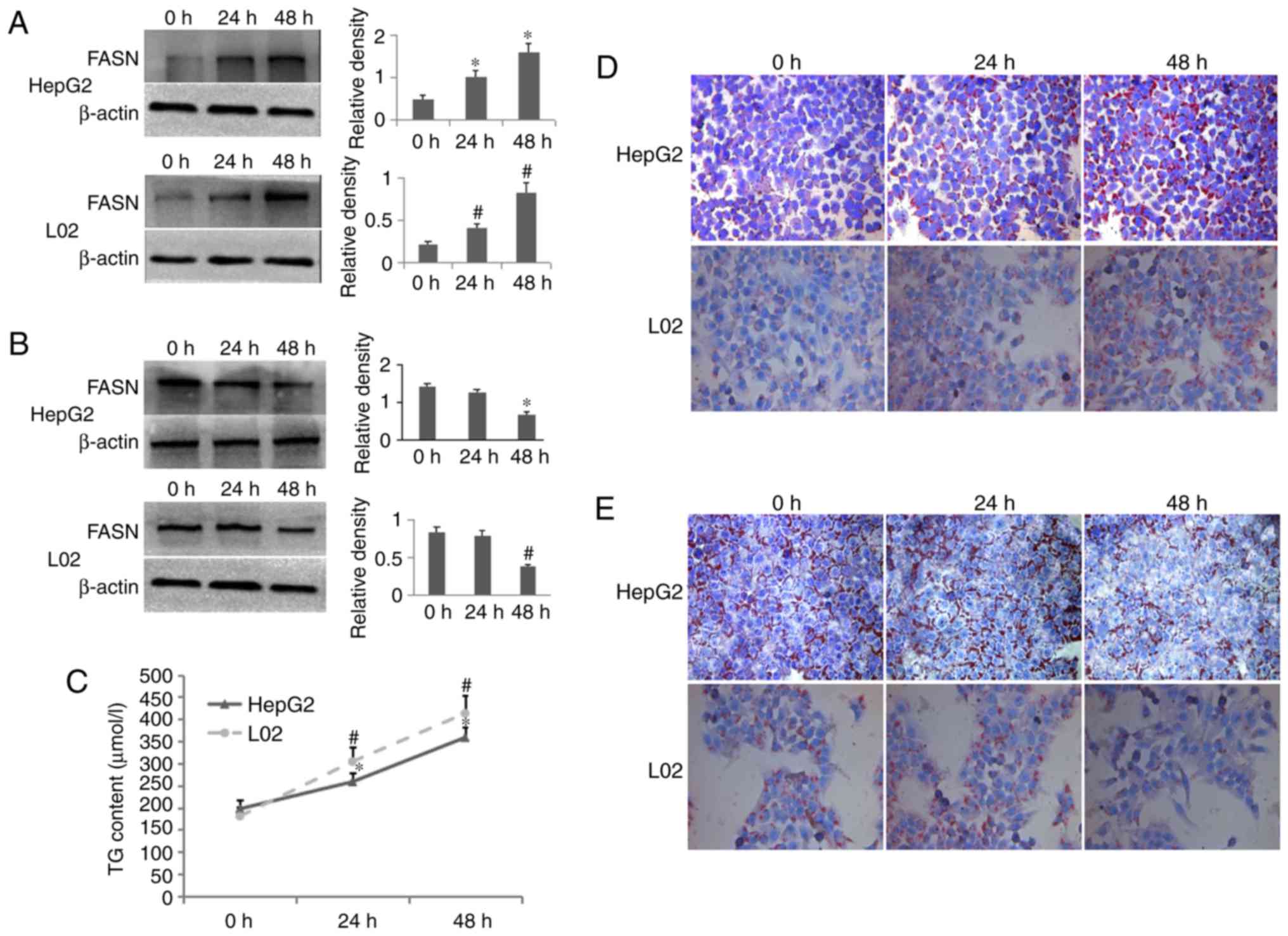
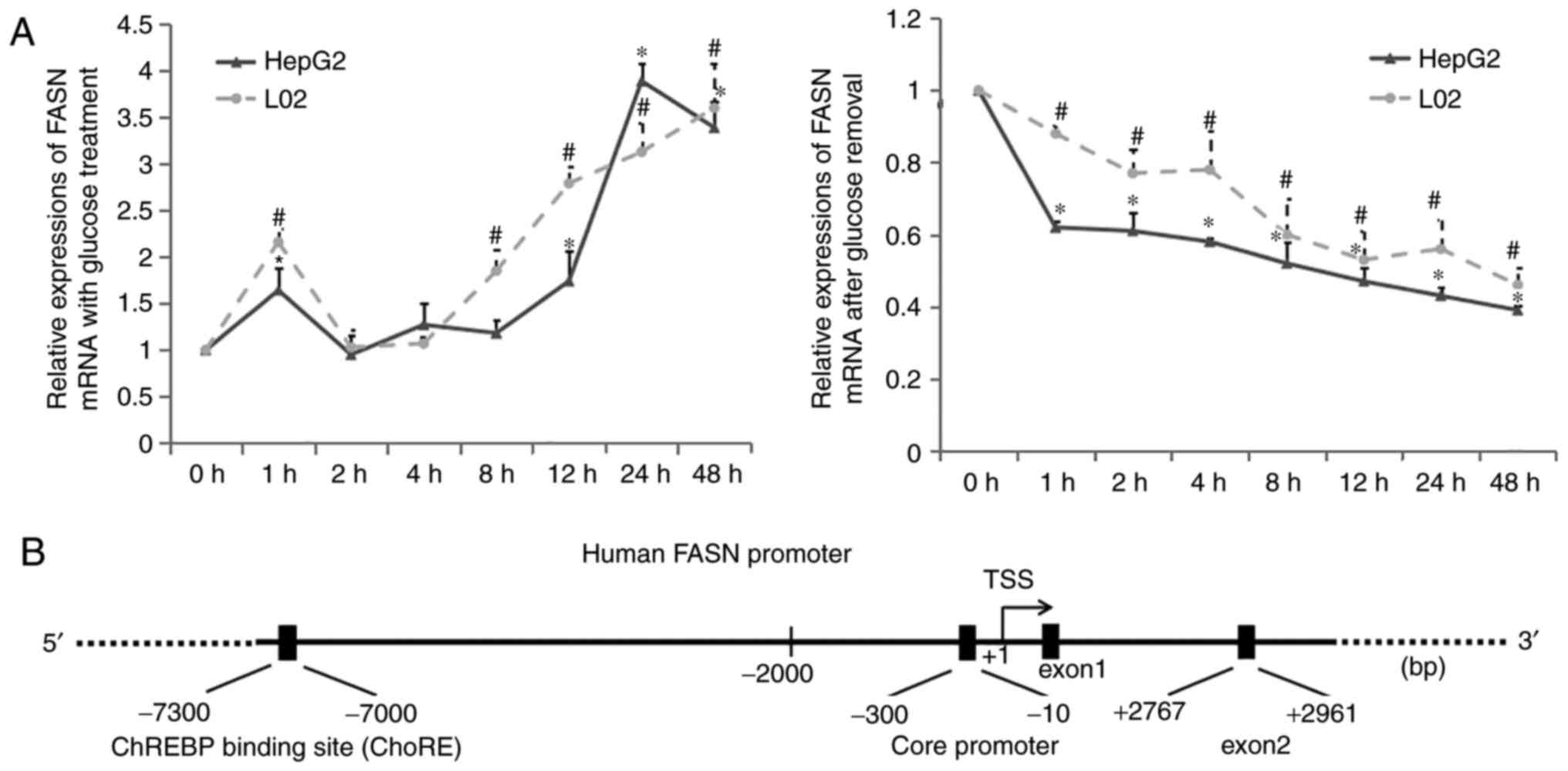
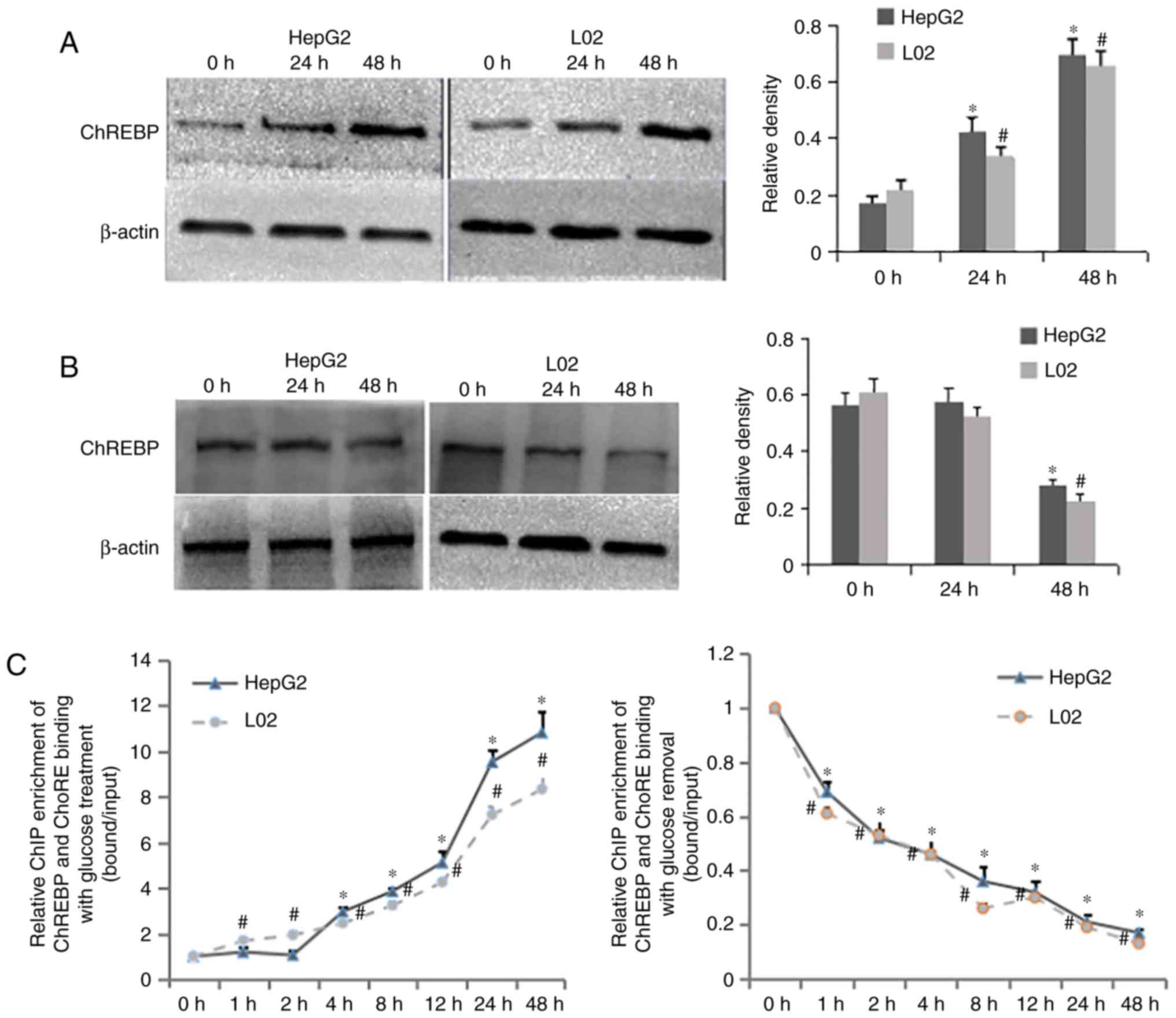
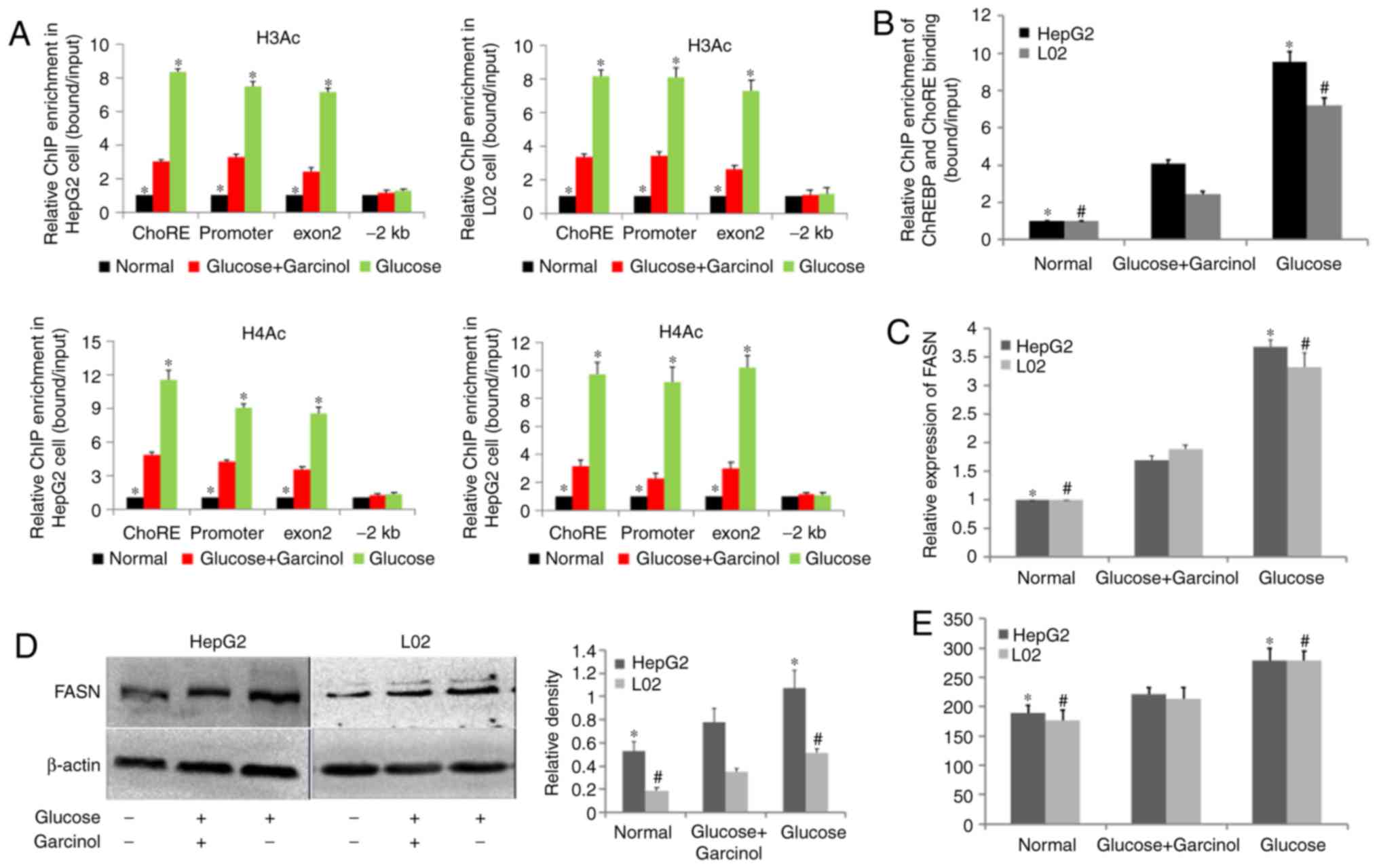
|
1
|
Farrell GC and Larter CZ: Nonalcoholic
fatty liver disease: From steatosis to cirrhosis. Hepatology.
43:S99–S112. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fan JG and Farrell GC: Epidemiology of
non-alcoholic fatty liver disease in China. J Hepatol. 50:204–210.
2009. View Article : Google Scholar
|
|
3
|
Ajmera VH, Gunderson EP, VanWagner LB,
Lewis CE, Carr JJ and Terrault NA: Gestational diabetes mellitus is
strongly associated with non-alcoholic fatty liver disease. Am J
Gastroenterol. 111:658–664. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lomonaco R, Bril F, Portillo-Sanchez P,
Ortiz-Lopez C, Orsak B, Biernacki D, Lo M, Suman A, Weber MH and
Cusi K: Metabolic impact of nonalcoholic steatohepatitis in obese
patients with type 2 diabetes. Diabetes Care. 39:632–638. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jeong YS, Kim D, Lee YS, Kim HJ, Han JY,
Im SS, Chong HK, Kwon JK, Cho YH, Kim WK, et al: Integrated
expression profiling and genome-wide analysis of ChREBP targets
reveals the dual role for ChREBP in glucose-regulated gene
expression. PLoS One. 6:e225442011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shiota M and Magnuson MA: Hepatic glucose
sensing: Does flux matter. J Clin Invest. 118:841–844. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xu X, So JS, Park JG and Lee AH:
Transcriptional control of hepatic lipid metabolism by SREBP and
ChREBP. Semin Liver Dis. 33:301–311. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sakiyama H, Wynn RM, Lee WR, Fukasawa M,
Mizuguchi H, Gardner KH, Repa JJ and Uyeda K: Regulation of nuclear
import/export of carbohydrate response element-binding protein
(ChREBP): Interaction of an alpha-helix of ChREBP with the 14-3-3
proteins and regulation by phosphorylation. J Biol Chem.
283:24899–24908. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Uyeda K and Repa JJ: Carbohydrate response
element binding protein, ChREBP, atranscription factor coupling
hepatic glucose utilization and lipid synthesis. Cell Metab.
4:107–110. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Benhamed F, Filhoulaud G, Caron S,
Lefebvre P, Staels B and Postic C: O-GlcNAcylation links ChREBP and
FXR to glucose-sensing. Front Endocrinol (Lausanne). 5:2302015.
|
|
11
|
Poupeau A and Postic C: Cross-regulation
of hepatic glucose metabolism via ChREBP and nuclear receptors.
Biochim Biophys Acta. 1812.995–1006. 2011.
|
|
12
|
Denechaud PD, Bossard P, Lobaccaro JM,
Millatt L, Staels B, Girard J and Postic C: ChREBP, but not LXRs,
is required for the induction of glucose-regulated genes in mouse
liver. J Clin Invest. 118:956–964. 2008.PubMed/NCBI
|
|
13
|
Denechaud PD, Dentin R, Girard J and
Postic C: Role of ChREBP in hepatic steatosis and insulin
resistance. FEBS Lett. 582:68–73. 2008. View Article : Google Scholar
|
|
14
|
Iizuka K: Recent progress on the role of
ChREBP in glucose and lipid metabolism. Endocr J. 60:543–555. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sahini N and Borlak J: Recent insights
into the molecular pathophysiology of lipid droplet formation in
hepatocytes. Prog Lipid Res. 54:86–112. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kawano Y and Cohen DE: Mechanisms of
hepatic triglyceride accumulation innon-alcoholic fatty liver
disease. J Gastroenterol. 48:434–441. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Karagianni P and Talianidis I:
Transcription factor networks regulating hepatic fatty acid
metabolism. Biochim Biophys Acta. 1851.2–8. 2015.
|
|
18
|
Wang Y, Viscarra J, Kim SJ and Sul HS:
Transcriptional regulation of hepatic lipogenesis. Nat Rev Mol Cell
Biol. 16:678–689. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ozkul Y and Galderisi U: The impact of
epigenetics on mesenchymal stem cell biology. J Cell Physiol.
231:2393–2401. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zoghbi HY and Beaudet AL: Epigenetics and
human disease. Cold Spring Harb Perspect Biol. 8:a0194972016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Martínez JA, Milagro FI, Claycombe KJ and
Schalinske KL: Epigenetics in adipose tissue, obesity, weight loss,
and diabetes. Adv Nutr. 5:71–81. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Stel J and Legler J: The role of
epigenetics in the latent effects of early life exposure to
obesogenic endocrine disrupting chemicals. Endocrinology.
156:3466–3472. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Youngson NA and Morris MJ: What obesity
research tells us about epigenetic mechanisms. Philos Trans R Soc
Lond B Biol Sci. 368:201103372013. View Article : Google Scholar :
|
|
24
|
Öst A and Pospisilik JA: Epigenetic
modulation of metabolic decisions. Curr Opin Cell Biol. 33:88–94.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lawrence M, Daujat S and Schneider R:
Lateral thinking: How histone modifications regulate gene
expression. Trends Genet. 32:42–56. 2016. View Article : Google Scholar
|
|
26
|
Harr JC, Gonzalez-Sandoval A and Gasser
SM: Histones and histone modifications in perinuclear chromatin
anchoring: From yeast to man. EMBO Rep. 17:139–155. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yu H, Wu J, Yang M, Guo J, Zheng L, Peng
M, Zhang Q, Xiang Y, Cao J and Shen W: Involvement of liver X
receptor alpha in histone modifications across the target fatty
acid synthase gene. Lipids. 47:249–257. 2012. View Article : Google Scholar
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−delta delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
29
|
Rufo C, Teran-Garcia M, Nakamura MT, Koo
SH, Towle HC and Clarke SD: Involvement of a unique
carbohydrate-responsive factor in the glucose regulation of rat
liver fatty-acid synthase gene transcription. J Biol Chem.
276:21969–21975. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Shechter D, Dormann HL, Allis CD and Hake
SB: Extraction, purification and analysis of histones. Nat Protoc.
2:1445–1457. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fan JG, Kim SU and Wong VW: New trends on
obesity and NAFLD in Asia. J Hepatol. 67:862–873. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hazlehurst JM, Woods C, Marjot T, Cobbold
JF and Tomlinson JW: Non-alcoholic fatty liver disease and
diabetes. Metabolism. 65:1096–1108. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Day CP and James OF: Steatohepatitis: A
tale of two ‘hits’. Gastroenterology. 114:842–845. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cordero P, Gomez-Uriz AM, Campion J,
Milagro FI and Martinez JA: Dietary supplementation with methyl
donors reduces fatty liver and modifies the fatty acid synthase DNA
methylation profile in rats fed an obesogenic diet. Genes Nutr.
8:105–113. 2013. View Article : Google Scholar :
|
|
35
|
Dorn C, Riener MO, Kirovski G, Saugspier
M, Steib K, Weiss TS, Gäbele E, Kristiansen G, Hartmann A and
Hellerbrand C: Expression of fatty acid synthase in nonalcoholic
fatty liver disease. Int J Clin Exp Pathol. 3:505–514.
2010.PubMed/NCBI
|
|
36
|
Jones SF and Infante JR: Molecular
pathways: Fatty acid synthase. Clin Cancer Res. 21:5434–5438. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kirk H, Cefalu WT, Ribnicky D, Liu Z and
Eilertsen KJ: Botanicals as epigenetic modulators for mechanisms
contributing to development of metabolic syndrome. Metabolism.
57:S16–S23. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Dawson MA and Kouzarides T: Cancer
epigenetics: From mechanism to therapy. Cell. 150:12–27. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tian Y, Wong VW, Chan HL and Cheng AS:
Epigenetic regulation of hepatocellular carcinoma in non-alcoholic
fatty liver disease. Semin Cancer Biol. 23:471–482. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Gołąbek K, Strzelczyk JK, Wiczkowski A and
Michalski M: Potential use of histone deacetylase inhibitors in
cancer therapy. Contemp Oncol (Pozn). 19:436–440. 2015.
|
|
41
|
Yi X, Jiang XJ, Li XY and Jiang DS:
Histone methyltransferases: Novel targets for tumor and
developmental defects. Am J Transl Res. 7:2159–2175. 2015.
|
|
42
|
Yen CY, Huang HW, Shu CW, Hou MF, Yuan SS,
Wang HR, Chang YT, Farooqi AA, Tang JY and Chang HW: DNA
methylation, histone acetylation and methylation of epigenetic
modifications as a therapeutic approach for cancers. Cancer Lett.
373:185–192. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Su X, Wellen KE and Rabinowitz JD:
Metabolic control of methylation and acetylation. Curr Opin Chem
Biol. 30:52–60. 2016. View Article : Google Scholar :
|
|
44
|
Warsito D, Lin Y, Gnirck AC, Sehat B and
Larsson O: Nuclearly translocated insulin-like growth factor 1
receptor phosphorylates histone H3 at tyrosine 41 and induces SNAI2
expression via Brg1 chromatin remodeling protein. Oncotarget.
7:42288–42302. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hazzalin CA and Mahadevan LC: Dynamic
acetylation of all lysine4-methylated histone H3 in the mouse
nucleus: Analysis at c-fos and c-jun. PLoS Biol. 3:e3932005.
View Article : Google Scholar
|
|
46
|
Tochiki KK, Maiarú M, Norris C, Hunt SP
and Géranton SM: The mitogen and stress-activated protein kinase 1
regulates the rapid epigenetic tagging of dorsal horn neurons and
nocifensive behaviour. Pain. 157:2594–2604. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ho CM, Donovan-Banfield IZ, Tan L, Zhang
T, Gray NS and Strang BL: Inhibition of IKKα by BAY61-3606 reveals
IKKα-dependent histone H3 phosphorylation in human cytomegalovirus
infected cells. PLoS One. 11:e01503392016. View Article : Google Scholar
|
|
48
|
Zippo A, De Robertis A, Serafini R and
Oliviero S: PIM1-dependent phosphorylation of histone H3 at serine
10 is required for MYC-dependent transcriptional activation and
oncogenic transformation. Nat Cell Biol. 9:932–944. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Burke SJ, Collier JJ and Scott DK: cAMP
prevents glucose-mediated modifications of histone H3 and
recruitment of the RNA polymerase II holoenzyme to the L-PK gene
promoter. J Mol Biol. 392:578–588. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Stoeckman AK, Ma L and Towle HC: Mlx is
the functional heteromeric partner of the carbohydrate response
element-binding protein in glucose regulation of lipogenic enzyme
genes. J Biol Chem. 279:15662–15669. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Ma L, Sham YY, Walters KJ and Towle HC: A
critical role for the loop region of the basic
helix-loop-helix/leucine zipper protein Mlx in DNAbinding and
glucose-regulated transcription. Nucleic Acids Res. 35:35–44. 2007.
View Article : Google Scholar
|
|
52
|
Furdas SD, Kannan S, Sippl W and Jung M:
Small molecule inhibitors of histone acetyltransferases as
epigenetic tools and drug candidates. Arch Pharm (Weinheim).
345:7–21. 2012. View Article : Google Scholar
|
|
53
|
Knutson SK, Chyla BJ, Amann JM, Bhaskara
S, Huppert SS and Hiebert SW: Liver-specific deletion of histone
deacetylase 3 disrupts metabolic transcriptional networks. EMBO J.
27:1017–1028. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Balasubramanyam K, Altaf M, Varier RA,
Swaminathan V, Ravindran A, Sadhale PP and Kundu TK:
Polyisoprenylated benzophenone, garcinol, a natural histone
acetyltransferase inhibitor, represses chromatin transcription and
alters global gene expression. J Biol Chem. 279:33716–33726. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Bricambert J, Miranda J, Benhamed F,
Girard J, Postic C and Dentin R: Salt-inducible kinase 2 links
transcriptional coactivator p300 phosphorylation to the prevention
of ChREBP-dependent hepatic steatosis in mice. J Clin Invest.
120:4316–4331. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
He Z, Jiang T, Wang Z, Levi M and Li J:
Modulation of carbohydrate response element-binding protein gene
expression in 3T3-L1 adipocytes and rat adipose tissue. Am J
Physiol Endocrinol Metab. 287:E424–E430. 2004. View Article : Google Scholar : PubMed/NCBI
|