expression levels of FASN
in the two hepatocyte lines in the presence of high glucose are
shown in Fig. 2A. The mRNA
expression of FASN was temporally increased at 1 h, compared with
that at 0 h, however, it returned to baseline over the subsequent
hours. When the HepG2 and L02 cells were incubated in high glucose
for 12 and 8 h, respectively, the mRNA expression of FASN was
gradually and stably increased with time. Following high glucose
removal, the increasing mRNA level of FASN was significantly
decreased with time in the HepG2 and L02 cells (Fig. 2A). All differences were of
statistical significance. These experiments were used to determine
suitable time-points for the following ChIP experiments.
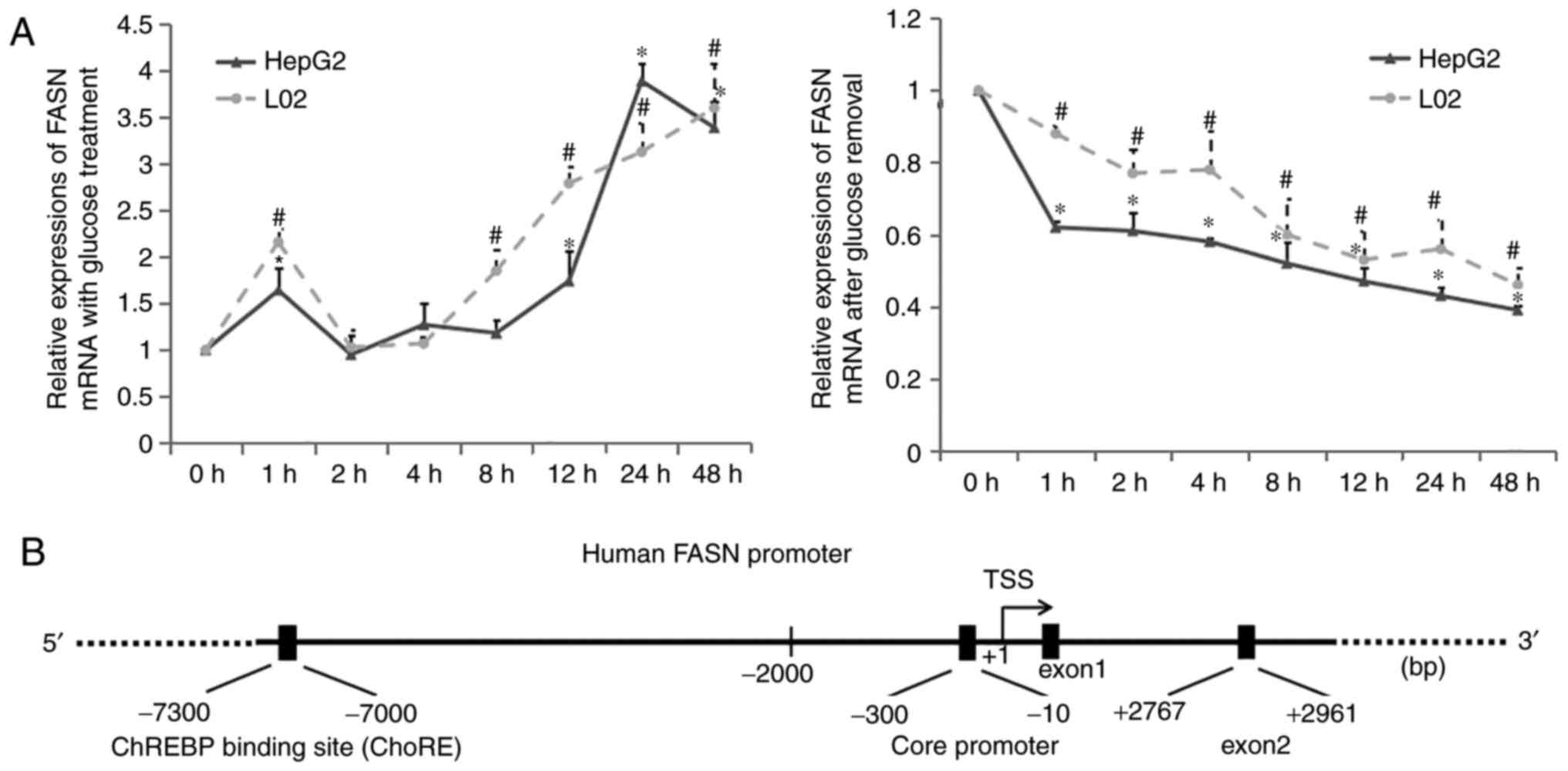 | Figure 2Association between histone
modifications and transcription of FASN in the presence of a high
glucose concentration and following its removal. Total RNA was
extracted and used for reverse transcription-quantitative
polymerase chain reaction analysis to examine the time mRNA
expression of FASN in HepG2 and L02 cells (A) under glucose
treatment or following removal; β-actin was used as an internal
control; *P<0.05, vs. 0 h in HepG2;
#P<0.05, vs. 0 h in L02. (B) Analysis of FASN
promoter regions around the TSS. Chromatin immunoprecipitation
assays were used to examine acetylated H3 and H4, and trimethylated
H3K4, H3K9 and H4K20, and phosphorylated H3S10 occupancy in the
FASN ChoRE, promoter, exon2, and −2 kb regions of (C) HepG2 and (D)
L02 cells treated with glucose, and (E) HepG2 and (F) L02 cells
following gluocse removal. Values at time 0 h were set as 1;
*P<0.05, vs. 0 h in HepG2 or L02. Data are expressed
as the mean ± standard deviation from at least three independent
experiments. FASN; fatty acid synthase; ChoRE, carbohydrate
response element; ChIP, chromatin immunoprecipitation. |
To elucidate the underlying epigenetic modifications
of FASN promoter regions under the stimulation of high glucose and
following its withdrawal, ChIP experiments were performed in the
hepatocyte lines. The promoter exhibits an extensive domain, which
consists of a core promoter region and regulatory regions. The
former is responsible for the transcription of genetic foundation
levels, whereas regulatory regions are able to respond to different
environmental factors or intracellular signals and then make the
corresponding adjustments for the selective transcription of genes.
Between the two domains, the core promoter is usually located
within −2,000 bp from the TSS, and the promoter sequence of the
human FASN gene exhibits high similarity with that of the rat
(29) (Fig. 2B). With the ChoRE and core
promoter locus as experimental groups, exon2 and the −2 kb locus
were selected as controls.
Compared with 0 h exposure to high glucose,
significant increases in H3 and H4 acetylation within the ChoRE,
promoter and exon2 regions were observed in the HepG2 cells
following exposure to high glucose for 12 h, and in the L02 cells
following exposure to high glucose for 8 h (Fig. 2C and D). Higher degrees of
phosphorylated H3S10 within the ChoRE, promoter and exon2 regions
were also observed in the HepG2 and L02 cells, compared with that
at 0 h (Fig. 2C and D). Depending
on the site and the degree of histone methylation, modifications
result in either gene activation or repression; increased H3K4
trimethylation was observed in the ChoRE, promoter and exon2
regions of the HepG2 and L02 cells following exposure to high
glucose, compared with that at 0 h (Fig. 2C and D). By contrast,
trimethylated H3K9 and H4K20 are repressive marks, and they
function through different mechanisms (30). Significant decreases in H3K9 and
H4K20 trimethylation were observed in the ChoRE, promoter and exon2
regions of the HepG2 and L02 cells following exposure to high
glucose, compared with that at 0 h (Fig. 2C and D).
When high glucose was removed, as previously
mentioned, for 1 h, it was observed that the increasing ranges of
H3 and H4 acetylation, H3S10 phosphorylation, and H3K4
trimethylation were decreased, whereas H3K9 trimethylation and
H4K20 trimethylation were increased in the FASN promoter regions of
the HepG2 and L02 cells, compared with the ranges at 0 h high
glucose removal (Fig. 2E and F).
Under high glucose treatment and following its removal, no changes
in epigenetic marks were detected within −2 kb in either
hepatocytes, compared with the 0 h controls. According to these
results, the levels of H3 and H4 acetylation, H3S10 phosphorylation
and H3K4 trimethylation were increased, but the trimethylation
levels of H3K9 and H4K20 were reduced in FASN under a high glucose
stimulus, which resulted in activation of the transcription of
FASN. Following the removal of glucose, the increasing levels of H3
and H4 acetylation, H3S10 phosphorylation and H3K4 trimethylation
levels all declined, whereas the levels of H3K9 and H4K20
trimethylation increased in FASN, which induced a decrease in the
increased transcription of FASN.
Involvement of ChREBP in histone
modifications at the FASN promoter regions
Subsequently, the present study attempted to
elucidate the mechanisms underlying histone modifications at the
FASN promoter regions in the hepatocyte lines. ChREBP is the main
molecule activated by the glucose signaling pathway and modulates
the induction of FASN by directly binding to ChoRE found in the
gene promoter. To determine the effect of ChREBP on the epigenetic
modifications at the FASN promoter regions of the hepatocyte lines,
the protein expression of ChREBP with high glucose incubation or
its removal was evaluated using western blot analysis. As shown in
Fig. 3A, the protein expression
of ChREBP was distinctly enhanced with the increase in high glucose
incubation time, which was accompanied by elevated intracellular
lipogenesis, as mentioned above. However, when high glucose was
removed for 48 h in the HepG2 and L02 cells, the protein level of
ChREBP was markedly decreased compared with that at 0 h glucose
removal, although no significant difference was found between 24
and 0 h glucose removal (Fig.
3B).
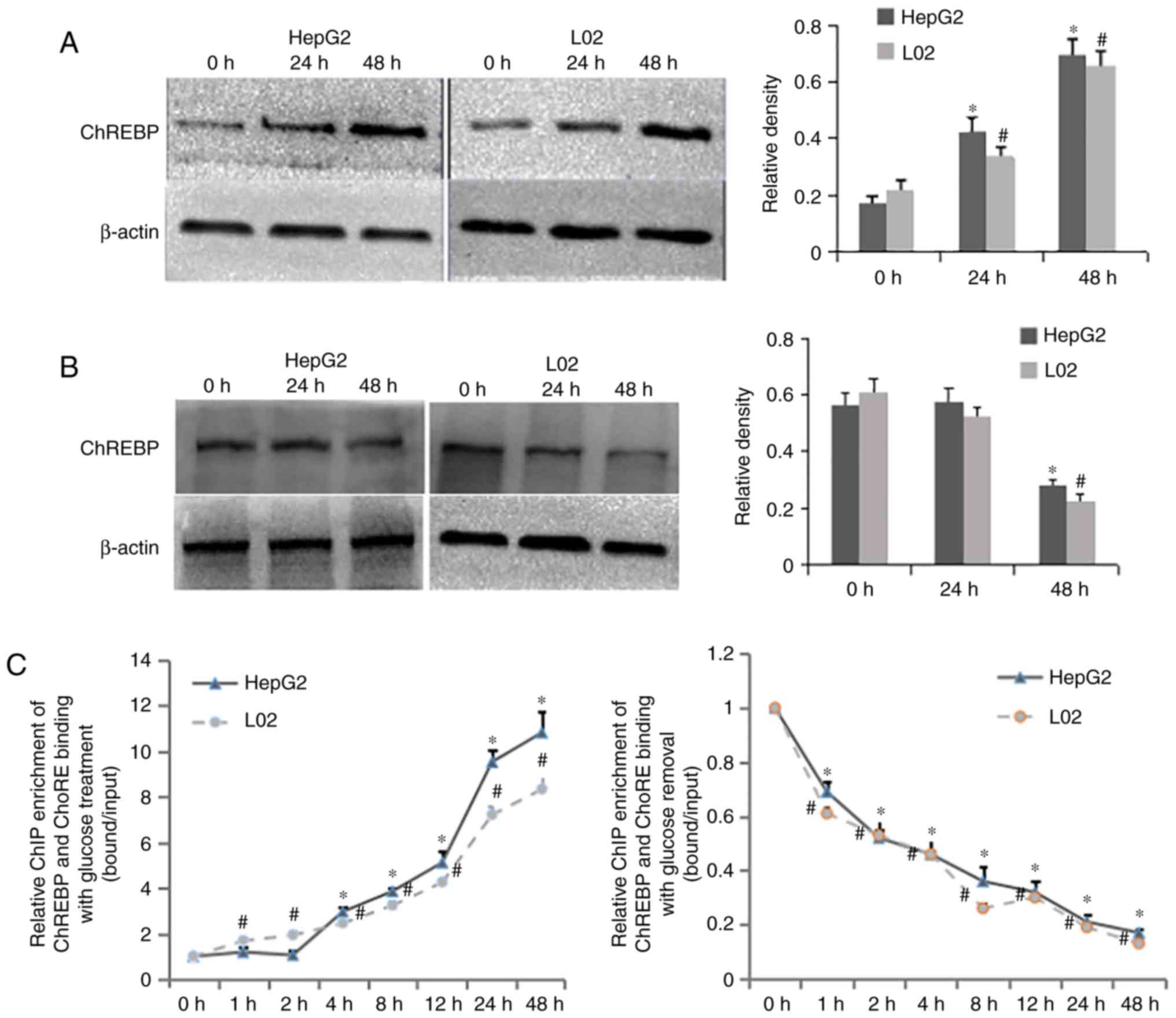 | Figure 3Involvement of ChREBP in histone
modifications at the FASN promoter regions. Protein levels of
ChREBP in HepG2 and L02 cells (A) with glucose treatment or (B)
following glucose removal were determined using western blot
analysis; β-actin was used as a loading control;
*P<0.05, vs. 0 h in HepG2; #P<0.05, vs.
0 h in L02. (C) ChIP-qPCR assays were used to examine the time
spectrum of ChREBP-ChoRE binding in HepG2 and L02 cells with
glucose treatment or removal; values at time 0 h were set as 1;
*P<0.05, vs. 0 h in HepG2; #P<0.05, vs.
0 h in L02. (D) Western blot analysis of protein levels of ChREBP
and FASN, and (E) Oil Red O staining of fat droplets in HepG2 and
L02 cells with siRNA-ChREBP plasmid transfection. (F) Western blot
analysis of protein levels of ChREBP and FASN, and (G) Oil Red O
staining of fat droplets in HepG2 and L02 cells with
ChREBP-overexpression plasmid transfection; β-actin was used as a
loading control; *P<0.05, vs. siRNA-ChREBP or
ChREBP-overexpression group in HepG2; #P<0.05, vs.
siRNA-ChREBP or ChREBP-overexpression group in L02. Magnification,
×200. (H) ChIP assays were used to investigate ChREBP-ChoRE
binding; values of normal controls were set as 1,
*P<0.05, vs. transfection group in HepG2;
#P<0.05, vs. transfection group in L02. Acetylated H3
and H4, trimethylated H3K4, H3K9 and H4K20, and phosphorylated
H3S10 occupancy in the FASN ChoRE, promoter, exon2, and −2 kb
regions of (I) HepG2 and (J) L02 cells with transfection of
siRNA-ChREBP and exposure to high glucose or (K) HepG2 and (L) L02
cells with transfection of ChREBP-overexpression, values of normal
controls were set as 1, *P<0.05, vs. transfection
group in HEepG2 or L02. Data are expressed as the mean ± standard
deviation from at least three independent experiments. FASN histone
hypo-acetylation, H3S10 hypo-phosphorylation, H3K4
hypo-trimethylation, and H3K9 and H4K20 hyper-trimethylation at the
ChoRE, promoter and exon2 regions were observed, ChREBP-ChoRE
binding, histone acetylation, phosphorylation of H3S10, and
trimethylation of H3K4 of the FASN promoter regions were increased,
and trimethylation of H3K9 and H4K20 were decreased in the
transfection group, compared with the three control groups. FASN;
fatty acid synthase; ChoRE, ChoRE, carbohydrate response element;
ChREBP, ChoRE binding protein; TSS, transcription start site; ChIP,
chromatin immunoprecipitation; siRNA, small interfering RNA. |
ChIP assays were also performed to examine the time
spectrum of the ChREBP-ChoRE binding. When stimulated with high
glucose, the ChREBP-ChoRE binding in the HepG2 and L02 cell lines
increased almost linearly (Fig.
3C), as did the mRNA expression of FASN. Following high glucose
removal, it was found that the increasing ChREBP-ChoRE binding
decreased linearly (Fig. 3C), and
this finding was consistent with a change in the mRNA expression
levels of FASN. These findings demonstrated that glucose affected
the transcription of FASN by regulating the expression and
transcriptional activity of ChREBP. ChREBP-ChoRE binding and the
expression of FASN also had a positive association.
The present study also investigated the functional
role of ChREBP in the establishment of histone modifications at the
FASN promoter regions by specifically knocking down or
overexpressing endogenous ChREBP in the HepG2 and L02 cells.
Following 48 h transfection with siRNA-ChREBP, the protein
expression levels of ChREBP and FASN were reduced in the HepG2 and
L02 cells, compared with levels in the normal group (Fig. 3D). Furthermore, Oil Red O staining
was used to examine the fat droplet levels in the three groups,
with no difference found between them (Fig. 3E). By contrast, the protein levels
of ChREBP and FASN were markedly increased in the HepG2 and L02
cells compared with those in the normal group following 48-h
transfection in the ChREBP-overexpression group (Fig. 3F). Using Oil Red O staining, it
appeared that the degree of lipid accumulation was enhanced in the
ChREBP-overexpression group, compared with that in the normal
group, which was consistent with the protein expression of ChREBP
and FASN between the two groups (Fig.
3G). The effect of the downregulation of ChREBP on histone
modifications of FASN was then determined, and the HepG2 or L02
cells were divided into four groups, including the normal control
(cultured in medium), positive control (exposed to high glucose for
12 or 8 h), transfection group (exposed to high glucose for 12 or 8
h following 48-h transfection with the siRNA-ChREBP plasmid) and
negative control (exposed to high glucose for 12 or 8 h following
48-h transfection with the GV102-scramble plasmid). The
ChREBP-ChoRE binding of FASN was weaker in the transfection group,
compared with that in the positive and negative controls in the two
cell lines (Fig. 3H). As shown in
Fig. 3I and J, FASN histone
hypo-acetylation, H3S10 hypo-phosphorylation, H3K4
hypo-trimethylation, and H3K9 and H4K20 hyper-trimethylation at the
ChoRE, promoter and exon2 regions were observed in the transfection
group, compared with the positive and negative controls.
The present study also examined the effect of the
upregulation of ChREBP on histone modifications of FASN, with
hepatocytes also divided into four groups: Normal control (cultured
in medium); positive control (exposed to high glucose for 48 h);
transfection group (transfected with the ChREBP-overexpression
plasmid for 48 h); and the negative control (transfected with the
scramble plasmid for 48 h). Concomitantly, the ChREBP-ChoRE
binding, histone acetylation, phosphorylation of H3S10 and
trimethylation of H3K4 in the FASN promoter regions were increased
in the transfection group, compared with the three control groups
(Fig. 3H, K and L). By contrast,
H3K9 and H4K20 trimethylation were decreased in the transfection
group, compared with the three control groups (Fig. 3I and J). Regardless of whether
transfection occurred with the siRNA-ChREBP or
ChREBP-overexpression plasmid, there was no change in the
epigenetic marks at −2 kb among the four groups. It was concluded
from the above results that ChREBP facilitated the acetylation of
H3 and H4, trimethylation of H3K4 and phosphorylation of H3S10, and
inhibited the trimethylation of H3K9 and H4K20 in the FASN promoter
regions. This facilitated FASN ChREBP-ChoRE binding and regulated
the transcription of FASN.
Hepatocellular steatosis is rescued by
inhibiting histone acetylation
On observing the increase in histone acetylation of
the FASN promoter regions in HepG2 and L02 cells under high glucose
stimulation, the present study investigated whether this change
affected the expression of FASN and hepatic lipid accumulation. The
hepatocytes were divided into three groups: Normal control group
(cultured in medium); glucose+garcinol group (treated with a high
glucose/garcinol cocktail for 24 h); and glucose group (treated
with high glucose for 24 h). The optimal garcinol concentration for
HepG2 and L02 cells was 5 µM as previously described
(27). The high glucose/garcinol
cocktail caused histone hypo-acetylation and decreased the
ChREBP-ChoRE binding of FASN compared with the glucose group;
however, this was increased compared with the normal group
(Fig. 4A and B). Total RNA and
protein were recovered for RT-qPCR and western blot analyses,
respectively. As shown in Fig. 4C and
D, the high glucose/garcinol cocktail failed to promote the
protein and mRNA expression of FASN relative to the glucose group;
however, it did increase the expression compared with the normal
group in the two hepatocyte lines. The results of the TG content
assay and Oil Red O staining were consistent with the expression of
FASN (Fig. 4E and F). Finally,
the protein expression of ChREBP in each group was examined.
Western blot analysis revealed that the expression levels of ChREBP
in the glucose+garcinol and glucose groups were almost the same but
were higher compared with that in the normal group (Fig. 4G). Taken together, these findings
that the inhibition of FASN histone acetylation led to no change in
the induction of ChREBP but led to a decrease in ChREBP-ChoRE
binding, and the subsequent alleviation of TG biosynthesis and
lipid accumulation in the HepG2 and L02 cells in response to high
glucose stimulation.
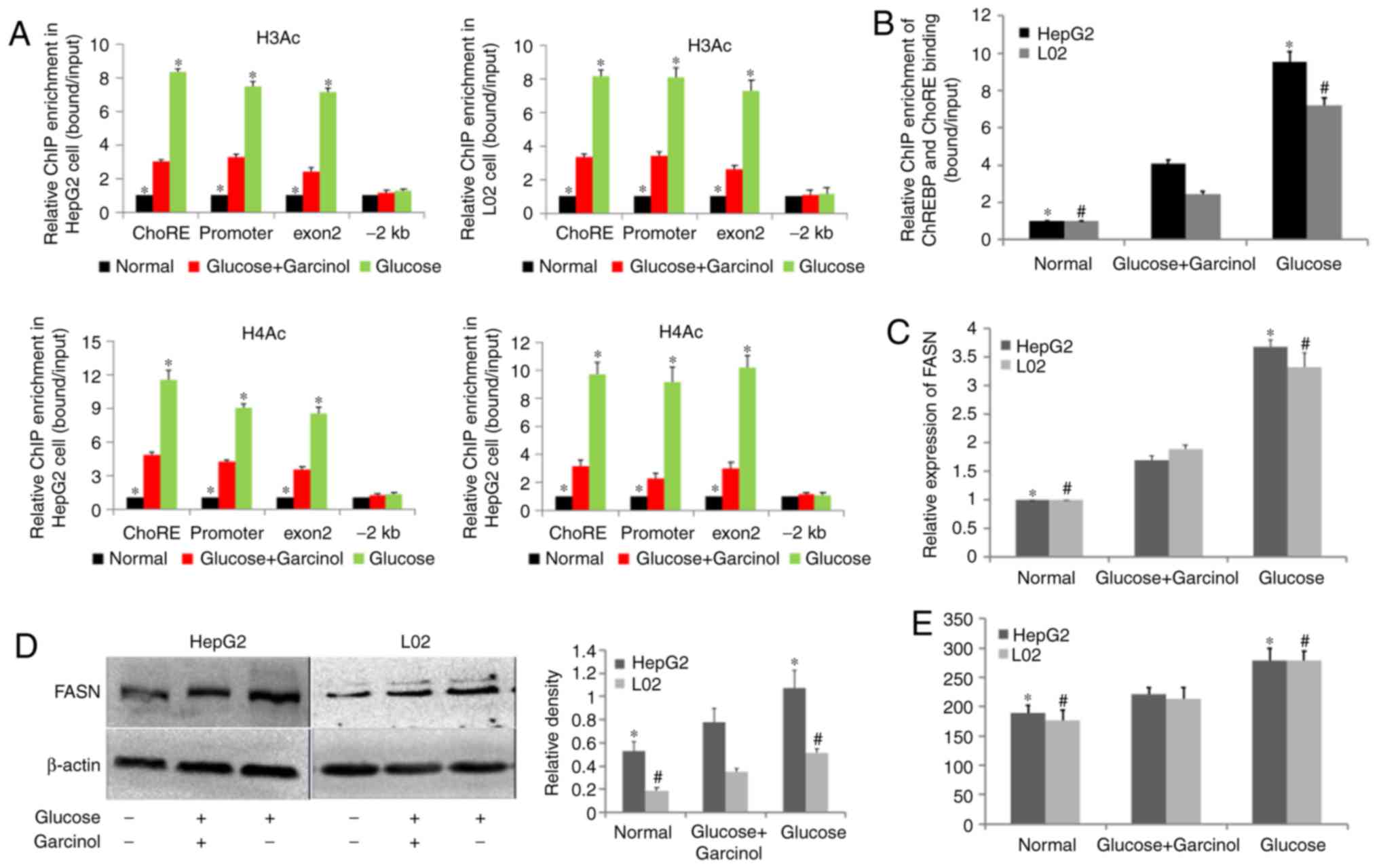 | Figure 4Hepatocellular steatosis is rescued
by inhibiting histone acetylation. Chromatin immunoprecipitation
assays were performed to measure (A) acetylated H3 and H4 occupancy
in the FASN ChoRE, promoter, exon2, and −2 kb regions, and (B)
ChREBP-ChoRE binding; values of the normal groups were set as 1.
Total protein and RNA were recovered for (C) western blot analysis
and (D) reverse transcription-quantitative polymerase chain
reaction analysis of FASN, respectively; β-actin was used as a
loading control. (E) TG contents were measured using a TG assay kit
and (F) fat droplets were evaluated using Oil Red O staining.
Magnification, ×200. (G) Protein levels of ChREBP were determined
using western blot analysis; β-actin was used as a loading control.
*P<0.05, vs. glucose+garcinol group in HepG2;
#P<0.05, vs. glucose+garcinol group in L02. Data are
expressed as the mean ± standard deviation from at least three
independent experiments. FASN; fatty acid synthase; TG,
triglyceride; ChoRE, carbohydrate response element; ChREBP, ChoRE
binding protein. |
Discussion
In the last decade, as the diets and lifestyles of
the Asian population have become rapidly westernized, NAFLD has
become one of the most common chronic liver diseases in Asia
(31). According to previous
studies, diabetes is a high-risk factor for NAFLD (3,4,32).
Therefore, the investigation of glucolipid metabolic disorder is
particularly important. In the well-recognized ‘double-hit’ theory,
aberrant lipid deposition in hepatocytes, which is also known as
hepatic steatosis, is considered to be the pathological basis of
NAFLD (15,16,33). FASN, as the key enzyme of de
novo lipogenesis, is an important downstream target gene for
the conversion of glucose to lipid. In addition, its transcription
is closely associated with the development of NAFLD (34–36). In the present study, a significant
enhancement of lipogenesis induced by high glucose was observed in
HepG2 and L02 cells, as measured by the expression of FASN and
lipid content. These pathological changes promote hepatic steatosis
and are consistent with a number of previous reports (9,11,18).
Gene expression can be regulated by any of the
following control steps: i) chromatin structure; ii) initiation of
transcription; iii) processing of the transcript; iv) transport to
the cytoplasm; v) translation of mRNA; and vi) stability of protein
activity. Among these steps, the chromatin structure is decisive in
selective gene expression. Epigenetic phenomena regulate chromatin
structure modifications and the initiation of transcription in a
manner that alters the availability of genes to the transcription
factors required for their expression (37). As structural proteins of
eukaryotic chromatin, histones combine with DNA to form the
nucleosome, which serves as the basic structural unit of chromatin.
Covalent modifications, including acetylation, methylation and
phosphorylation, can occur on the amino acid residues in the
N-terminal of histones, which are catalyzed by enzymes to alter the
chromatin structure, which result in the activation or suppression
of gene transcription. Dysregulation of the histone modification of
a critical gene has been known to initiate cancer development and
contribute to cancer progression (38–41). However, investigations of histone
modifications on NAFLD and associated metabolic syndromes remain in
their infancy. The present study revealed for the first time, to
the best of our knowledge, the association between histone
modifications and the expression of FASN in the presence of high
glucose and following its withdrawal. It was found that H3 and H4
hyper-acetylation, H3K4 hyper-trimethylation and H3S10
hyper-phosphorylation within the FASN promoter regions were linked
to the activation of FASN and lipogenesis in HepG2 and L02 cells.
By contrast, H3K9 and H4K20 hyper-trimethylation were negatively
associated with the transcription of FASN in the HepG2 and L02
cells. These findings are consistent with previous studies on
histone modifications: Histone acetylation, including H2A, H2B, H3
and H4, is mainly involved in the activation of gene transcription.
By contrast, histone methylation and phosphorylation are involved
in the activation and suppression of gene transcription according
to modification on different residue sites of amino acids. For
example, the methylation of H3K4 and phosphorylation of H3S10 are
involved in gene activation, whereas the methylation of H3K9 and
H4K20 leads to the suppression of gene transcription (25,42–44). The phosphorylation of different
histones and amino sites is crucial in gene transcription, the
condensation status of the chromatin and DNA repair; it is also
involved in the regulation of vital activities, including cell
growth and division, together with histone acetylation and
methylation. At present, reports on histone phosphorylation have
mainly focused on serine 10 of H3. In 1991, Hazzalin and Mahadevan
(45) found, for the first time,
that the phosphorylation of H3 was correlated with the activity of
gene transcription. The phosphorylation of H3 generated under
external stimuli is rapid and transient and can affect
phosphorylated H3 in a different manner from that detected in
normal cell division. In addition, the level of this H3
phosphorylation is positively correlated with the level of
acetylation. Numerous studies have confirmed that the
phosphorylation of H3Sl0 induced by various external stimuli
involves multiple protein kinase signaling pathways, including
mitogen and stress-activated kinase 1, ribosomal protein S6 kinase
2, inhibitor of nuclear factor-κB kinase-α, cAMP-dependent protein
kinase C, Fyn and PIM1 (46–48). These kinases are involved in
regulating the phosphorylation of H3S10 under different stimuli to
induce specific gene transcription.
ChREBP is expressed at high levels in metabolically
active organs, including the liver, fat, kidney, small intestine
and muscles. Xylulose-5-phosphate, a metabolic product derived from
the pentose phosphate signaling pathway, causes the
dephosphorylation of ChREBP to promote its nuclear translocation,
so that it can be combined with Mlx to form a heterodimer, which
finally binds to ChoRE in the promoter region of a target gene to
exert the transcriptional activity of ChREBP (5). Global or liver-specific deletion of
ChREBP has been found to markedly ameliorate fatty liver disease
and improve overall glucose tolerance and insulin sensitivity in
ob/ob mice, potentially via a decrease in de novo
lipogenesis (13). The
overexpression of ChREBP in the liver was reported to increase
hepatic steatosis associated with an increased expression of genes
regulating fatty acid and TG synthesis in the liver (5). Therefore, ChREBP is a key
transcription factor in glucolipid metabolism (7,10).
Prior to/when ChREBP initiates the transcription of a target gene,
it requires accompanied changes in the local chromatin structure of
the gene. ChREBP has been shown to bind in a glucose- and
cAMP-dependent manner to ChoREs located in the promoter/enhancer
regions of hepatic lipogenic genes, including FASN (49). To further determine the epigenetic
mechanisms underlying histone modifications at hepatic FASN
promoter regions under high glucose, the present study examined the
involvement of ChREBP in FASN histone modifications. The
corresponding results indicated that the expression level of ChREBP
and the occupancy of ChREBP-ChoRE binding at the FASN promoter were
positively associated with the expression of FASN during the
stimulation of high glucose and following its removal. In addition,
the inhibition of ChREBP caused a decline in the levels of
ChREBP-ChoRE binding, acetylation of H3 and H4, trimethylation of
H3K4 and phosphorylation of H3S10, whereas an increase in the
trimethylation levels of H3K9 and H4K20 in the FASN promoter
regions occurred, leading to the suppression of FASN. In addition,
when ChREBP was overexpressed, the levels of ChREBP-ChoRE binding,
histone acetylation, trimethylation of H3K4 and phosphorylation of
H3S10 increased, whereas the modification in the trimethylation
levels of H3K9 and H4K20 decreased within the FASN promoter
regions, resulting in high expression levels of FASN, and
contributing to increased hepatocellular lipogenesis and hepatic
steatosis. Therefore, the present study found that histone
modifications in FASN promoter regions were closely associated with
ChREBP. Studies have shown that ChREBP accurately regulates the
expression of glucose-sensitive genes by forming a complex with
transcription factor p300, coactivator CREBP binding protein (CBP),
and RNA polymerase II, and being heterodimerized with Mlx (50,51). However, the components of this
complex remain to be fully elucidated. p300 and CBP are proteins
functioning as histone-modifying enzymes (52) and are key to open the structure of
chromatin. They affect the affinity between histones and DNA by
modifying the histone residues, which exposes ChoRE in the promoter
of target genes. The heterodimer ChREBP/Mlx of the transcription
complex identifies and specifically binds to exposed ChoRE to
promote the modification of histone residues by histone-modifying
enzymes, and to disassemble the chromatin structure of target
genes, eventually leading to the activation of gene transcription.
The present study confirmed that glucose affected the transcription
of FASN by regulating the expression and transcriptional activity
of ChREBP. Furthermore, ChREBP induced FASN ChREBP-ChoRE binding to
accelerate the transcription of FASN by facilitating the
acetylation of H3 and H4, trimethylation of H3K4 and
phosphorylation of H3S10, and inhibiting the trimethylation of H3K9
and H4K20 in the FASN promoter regions.
As histone modifications are reversible, novel
therapeutic strategies to modulate epigenetic aberrancies are being
investigated extensively (38,53). Histone acetylation is catalyzed by
HAT directly at lysine residues of the N-terminal of histone,
enabling local chromatin to be dissected from the histone octamer
to facilitate the binding between various transcription factors and
DNA regulatory factors, resulting in gene transcription. HDAC
decreases the acetylation level of histone to make the association
between DNA and the histone octamer closer to further intensify
local chromatin contraction and to also inhibit the binding between
transcription factors and DNA regulatory factors, which suppress
gene transcription. The altered expression and activity of specific
histone acetylation-modifying enzymes have been reported to affect
phenotypic gene expression in metabolic disease (53). It is known that p300 and CBP are
important components of the HAT family members, and garcinol, an
organic matter extracted from Indian Garcinia, has the
capability to penetrate through cytomembranes to prevent the
activity of p300/CBP- and p300/CBP-associated factors (52,54,55). Therefore, the present study used
garcinol, as an HAT inhibitor, to further observe the latent impact
of histone acetylation on FASN transcription regulated by ChREBP. A
previous study showed that glucose, as an endogenous ligand of LXR,
upregulated the expression of ChREBP by activating LXR. Glucose was
metabolized to xylulose-5-phosphate, which led the
dephosphorylation of ChREBP to increase the transcriptional
activity of ChREBP, by promoting its translocation from the
cytoplasm to the nucleus and its binding to ChoRE in promoters
(56). In addition, it has been
shown that glucose-activated p300 acetylated ChREBP on Lys672 and
increased its transcriptional activity by enhancing its nuclear
translocation and recruitment to its target gene promoters, which
resulted in increased hepatic lipid synthesis (55). Therefore, the transcriptional
activity of ChREBP requires the nuclear translocation and the
specific binding of ChREBP-ChoRE in the promoter region of a target
gene. The present study found that, when L02 and HepG2 cells were
stimulated by a high glucose/garcinol cocktail, garcinol partially
antagonized the positive regulation of glucose on the
transcriptional activity of ChREBP by inhibiting the effects of
p300 and CBP. Therefore, the high glucose/garcinol cocktail reduced
ChREBP-ChoRE binding and the acetylation of H3 and H4, and caused
the downregulated expression of FASN, compared with that following
high glucose treatment alone, although the values from the cocktail
group were higher than those from the normal group. As garcinol was
unable to interfere with the regulation of glucose on the
expression of ChREBP, no significant difference was found in the
protein level of ChREBP between the cocktail and the high glucose
groups, although both were increased compared with that in the
normal group. It was concluded that the biological function of
ChREBP protein was dependent on its expression level and its
transcriptional activity. In conclusion, these findings indicated
that the inhibition of histone acetylation under high glucose
treatment failed to suppress the expression of ChREBP, however, it
inhibited the transcriptional activation of ChREBP by reducing the
specific binding of ChREBP-ChoRE in the FASN promoter region, which
resulted in the transcriptional repression of FASN and alleviation
of hepatocyte fatty degeneration. This process demonstrated that
the transcriptional activity of ChREBP is required for the
regulation of FASN transcription as histone acetylation leads to
conformational transformation in FASN chromatin, indicating that
histone acetylation is a crucial mechanism involved in the
transcription of FASN modulated by ChREBP.
In conclusion, the results of the present study
suggested that ChREBP-mediated histone modifications at the FASN
promoter regions in HepG2 and L02 cells were manifested by H3 and
H4 hyper-acetylation, H3K4 hyper-trimethylation, H3S10
hyper-phosphorylation and H3K9 and H4K20 hypo-trimethylation, which
resulted in an active chromatin structure increasing the
transcription of FASN in response to high glucose. Consistent with
this finding, the hepatocytes showed enhanced potential for
lipogenesis in response to the FASN histone modifications
described. Consequently, histone acetylation was provided as a
novel theoretical basis for glucolipid metabolism disorder, and for
the pathogenesis and therapeutic targets of NAFLD.
Acknowledgments
Not applicable.
Abbreviations:
|
NAFLD
|
non-alcoholic fatty liver disease
|
|
ChREBP
|
carbohydrate responsive element
binding protein
|
|
FASN
|
fatty acid synthase
|
|
HAT
|
histone acetyltransferase
|
|
HDAC
|
histone deacetylase
|
|
ChoRE
|
carbohydrate response element
|
|
ChIP
|
chromatin immunoprecipitation
|
Funding
The present study was funded by the National Natural
Science Foundation of China (grant nos. 81270494 and 81500443).
Availability of data and material
The analyzed data sets generated during the study
are available from the corresponding author on reasonable
request.
Authors’ contributions
CC, HY and WS designed the experiments. CC, GH and
XY performed the experiments. CC, XD and YZ analyzed the results.
CC wrote the manuscript. All the authors read and approved the
final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors confirm that they have no competing
interests.
References
|
1
|
Farrell GC and Larter CZ: Nonalcoholic
fatty liver disease: From steatosis to cirrhosis. Hepatology.
43:S99–S112. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fan JG and Farrell GC: Epidemiology of
non-alcoholic fatty liver disease in China. J Hepatol. 50:204–210.
2009. View Article : Google Scholar
|
|
3
|
Ajmera VH, Gunderson EP, VanWagner LB,
Lewis CE, Carr JJ and Terrault NA: Gestational diabetes mellitus is
strongly associated with non-alcoholic fatty liver disease. Am J
Gastroenterol. 111:658–664. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lomonaco R, Bril F, Portillo-Sanchez P,
Ortiz-Lopez C, Orsak B, Biernacki D, Lo M, Suman A, Weber MH and
Cusi K: Metabolic impact of nonalcoholic steatohepatitis in obese
patients with type 2 diabetes. Diabetes Care. 39:632–638. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jeong YS, Kim D, Lee YS, Kim HJ, Han JY,
Im SS, Chong HK, Kwon JK, Cho YH, Kim WK, et al: Integrated
expression profiling and genome-wide analysis of ChREBP targets
reveals the dual role for ChREBP in glucose-regulated gene
expression. PLoS One. 6:e225442011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shiota M and Magnuson MA: Hepatic glucose
sensing: Does flux matter. J Clin Invest. 118:841–844. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xu X, So JS, Park JG and Lee AH:
Transcriptional control of hepatic lipid metabolism by SREBP and
ChREBP. Semin Liver Dis. 33:301–311. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sakiyama H, Wynn RM, Lee WR, Fukasawa M,
Mizuguchi H, Gardner KH, Repa JJ and Uyeda K: Regulation of nuclear
import/export of carbohydrate response element-binding protein
(ChREBP): Interaction of an alpha-helix of ChREBP with the 14-3-3
proteins and regulation by phosphorylation. J Biol Chem.
283:24899–24908. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Uyeda K and Repa JJ: Carbohydrate response
element binding protein, ChREBP, atranscription factor coupling
hepatic glucose utilization and lipid synthesis. Cell Metab.
4:107–110. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Benhamed F, Filhoulaud G, Caron S,
Lefebvre P, Staels B and Postic C: O-GlcNAcylation links ChREBP and
FXR to glucose-sensing. Front Endocrinol (Lausanne). 5:2302015.
|
|
11
|
Poupeau A and Postic C: Cross-regulation
of hepatic glucose metabolism via ChREBP and nuclear receptors.
Biochim Biophys Acta. 1812.995–1006. 2011.
|
|
12
|
Denechaud PD, Bossard P, Lobaccaro JM,
Millatt L, Staels B, Girard J and Postic C: ChREBP, but not LXRs,
is required for the induction of glucose-regulated genes in mouse
liver. J Clin Invest. 118:956–964. 2008.PubMed/NCBI
|
|
13
|
Denechaud PD, Dentin R, Girard J and
Postic C: Role of ChREBP in hepatic steatosis and insulin
resistance. FEBS Lett. 582:68–73. 2008. View Article : Google Scholar
|
|
14
|
Iizuka K: Recent progress on the role of
ChREBP in glucose and lipid metabolism. Endocr J. 60:543–555. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sahini N and Borlak J: Recent insights
into the molecular pathophysiology of lipid droplet formation in
hepatocytes. Prog Lipid Res. 54:86–112. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kawano Y and Cohen DE: Mechanisms of
hepatic triglyceride accumulation innon-alcoholic fatty liver
disease. J Gastroenterol. 48:434–441. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Karagianni P and Talianidis I:
Transcription factor networks regulating hepatic fatty acid
metabolism. Biochim Biophys Acta. 1851.2–8. 2015.
|
|
18
|
Wang Y, Viscarra J, Kim SJ and Sul HS:
Transcriptional regulation of hepatic lipogenesis. Nat Rev Mol Cell
Biol. 16:678–689. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ozkul Y and Galderisi U: The impact of
epigenetics on mesenchymal stem cell biology. J Cell Physiol.
231:2393–2401. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zoghbi HY and Beaudet AL: Epigenetics and
human disease. Cold Spring Harb Perspect Biol. 8:a0194972016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Martínez JA, Milagro FI, Claycombe KJ and
Schalinske KL: Epigenetics in adipose tissue, obesity, weight loss,
and diabetes. Adv Nutr. 5:71–81. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Stel J and Legler J: The role of
epigenetics in the latent effects of early life exposure to
obesogenic endocrine disrupting chemicals. Endocrinology.
156:3466–3472. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Youngson NA and Morris MJ: What obesity
research tells us about epigenetic mechanisms. Philos Trans R Soc
Lond B Biol Sci. 368:201103372013. View Article : Google Scholar :
|
|
24
|
Öst A and Pospisilik JA: Epigenetic
modulation of metabolic decisions. Curr Opin Cell Biol. 33:88–94.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lawrence M, Daujat S and Schneider R:
Lateral thinking: How histone modifications regulate gene
expression. Trends Genet. 32:42–56. 2016. View Article : Google Scholar
|
|
26
|
Harr JC, Gonzalez-Sandoval A and Gasser
SM: Histones and histone modifications in perinuclear chromatin
anchoring: From yeast to man. EMBO Rep. 17:139–155. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yu H, Wu J, Yang M, Guo J, Zheng L, Peng
M, Zhang Q, Xiang Y, Cao J and Shen W: Involvement of liver X
receptor alpha in histone modifications across the target fatty
acid synthase gene. Lipids. 47:249–257. 2012. View Article : Google Scholar
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−delta delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
29
|
Rufo C, Teran-Garcia M, Nakamura MT, Koo
SH, Towle HC and Clarke SD: Involvement of a unique
carbohydrate-responsive factor in the glucose regulation of rat
liver fatty-acid synthase gene transcription. J Biol Chem.
276:21969–21975. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Shechter D, Dormann HL, Allis CD and Hake
SB: Extraction, purification and analysis of histones. Nat Protoc.
2:1445–1457. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fan JG, Kim SU and Wong VW: New trends on
obesity and NAFLD in Asia. J Hepatol. 67:862–873. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hazlehurst JM, Woods C, Marjot T, Cobbold
JF and Tomlinson JW: Non-alcoholic fatty liver disease and
diabetes. Metabolism. 65:1096–1108. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Day CP and James OF: Steatohepatitis: A
tale of two ‘hits’. Gastroenterology. 114:842–845. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cordero P, Gomez-Uriz AM, Campion J,
Milagro FI and Martinez JA: Dietary supplementation with methyl
donors reduces fatty liver and modifies the fatty acid synthase DNA
methylation profile in rats fed an obesogenic diet. Genes Nutr.
8:105–113. 2013. View Article : Google Scholar :
|
|
35
|
Dorn C, Riener MO, Kirovski G, Saugspier
M, Steib K, Weiss TS, Gäbele E, Kristiansen G, Hartmann A and
Hellerbrand C: Expression of fatty acid synthase in nonalcoholic
fatty liver disease. Int J Clin Exp Pathol. 3:505–514.
2010.PubMed/NCBI
|
|
36
|
Jones SF and Infante JR: Molecular
pathways: Fatty acid synthase. Clin Cancer Res. 21:5434–5438. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kirk H, Cefalu WT, Ribnicky D, Liu Z and
Eilertsen KJ: Botanicals as epigenetic modulators for mechanisms
contributing to development of metabolic syndrome. Metabolism.
57:S16–S23. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Dawson MA and Kouzarides T: Cancer
epigenetics: From mechanism to therapy. Cell. 150:12–27. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tian Y, Wong VW, Chan HL and Cheng AS:
Epigenetic regulation of hepatocellular carcinoma in non-alcoholic
fatty liver disease. Semin Cancer Biol. 23:471–482. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Gołąbek K, Strzelczyk JK, Wiczkowski A and
Michalski M: Potential use of histone deacetylase inhibitors in
cancer therapy. Contemp Oncol (Pozn). 19:436–440. 2015.
|
|
41
|
Yi X, Jiang XJ, Li XY and Jiang DS:
Histone methyltransferases: Novel targets for tumor and
developmental defects. Am J Transl Res. 7:2159–2175. 2015.
|
|
42
|
Yen CY, Huang HW, Shu CW, Hou MF, Yuan SS,
Wang HR, Chang YT, Farooqi AA, Tang JY and Chang HW: DNA
methylation, histone acetylation and methylation of epigenetic
modifications as a therapeutic approach for cancers. Cancer Lett.
373:185–192. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Su X, Wellen KE and Rabinowitz JD:
Metabolic control of methylation and acetylation. Curr Opin Chem
Biol. 30:52–60. 2016. View Article : Google Scholar :
|
|
44
|
Warsito D, Lin Y, Gnirck AC, Sehat B and
Larsson O: Nuclearly translocated insulin-like growth factor 1
receptor phosphorylates histone H3 at tyrosine 41 and induces SNAI2
expression via Brg1 chromatin remodeling protein. Oncotarget.
7:42288–42302. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hazzalin CA and Mahadevan LC: Dynamic
acetylation of all lysine4-methylated histone H3 in the mouse
nucleus: Analysis at c-fos and c-jun. PLoS Biol. 3:e3932005.
View Article : Google Scholar
|
|
46
|
Tochiki KK, Maiarú M, Norris C, Hunt SP
and Géranton SM: The mitogen and stress-activated protein kinase 1
regulates the rapid epigenetic tagging of dorsal horn neurons and
nocifensive behaviour. Pain. 157:2594–2604. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ho CM, Donovan-Banfield IZ, Tan L, Zhang
T, Gray NS and Strang BL: Inhibition of IKKα by BAY61-3606 reveals
IKKα-dependent histone H3 phosphorylation in human cytomegalovirus
infected cells. PLoS One. 11:e01503392016. View Article : Google Scholar
|
|
48
|
Zippo A, De Robertis A, Serafini R and
Oliviero S: PIM1-dependent phosphorylation of histone H3 at serine
10 is required for MYC-dependent transcriptional activation and
oncogenic transformation. Nat Cell Biol. 9:932–944. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Burke SJ, Collier JJ and Scott DK: cAMP
prevents glucose-mediated modifications of histone H3 and
recruitment of the RNA polymerase II holoenzyme to the L-PK gene
promoter. J Mol Biol. 392:578–588. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Stoeckman AK, Ma L and Towle HC: Mlx is
the functional heteromeric partner of the carbohydrate response
element-binding protein in glucose regulation of lipogenic enzyme
genes. J Biol Chem. 279:15662–15669. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Ma L, Sham YY, Walters KJ and Towle HC: A
critical role for the loop region of the basic
helix-loop-helix/leucine zipper protein Mlx in DNAbinding and
glucose-regulated transcription. Nucleic Acids Res. 35:35–44. 2007.
View Article : Google Scholar
|
|
52
|
Furdas SD, Kannan S, Sippl W and Jung M:
Small molecule inhibitors of histone acetyltransferases as
epigenetic tools and drug candidates. Arch Pharm (Weinheim).
345:7–21. 2012. View Article : Google Scholar
|
|
53
|
Knutson SK, Chyla BJ, Amann JM, Bhaskara
S, Huppert SS and Hiebert SW: Liver-specific deletion of histone
deacetylase 3 disrupts metabolic transcriptional networks. EMBO J.
27:1017–1028. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Balasubramanyam K, Altaf M, Varier RA,
Swaminathan V, Ravindran A, Sadhale PP and Kundu TK:
Polyisoprenylated benzophenone, garcinol, a natural histone
acetyltransferase inhibitor, represses chromatin transcription and
alters global gene expression. J Biol Chem. 279:33716–33726. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Bricambert J, Miranda J, Benhamed F,
Girard J, Postic C and Dentin R: Salt-inducible kinase 2 links
transcriptional coactivator p300 phosphorylation to the prevention
of ChREBP-dependent hepatic steatosis in mice. J Clin Invest.
120:4316–4331. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
He Z, Jiang T, Wang Z, Levi M and Li J:
Modulation of carbohydrate response element-binding protein gene
expression in 3T3-L1 adipocytes and rat adipose tissue. Am J
Physiol Endocrinol Metab. 287:E424–E430. 2004. View Article : Google Scholar : PubMed/NCBI
|