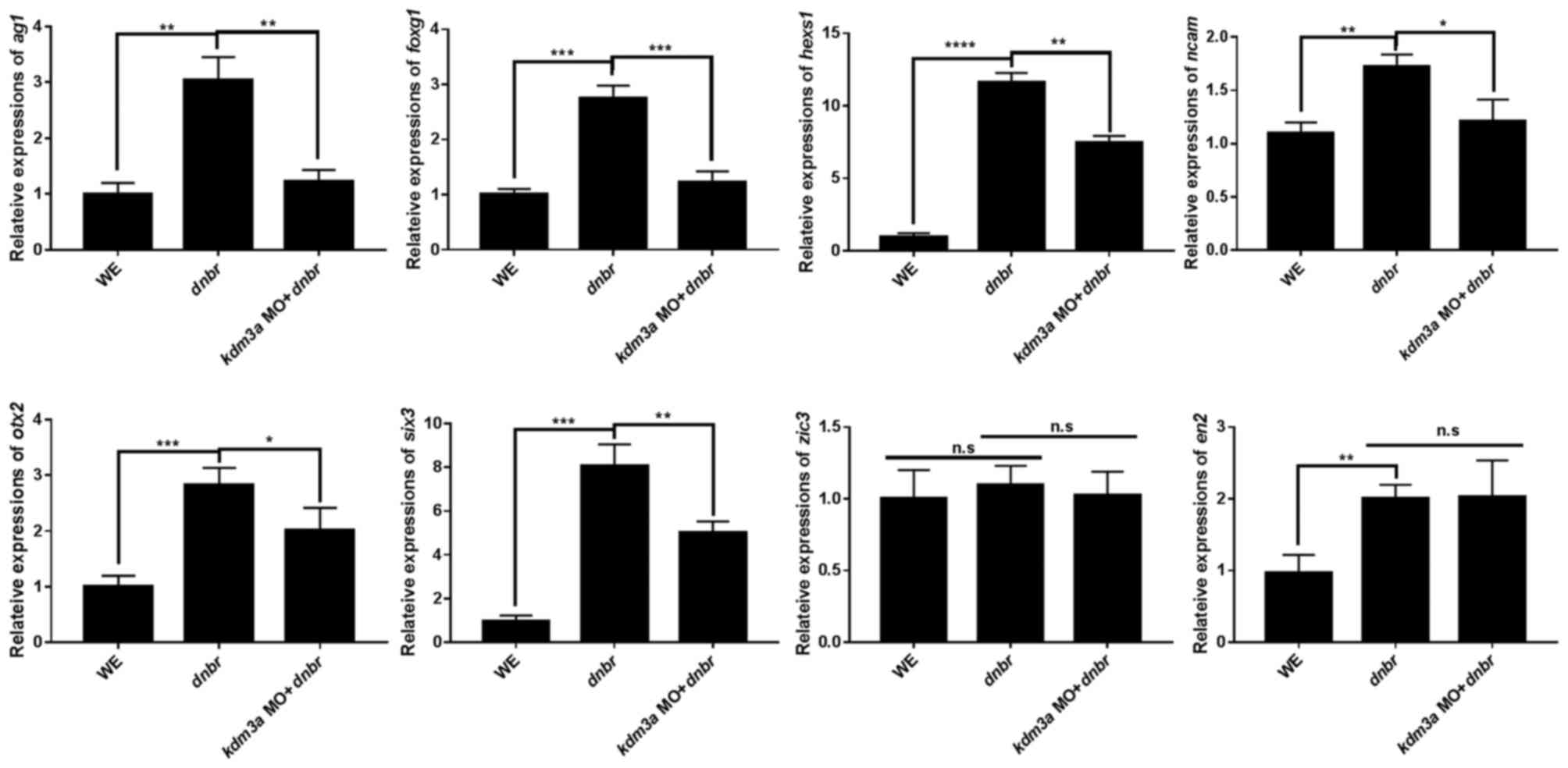
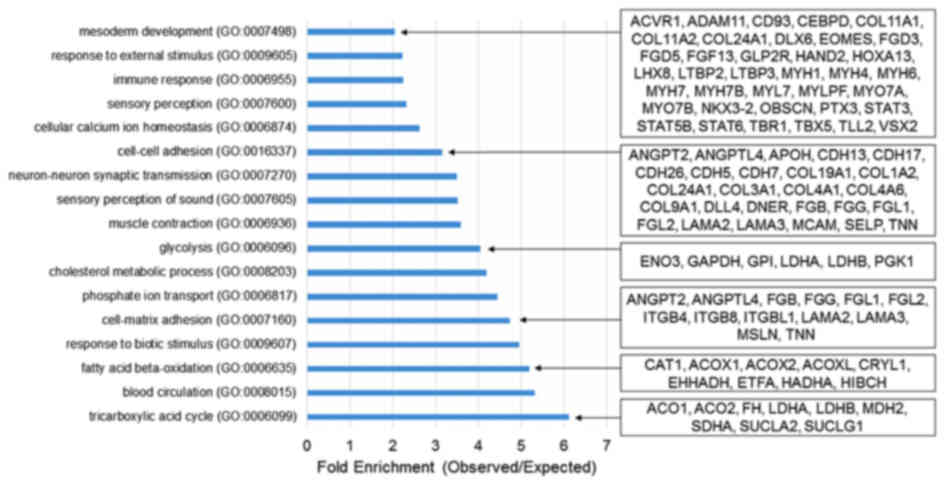
|
1
|
Meissner A: Epigenetic modifications in
pluripotent and differentiated cells. Nat Biotechnol. 28:1079–1088.
2010. View Article : Google Scholar
|
|
2
|
Mohn F and Schübeler D: Genetics and
epigenetics: Stability and plasticity during cellular
differentiation. Trends Genet. 25:129–136. 2009. View Article : Google Scholar
|
|
3
|
Mattout A and Meshorer E: Chromatin
plasticity and genome organization in pluripotent embryonic stem
cells. Curr Opin Cell Biol. 22:334–341. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jenuwein T: The epigenetic magic of
histone lysine methylation. FEBS J. 273:3121–3135. 2006. View Article : Google Scholar
|
|
5
|
Black JC, Van Rechem C and Whetstine JR:
Histone lysine methylation dynamics: Establishment, regulation, and
biological impact. Mol Cell. 48:491–507. 2012. View Article : Google Scholar
|
|
6
|
Bernstein BE, Mikkelsen TS, Xie X, Kamal
M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al:
A bivalent chromatin structure marks key developmental genes in
embryonic stem cells. Cell. 125:315–326. 2006. View Article : Google Scholar
|
|
7
|
Tachibana M, Sugimoto K, Nozaki M, Ueda J,
Ohta T, Ohki M, Fukuda M, Takeda N, Niida H, Kato H, et al: G9a
histone meth-yltransferase plays a dominant role in euchromatic
histone H3 lysine 9 methylation and is essential for early
embryogenesis. Genes Dev. 16:1779–1791. 2002. View Article : Google Scholar
|
|
8
|
Klose RJ, Kallin EM and Zhang Y:
JmjC-domain-containing proteins and histone demethylation. Nat Rev
Genet. 7:715–727. 2006. View Article : Google Scholar
|
|
9
|
Fodor BD, Kubicek S, Yonezawa M,
O'Sullivan RJ, Sengupta R, Perez-Burgos L, Opravil S, Mechtler K,
Schotta G and Jenuwein T: Jmjd2b antagonizes H3K9 trimethylation at
peri-centric heterochromatin in mammalian cells. Genes Dev.
20:1557–1562. 2006. View Article : Google Scholar :
|
|
10
|
Wissmann M, Yin N, Müller JM, Greschik H,
Fodor BD, Jenuwein T, Vogler C, Schneider R, Günther T, Buettner R,
et al: Cooperative demethylation by JMJD2C and LSD1 promotes
androgen receptor-dependent gene expression. Nat Cell Biol.
9:347–353. 2007. View Article : Google Scholar
|
|
11
|
Yamane K, Toumazou C, Tsukada Y,
Erdjument-Bromage H, Tempst P, Wong J and Zhang Y: JHDM2A, a
JmjC-containing H3K9 demethylase, facilitates transcription
activation by androgen receptor. Cell. 125:483–495. 2006.
View Article : Google Scholar
|
|
12
|
Knebel J, De Haro L and Janknecht R:
Repression of transcription by TSGA/Jmjd1a, a novel interaction
partner of the ETS protein ER71. J Cell Biochem. 99:319–329. 2006.
View Article : Google Scholar
|
|
13
|
Cai C, Yuan X and Balk SP: Androgen
receptor epigenetics. Transl Androl Urol. 2:148–157. 2013.
|
|
14
|
Inagaki T, Tachibana M, Magoori K, Kudo H,
Tanaka T, Okamura M, Naito M, Kodama T, Shinkai Y and Sakai J:
Obesity and metabolic syndrome in histone demethylase JHDM2a-
deficient mice. Genes Cells. 14:991–1001. 2009. View Article : Google Scholar
|
|
15
|
Kuroki S, Matoba S, Akiyoshi M, Matsumura
Y, Miyachi H, Mise N, Abe K, Ogura A, Wilhelm D, Koopman P, et al:
Epigenetic regulation of mouse sex determination by the histone
demethylase Jmjd1a. Science. 341:1106–1109. 2013. View Article : Google Scholar
|
|
16
|
Okada Y, Scott G, Ray MK, Mishina Y and
Zhang Y: Histone demethylase JHDM2A is critical for Tnp1 and Prm1
transcription and spermatogenesis. Nature. 450:119–123. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tateishi K, Okada Y, Kallin EM and Zhang
Y: Role of Jhdm2a in regulating metabolic gene expression and
obesity resistance. Nature. 458:757–761. 2009. View Article : Google Scholar
|
|
18
|
Beyer S, Kristensen MM, Jensen KS,
Johansen JV and Staller P: The histone demethylases JMJD1A and
JMJD2B are transcriptional targets of hypoxia-inducible factor HIF.
J Biol Chem. 283:36542–36552. 2008. View Article : Google Scholar
|
|
19
|
Ueda J, Ho JC, Lee KL, Kitajima S, Yang H,
Sun W, Fukuhara N, Zaiden N, Chan SL, Tachibana M, et al: The
hypoxia-inducible epigenetic regulators Jmjd1a and G9a provide a
mechanistic link between angiogenesis and tumor growth. Mol Cell
Biol. 34:3702–3720. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wade MA, Jones D, Wilson L, Stockley J,
Coffey K, Robson CN and Gaughan L: The histone demethylase enzyme
KDM3A is a key estrogen receptor regulator in breast cancer.
Nucleic Acids Res. 43:196–207. 2015. View Article : Google Scholar
|
|
21
|
Yamada D, Kobayashi S, Yamamoto H,
Tomimaru Y, Noda T, Uemura M, Wada H, Marubashi S, Eguchi H,
Tanemura M, et al: Role of the hypoxia-related gene, JMJD1A, in
hepatocellular carcinoma: Clinical impact on recurrence after
hepatic resection. Ann Surg Oncol. 19(Suppl 3): S355–S364. 2012.
View Article : Google Scholar
|
|
22
|
Wang J, Park JW, Drissi H, Wang X and Xu
RH: Epigenetic regulation of miR-302 by JMJD1C inhibits neural
differentiation of human embryonic stem cells. J Biol Chem.
289:2384–2395. 2014. View Article : Google Scholar :
|
|
23
|
Laurent B, Ruitu L, Murn J, Hempel K,
Ferrao R, Xiang Y, Liu S, Garcia BA, Wu H, Wu F, et al: A specific
LSD1/KDM1A isoform regulates neuronal differentiation through H3K9
demethylation. Mol Cell. 57:957–970. 2015. View Article : Google Scholar
|
|
24
|
Huang C, Chen J, Zhang T, Zhu Q, Xiang Y,
Chen CD and Jing N: The dual histone demethylase KDM7A promotes
neural induction in early chick embryos. Dev Dyn. 239:3350–3357.
2010. View Article : Google Scholar
|
|
25
|
Huang C, Xiang Y, Wang Y, Li X, Xu L, Zhu
Z, Zhang T, Zhu Q, Zhang K, Jing N, et al: Dual-specificity histone
demethylase KIAA1718 (KDM7A) regulates neural differentiation
through FGF4. Cell Res. 20:154–165. 2010. View Article : Google Scholar
|
|
26
|
Tsukada Y, Ishitani T and Nakayama KI:
KDM7 is a dual demethylase for histone H3 Lys 9 and Lys 27 and
functions in brain development. Genes Dev. 24:432–437. 2010.
View Article : Google Scholar
|
|
27
|
Lipchina I, Studer L and Betel D: The
expanding role of miR-302-367 in pluripotency and reprogramming.
Cell Cycle. 11:1517–1523. 2012. View Article : Google Scholar
|
|
28
|
Hemmati-Brivanlou A, Kelly OG and Melton
DA: Follistatin, an antagonist of activin, is expressed in the
Spemann organizer and displays direct neuralizing activity. Cell.
77:283–295. 1994. View Article : Google Scholar
|
|
29
|
Sasai Y, Lu B, Steinbeisser H, Geissert D,
Gont LK and De Robertis EM: Xenopus chordin: A novel dorsalizing
factor activated by organizer-specific homeobox genes. Cell.
79:779–790. 1994. View Article : Google Scholar
|
|
30
|
Lorenz TC: Polymerase chain reaction:
Basic protocol plus troubleshooting and optimization strategies. J
Vis Exp. 63:e39982012.
|
|
31
|
Jones CM and Smith JC: Wholemount in situ
hybridization to Xenopus embryos. Methods Mol Biol. 461:697–702.
2008. View Article : Google Scholar
|
|
32
|
Yuan JS, Reed A, Chen F and Stewart CN Jr:
Statistical analysis of real-time PCR data. BMC Bioinformatics.
7:852006. View Article : Google Scholar
|
|
33
|
Rozen S and Skaletsky H: Primer3 on the
WWW for general users and for biologist programmers. Methods Mol
Biol. 132:365–386. 2000.
|
|
34
|
Yoon J, Kim JH, Lee OJ, Yu SB, Kim JI, Kim
SC, Park JB, Lee JY and Kim J: xCITED2 Induces Neural Genes in
Animal Cap Explants of Xenopus Embryos. Exp Neurobiol. 20:123–129.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Session AM, Uno Y, Kwon T, Chapman JA,
Toyoda A, Takahashi S, Fukui A, Hikosaka A, Suzuki A, Kondo M, et
al: Genome evolution in the allotetraploid frog Xenopus laevis.
Nature. 538:336–343. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li H and Durbin R: Fast and accurate short
read alignment with Burrows-Wheeler transform. Bioinformatics.
25:1754–1760. 2009. View Article : Google Scholar
|
|
37
|
Mi H, Huang X, Muruganujan A, Tang H,
Mills C, Kang D and Thomas PD: PANTHER version 11: Expanded
annotation data from Gene Ontology and Reactome pathways, and data
analysis tool enhancements. Nucleic Acids Res. 45(D1): D183–D189.
2017. View Article : Google Scholar
|
|
38
|
Altschul SF, Gish W, Miller W, Myers EW
and Lipman DJ: Basic local alignment search tool. J Mol Biol.
215:403–410. 1990. View Article : Google Scholar
|
|
39
|
Betancur P, Bronner-Fraser M and
Sauka-Spengler T: Assembling neural crest regulatory circuits into
a gene regulatory network. Annu Rev Cell Dev Biol. 26:581–603.
2010. View Article : Google Scholar
|
|
40
|
Sauka-Spengler T and Bronner-Fraser M: A
gene regulatory network orchestrates neural crest formation. Nat
Rev Mol Cell Biol. 9:557–568. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ishii M, Merrill AE, Chan YS, Gitelman I,
Rice DP, Sucov HM and Maxson RE Jr: Msx2 and Twist cooperatively
control the development of the neural crest-derived skeletogenic
mesenchyme of the murine skull vault. Development. 130:6131–6142.
2003. View Article : Google Scholar
|
|
42
|
Huang C, Kratzer M-C, Wedlich D and Kashef
J: E-cadherin is required for cranial neural crest migration in
Xenopus laevis. Dev Biol. 411:159–171. 2016. View Article : Google Scholar
|
|
43
|
Bronner-Fraser M: Neural crest cell
formation and migration in the developing embryo. FASEB J.
8:699–706. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chen ZF and Behringer RR: twist is
required in head mesenchyme for cranial neural tube morphogenesis.
Genes Dev. 9:686–699. 1995. View Article : Google Scholar
|
|
45
|
Lin H, Zhu X, Chen G, Song L, Gao L, Khand
AA, Chen Y, Lin G and Tao Q: KDM3A-mediated demethylation of
histone H3 lysine 9 facilitates the chromatin binding of Neurog2
during neurogenesis. Development. 144:3674–3685. 2017. View Article : Google Scholar
|
|
46
|
Bellmeyer A, Krase J, Lindgren J and
LaBonne C: The protoon-cogene c-myc is an essential regulator of
neural crest formation in xenopus. Dev Cell. 4:827–839. 2003.
View Article : Google Scholar
|
|
47
|
Monsoro-Burq A-H, Wang E and Harland R:
Msx1 and Pax3 cooperate to mediate FGF8 and WNT signals during
Xenopus neural crest induction. Dev Cell. 8:167–178. 2005.
View Article : Google Scholar
|
|
48
|
Ishimura A, Maeda R, Takeda M, Kikkawa M,
Daar IO and Maéno M: Involvement of BMP-4/msx-1 and FGF pathways in
neural induction in the Xenopus embryo. Dev Growth Differ.
42:307–316. 2000. View Article : Google Scholar
|
|
49
|
Basch ML, Bronner-Fraser M and
García-Castro MI: Specification of the neural crest occurs during
gastrulation and requires Pax7. Nature. 441:218–222. 2006.
View Article : Google Scholar
|
|
50
|
Mayor R, Young R and Vargas A: Development
of neural crest in Xenopus. Curr Top Dev Biol. 43:85–113. 1999.
View Article : Google Scholar
|
|
51
|
Dyson S and Gurdon JB: Activin signalling
has a necessary function in Xenopus early development. Curr Biol.
7:81–84. 1997. View Article : Google Scholar
|
|
52
|
Aberger F, Weidinger G, Grunz H and
Richter K: Anterior specification of embryonic ectoderm: The role
of the Xenopus cement gland-specific gene XAG-2. Mech Dev.
72:115–130. 1998. View Article : Google Scholar
|
|
53
|
Tereshina MB, Zaraisky AG and Novoselov
VV: Ras-dva, a member of novel family of small GTPases, is required
for the anterior ectoderm patterning in the Xenopus laevis embryo.
Development. 133:485–494. 2006. View Article : Google Scholar
|
|
54
|
Nakata K, Nagai T, Aruga J and Mikoshiba
K: Xenopus Zic3, a primary regulator both in neural and neural
crest development. Proc Natl Acad Sci USA. 94:11980–11985. 1997.
View Article : Google Scholar
|
|
55
|
Doniach T and Musci TJ: Induction of
anteroposterior neural pattern in Xenopus: Evidence for a
quantitative mechanism. Mech Dev. 53:403–413. 1995. View Article : Google Scholar
|
|
56
|
Slack JM, Darlington BG, Gillespie LL,
Godsave SF, Isaacs HV and Paterno GD: Mesoderm induction by
fibroblast growth factor in early Xenopus development. Philos Trans
R Soc Lond B Biol Sci. 327:75–84. 1990. View Article : Google Scholar
|
|
57
|
Tsukada Y, Fang J, Erdjument-Bromage H,
Warren ME, Borchers CH, Tempst P and Zhang Y: Histone demethylation
by a family of JmjC domain-containing proteins. Nature.
439:811–816. 2006. View Article : Google Scholar
|
|
58
|
Shpargel KB, Starmer J, Yee D, Pohlers M
and Magnuson T: KDM6 demethylase independent loss of histone H3
lysine 27 trimethylation during early embryonic development. PLoS
Genet. 10:e10045072014. View Article : Google Scholar :
|
|
59
|
Kinoshita T, Haruta Y, Sakamoto C and
Imaoka S: Antagonistic role of XESR1 and XESR5 in mesoderm
formation in Xenopus laevis. Int J Dev Biol. 55:25–31. 2011.
View Article : Google Scholar
|