Introduction
Oral squamous cell carcinoma (OSCC), which is the
sixth most common type of human cancer worldwide, is an aggressive
epithelial malignancy (1). Each
year >300,000 new cases are reported and ~9,000 individuals
succumb to this disease in the United States. The 5-year survival
rate is ~50%, even though there have been some advancements in
therapeutic regimens (2). The
lack of characteristic symptoms in the early stages and the absence
of non-invasive screening tests leads to high mortality in patients
with OSCC (3). In addition, local
tumor recurrence and metastasis are key factors in the development
of OSCC (4). Therefore, there is
an urgent need to identify and develop novel strategies for the
clinical diagnosis and treatment of patients with OSCC.
SRY-box 4 (SOX4) is a 47-kDa protein and a member of
the SOX family of transcription factors, which has an important
role in embryonic development and cell-fate determination during
organogenesis (5). SOX4 has been
reported to be upregulated in various types of cancer, and SOX4
aberrant expression may regulate cellular transformation, cell
survival and metastasis (6-8).
Numerous studies have reported that SOX4 acts as an oncogene in
various types of cancer, including prostate cancer, hepatocellular
carcinoma and lung cancer (9-11).
In addition, SOX4 has been demonstrated to induce
epithelial-mesenchymal transition (EMT) (12,13), which serves a key role in tumor
metastasis and is associated with tumor recurrence (14).
MicroRNAs (miRNAs/miRs) are a class of small
non-coding RNAs (length, 21-23 nucleotides), which negatively
modulate gene expression at the post-transcriptional level through
inhibiting translation or inducing RNA degradation (15,16). miRNAs are associated with various
physiological and pathological processes, including cellular
metabolism, proliferation, differentiation and death (17). Accumulating evidence has revealed
that miRNAs are dysregulated and have key roles in several types of
cancer, and can act as tumor suppressor genes and potent oncogenes
(18). Notably, miRNA
dysregulation is considered to be associated with the occurrence
and development of OSCC (19).
Previous studies have demonstrated that miR-199a-5p serves a key
role in tumor development via modulation of tumor suppressor genes
or oncogenes in various types of cancer, including hepatocellular
cancer, gastric cancer and ovarian cancer (20-22). He et al revealed that
miR-199a-5p is upregulated in gastric cancer tissues and functions
as an oncogene in gastric cancer (22). Furthermore, upregulation of
miR-199a-5p has been recognized as a signature for poor prognosis
in uveal melanoma and it is associated with a high metastatic risk
(23). Conversely, Zhong et
al reported that miR-199a-5p is downregulated in prostate
adenocarcinoma, and overexpression of miR-199a-5p attenuates cell
motility, proliferation and tumor angiogenesis in prostate
adenocarcinoma cell lines (24).
In addition, miR-199a-5p has been identified to function as an
antitumor miRNA in head and neck squamous cell carcinoma cell lines
(25). However, the molecular
mechanism underlying the role of miR-199a-5p in the development of
OSCC remains unclear.
The present study performed reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) to
detect the expression levels of miR-199a-5p in OSCC tissues and
explored the molecular mechanism underlying the biological function
of this miRNA. The results revealed that miR-199a-5p is
downregulated in human OSCC tissues and cell lines, and its
expression in OSCC tissues with lymph node metastasis is lower than
that in tissues without lymph node metastasis. In addition, the
present findings revealed that miR-199a-5p inhibits cell invasion
and migration in OSCC cells through targeting the oncogene SOX4,
thus suggesting that miR-140-5p may possess potential value in the
clinical diagnosis and treatment of OSCC.
Materials and methods
Patients and tissue samples
The OSCC tissues and adjacent normal oral epithelial
tissues were collected from 40 patients with OSCC at the Department
of Oral and Maxillofacial Surgery, The First Affiliated Hospital of
Xinxiang Medical University (Weihui, China) between November 2015
and January 2016. The patients enrolled in this study did not
receive radiotherapy or chemotherapy prior to surgical resection.
All tissue samples were confirmed by pathological examination. All
OSCC tissues and corresponding normal oral epithelial tissues were
frozen in liquid nitrogen and stored at −80°C until further use.
Patient characteristics are presented in Table I. The present study was approved
by the Ethics Committee of The First Affiliated Hospital of
Xinxiang Medical University and all patients provided written
informed consent.
 | Table IAssociation between miR-199a-5p
expression and the clinicopathological features of patients with
oral squamous cell carcinoma. |
Table I
Association between miR-199a-5p
expression and the clinicopathological features of patients with
oral squamous cell carcinoma.
| Clinical
parameters | All cases
(n=40) | miR-199-5p
expression
| P-value |
|---|
| High (n=16) | Low (n=24) |
|---|
| Sex | | | | 0.685 |
| Male | 26 | 11 | 15 | |
| Female | 14 | 5 | 9 | |
| Age (years) | | | | 0.408 |
| ≥50 | 27 | 12 | 15 | |
| <50 | 13 | 4 | 9 | |
| Site | | | | 0.604 |
| Buccal mucosa | 22 | 8 | 14 | |
| Non-buccal
mucosa | 18 | 8 | 10 | |
| Alcohol use | | | | 0.505 |
| Yes | 25 | 9 | 16 | |
| No | 15 | 7 | 8 | |
| Smoking | | | | 0.897 |
| Yes | 22 | 9 | 13 | |
| No | 18 | 7 | 11 | |
| cTNM stage | | | | 0.064 |
| I + II | 9 | 6 | 3 | |
| III + IV | 31 | 10 | 21 | |
| Tumor size
(cm) | | | | 0.020a |
| ≥2 | 19 | 4 | 15 | |
| <2 | 21 | 12 | 9 | |
|
Differentiation | | | | 0.037a |
| Well and
moderate | 23 | 6 | 17 | |
| Poor | 17 | 10 | 7 | |
| Lymph node
metastasis | | | | 0.003b |
| Present | 28 | 7 | 21 | |
| Absent | 12 | 9 | 3 | |
Cell culture
The human OSCC cell lines HSC-2, CAL-27, Tca8113 and
SCC-4 were obtained from the American Type Culture Collection
(Manassas, VA, USA). The cells were grown in RPMI 1640 medium
(Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany), 100 IU/ml penicillin and 100 mg/ml
streptomycin at 37°C in a humidified atmosphere containing 5%
CO2. The normal human oral keratinocytes (HOK)
(ScienCell Research Laboratories, Inc., San Diego, CA, USA) were
grown in oral keratinocyte medium (Invitrogen; Thermo Fisher
Scientific, Inc.).
Cell transfection
miR-199a-5p mimics/inhibitor and the corresponding
negative control (NC) sequences (mimics/inhibitor NC) were
synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China).
Small interfering (si)RNA sequences, si-SOX4 and NC si-Scramble,
were purchased from Ambion (Thermo Fisher Scientific, Inc.). The
oligonucleotides sequences as the following: miR-199a-5p mimics,
sense 5′-CCCAGUGUUCAGACUACCUGUUC-3′; mimics NC, sense
5′-CGGTGUGUUCAGACUACCUGUUC-3′; miR-199a-5p inhibitor, sense
5′-AACAGGTAGTCTGAACACT-3′; inhibitor NC, sense:
5′-TAACACGTCTATACGCCCA-3′; si-SOX4-1,
5′-GCAAACGCTGGAAGCTGCTCAAAGA-3′; si-SOX4-2,
5′-TGGAAGCTGCTCAAAGACAGCGACA-3′; si-SOX4-3,
5′-GGAAGCTGCTCAAAGACAGCGACAA-3′; and si-Scramble,:
5′-TGGGTCGACTCAGAACGACGAAACA-3′. For plasmid construction
(pcDNA-SOX4), the SOX4 3′-untranslated region (UTR) was cloned into
the XhoI and KpnI sites of the pcDNA3.1 expression
vector (Invitrogen; Thermo Fisher Scientific, Inc.); an empty
pcDNA3.1 vector (pcDNA-vector) used as a control. For transfection,
SCC-4 or HSC-2 cells were plated in 6-well plates (2×106
cells/well) and were transfected with miR-199a-5p mimics/inhibitor,
mimics/inhibitor NC, si-SOX4/si-Scramble or pcDNA-SOX4/pcDNA-vector
at a final concentration of 100 nM using Lipofectamine®
2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. Subsequently, cells were cultured at 37°C
in an incubator containing 5% CO2 for 48 h after
trans-fection, after which, cells underwent subsequent experiments.
Transfection efficiency was assessed using RT-qPCR.
RT-qPCR
Total RNA was isolated from tissues and cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocol. TaqMan Gene
Expression Assay and TaqMan MicroRNA Reverse Transcription kits
(Applied Biosystems; Thermo Fisher Scientific, Inc.) were used for
RT prior to SOX-4 and miRNA detection, according to the
manufacturer's protocol. PCR was performed on an ABI PRISM 7300
sequence detection system using a SYBR-Green I Real-Time PCR kit
(both Applied Biosystems; Thermo Fisher Scientific, Inc.). PCR
amplification was conducted at 95°C for 3 min, followed by 35
cycles comprising 95°C for 10 sec, 60°C for 30 sec and 72°C for 30
sec. U6 served as an endogenous control for miR-199a-5p and GAPDH
served as a reference control for SOX4. The following primers were
used: miR-199a-5p, forward 5′-TCCCAGTGTTCAGACTACC-3′, reverse
5′-TTTGGCACTAGCACATT-3′; SOX4, forward 5′-CCAGTTCTTGCACGCTGTTT-3′,
reverse 5′-TGTTGCAAGGTAGGAAGCCA-3′; U6, forward
5′-CTCGCTTCGGCAGCACA-3′, reverse 5′-AACGCTTCACGAATTTGCGT-3′; and
GAPDH, forward 5′-AACGTGTCAGTGGTGGACCTG-3′ and reverse
5′-AGTGGGTGTCGCTGTTGAAGT-3′. Fold changes were determined using the
relative quantification (2−ΔΔCq) method (26).
Cell invasion assay
Cell invasion assays were performed using 24-well
Transwell chambers with polycarbonate membranes (pore size, 8.0
µm; Corning Inc., Corning, NY, USA). Briefly, SCC-4 or HSC-2
cells (2×106 cells/well) were plated into the upper
chambers, which were precoated with Matrigel (BD Biosciences,
Franklin Lakes, NJ, USA). After 48 h at 37°C, non-invading cells on
the upper side of the membrane were removed and invasive cells on
the lower surface of the membrane were fixed with 20% methanol at
4°C for 30 min and stained with 0.1% crystal violet (Sigma-Aldrich;
Merck KGaA). The number of invasive cells was counted from six
random fields in each well using an IX71 inverted microscope
(Olympus Corporation, Tokyo, Japan); images were then captured
(magnification, ×200).
Cell migration assay
A wound-healing assay was performed to analyze cell
migration. Briefly, SCC-4 or HSC-2 cells (2×106
cells/well) were plated into 6-well plates. After 48 h at 37°C, the
migration status was examined by measuring the movement of cells
into a scraped area, which was created using a micropipette tip.
After 0 or 48 h, cells were fixed in 4% paraformaldehyde (cat. no.
15714; Electron Microscopy Sciences) for 1 h at room temperature,
and were observed under a microscope. The wounds were analyzed in
six random fields using an IX71 inverted microscope (Olympus
Corporation; magnification, ×200). The percentage of closure of the
denuded area was calculated using ImageJ software (version 1.44;
National Institutes of Health, Bethesda, MD, USA).
Cell viability analysis
Cell viability was evaluated using the Cell Counting
Kit-8 (CCK-8) assay, according to the manufacturer's protocol.
Briefly, SCC-4 or HSC-2 cells were seeded into 96-well plates at an
initial density of 5×104 cells/well, and were cultured
in 100 µl RPMI-1640 medium supplemented with 10% FBS. At the
specified time points (24, 48 and 72 h), 10 µl CCK-8
solution (Dojindo Molecular Technologies, Inc., Rockville, MD, USA)
was added to each well. Subsequently, the plate was incubated for 1
h at 37°C in an atmosphere containing 5% CO2 and the
absorbance rate was measured at 450 nm using a microplate reader
(Bio-Rad Laboratories, Inc., Hercules, CA, USA). Experiments were
performed in triplicate.
Western blot analysis
SCC-4 or HSC-2 cells were lysed as described
previously (2) and the protein
concentrations were measured using a bicinchoninic acid protein
assay kit (Pierce; Thermo Fisher Scientific, Inc.). Protein samples
(30 µg) were then separated by 10% SDS-PAGE (Sigma-Aldrich;
Merck KGaA) and were transferred onto polyvinylidene difluoride
(PVDF) membranes (GE Healthcare Life Sciences, Little Chalfont,
UK). The PVDF membranes were blocked in 5% nonfat milk
(Sigma-Aldrich; Merck KGaA) at room temperature for 1 h and were
then incubated with the following primary antibodies at 4°C
overnight: SOX-4 (1:500; cat. no. ab80261; Abcam, Cambridge, MA
USA), E-cadherin (1:1,000; cat. no. sc-7870; Santa Cruz
Biotechnology, Inc., Dallas, TX. USA), N-cadherin (1:1,000; cat.
no. ab18203; Abcam), fibronectin (1:1,000; cat. no. sc-18825; Santa
Cruz Biotechnology, Inc.) and vimentin (1:1,000; cat. no. sc-5565;
Santa Cruz Biotechnology, Inc.). β-actin (1:750; cat. no. A5060;
Sigma-Aldrich; Merck KGaA) served as an internal control. Membranes
were then incubated with horseradish peroxidase-conjugated goat
anti-rabbit or goat anti-mouse IgG secondary antibodies [dilution
1:10,000; cat. nos. GAR0072 and GAM0072; MultiSciences (Lianke)
Biotech, Co., Ltd., Hangzhou, China] at room temperature for 2 h.
Subsequently, the protein bands were scanned on X-ray film and were
visualized using the enhanced chemiluminescence detection system
(PerkinElmer, Inc., Waltham, MA, USA). The relative intensity of
each band was semi-quantified using Alpha Imager software version
2000 (ProteinSimple, San Jose, CA, USA). The measurements were
conducted independently at least three times with similar
results.
Luciferase reporter assay
Bioinformatics analysis (TargetScan; www.targetscan.org/vert_72) was used to predict
the putative target genes of miR-199a-5p in OSCC cells. The SOX4
3′-UTR encompassing the target sequence for miR-199a-5p was
amplified by PCR and cloned into the psi-CHECK-2 Vector (Promega
Corporation, Madison, WI, USA) (wild type psi-CHECK-2-SOX4-3′-UTR).
Site-directed mutagenesis of the miR-199a-5p target site in the
SOX4 3′-UTR (psi-CHECK-2-SOX4-mut-3′-UTR) was performed using the
QuikChange II Site-Directed Mutagenesis kit (Agilent Technologies,
Inc., Santa Clara, CA, USA). SCC-4 cells (5×104
cells/well) were seeded into 96-well plates and the cells were
co-transfected with 400 ng psi-CHECK-2-SOX4-3′-UTR or
psi-CHECK-2-SOX4-mut-3′-UTR, and 50 ng miR-199a-5p mimics/inhibitor
or the corresponding NC using Lipofectamine® 2000
reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The ratio of
firefly to Renilla luciferase was used to normalize relative
firefly luciferase activity 48 h post-transfection, and luciferase
activity was measured using the Dual-Luciferase®
Reporter Assay system (Promega Corporation), according to the
manufacturer's protocol.
Statistical analysis
All statistical analyses were carried out using SPSS
software (version 18.0; SPSS, Inc., Chicago, IL, USA). Data are
presented as the means ± standard deviation. Comparisons between
two groups were made using Student's t-test. A one-way analysis of
variance followed by Tukey's post-hoc multiple comparisons test was
used to determine statistically differences among multiple groups.
The associations between miR-199a-5p level and clinicopathological
features were analyzed using χ2 test. Survival analysis
was conducted using the Kaplan-Meier method followed by a log-rank
(Mantel-Cox) test. The correlation between miR-199a-5p and SOX4
expression was assessed by Spearman's analyses. P<0.05 was
considered to indicate a statistically significant difference.
Results
miR-199a-5p is downregulated in human
OSCC tissues and cell lines
To investigate the association between miR-199a-5p
expression and OSCC, RT-qPCR was used to detect miR-199a-5p
expression in 40 paired OSCC tissues and adjacent normal oral
epithelial tissues. As shown in Fig.
1A, miR-199a-5p expression was markedly downregulated in OSCC
tissues compared with in normal oral epithelial tissues
(P<0.01). Furthermore, miR-199a-5p expression was reduced in
OSCC tissues with lymph node metastasis (n=28) compared with in
tissues without lymph node metastasis (n=12) (P<0.01; Fig. 1B). This study further analyzed the
association between miR-199a-5p and the clinicopathological
features of patients with OSCC (Table
I). To determine the clinical value of miR-199a-5p, the mean
expression level of miR-199a-5p was used as a cut-off value to
divide 40 patients with OSCC into two groups: The miR-199a-5p high
expression group and the miR-199a-5p low expression group. The
results revealed that low miR-199a-5p expression in OSCC was
associated with lymph node metastasis (P=0.003), differentiation
(P=0.037) and tumor size (cm) (P=0.020) (Table I). However, miR-199a-5p expression
was not associated with sex, age (years), tumor site, alcohol use,
smoking and clinical Tumor-Node-Metastasis stage (27) (Table
I). Kaplan-Meier analysis indicated that OSCC patients with low
miR-199a-5p expression (n=24) had a poorer overall survival rate
compared with patients with high miR-199a-5p expression (n=16)
(P<0.01; Fig. 1C). In
addition, this study further confirmed that miR-199a-5p was
markedly downregulated in the OSCC cell lines HSC-2, CAL-27,
Tca8113 and SCC-4 compared with in HOK cells (P<0.01; Fig. 1D). The two OSCC cell lines, HSC-2
and SCC-4, with the lowest expression levels of miR-199a-5p, were
selected for subsequent analysis in the present study. These data
indicated that miR-199a-5p may serve an important role in the
development of OSCC.
Overexpression of miR-199a-5p suppresses
cell invasion and migration in vitro
To further elucidate the role of miR-199a-5p in OSCC
cells, SCC-4 and HSC-2 cells were transfected with miR-199a-5p
mimics or mimics NC, and cell invasion and migration were evaluated
using Transwell invasion and wound-healing assays, respectively.
This study initially evaluated the transfection efficiency of
miR-199a-5p mimics and miR-199a-5p inhibitor in SCC-4 and HSC-2
cells. The results revealed that miR-199a-5p expression was
markedly upregulated in SCC-4 and HSC-2 cells transfected with
miR-199a-5p mimics compared with in cell transfected with mimics
NC, whereas transfection with miR-199a-5p inhibitor downregulated
miR-199a-5p expression compared with inhibitor NC (P<0.01;
Fig. 2A and B). Subsequently, the
results revealed that miR-199a-5p overexpression significantly
inhibited the invasion of SCC-4 and HSC-2 cells compared with
mimics NC (P<0.01; Fig. 2C and
D). Furthermore, the results further confirmed that
overexpression of miR-199a-5p markedly suppressed cell migration
compared with mimics NC (P<0.01; Fig. 2E and F). These data suggested that
miR-199a-5p may exert suppressive effects on cell migration and
invasion in OSCC.
Tumor cell invasion and metastasis are tightly
correlated with various processes, including EMT. During EMT,
epithelial tumors progress to more aggressive metastatic tumors,
alongside which the expression of mesenchymal protein markers,
including N-cadherin and vimentin, is upregulated, whereas the
epithelial protein marker E-cadherin is downregulated (14,28-29). It has been well documented that
miRNAs regulate tumorigenesis via modulating EMT in various types
of cancer, including OSCC (30-32). The present results demonstrated
that miR-199a-5p upregulated the expression levels of E-cadherin,
and downregulated N-cadherin, vimentin and fibronectin in SCC-4 and
HSC-2 cells (Fig. 2G). Taken
together, these results suggested that miR-199a-5p may inhibit cell
invasion and migration in OSCC cells via modulating EMT.
miR-199a-5p inhibits SOX4 expression by
directly targeting its 3′-UTR in OSCC cells
Bioinformatics analysis revealed that SOX4 may be a
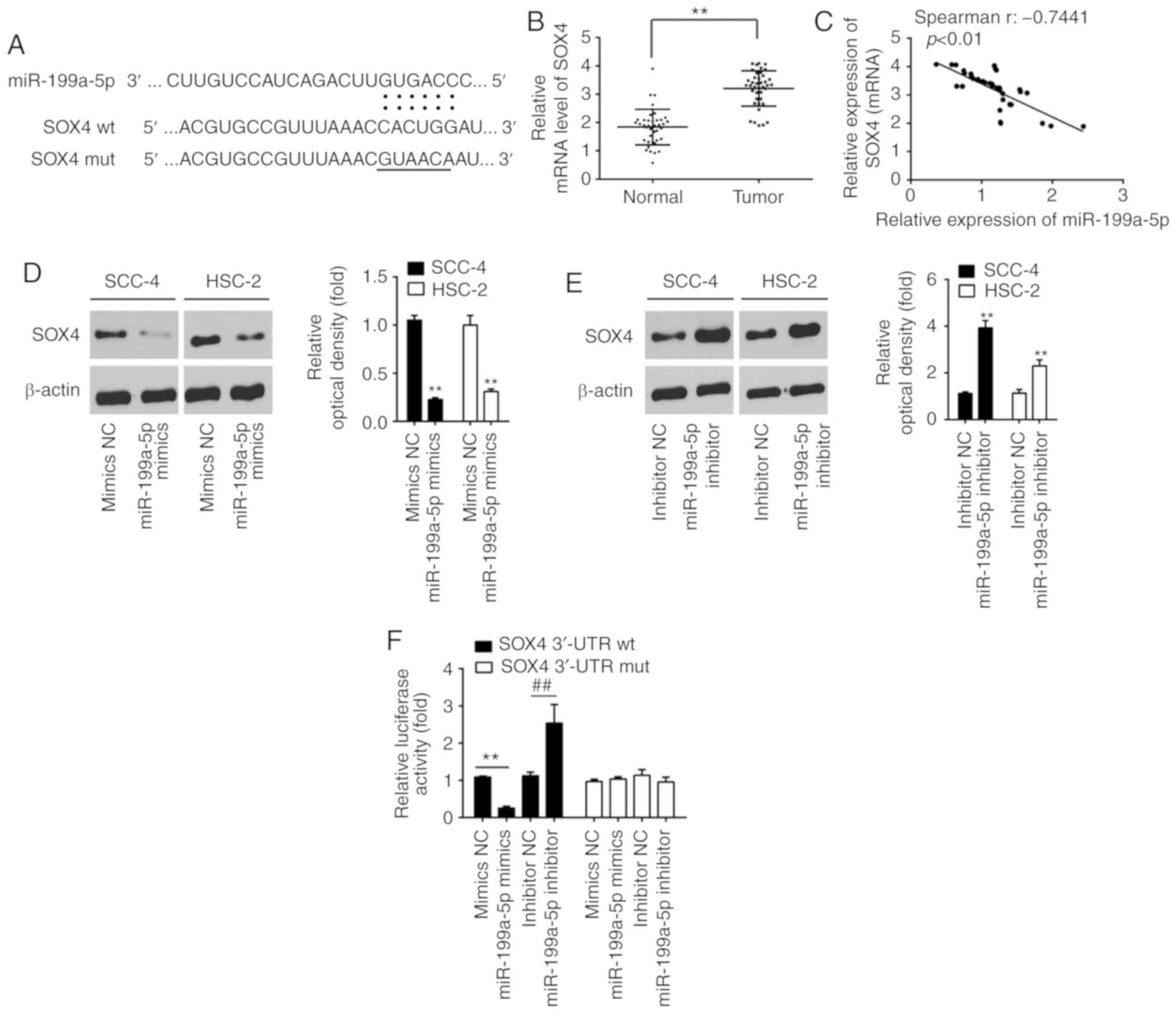
potential target gene of miR-199a-5p in OSCC cells (Fig. 3A). Previous studies have reported
that SOX4 functions as an oncogene in numerous types of cancer,
such as prostate cancer, lung cancer and hepatocellular carcinoma
(9-11). Additionally, SOX4 has been
identified to serve a key role in EMT and may induce cancer cell
metastasis (1,13). Therefore, to further investigate
the association between miR-199a-5p and SOX4 in OSCC, SOX4
expression was detected in OSCC tissues and adjacent normal oral
epithelial tissues. As shown in Fig.
3B, SOX4 expression was significantly upregulated in tumor
tissues compared with in normal tissues (P<0.01). Correlation
analysis revealed that there was a significant negative correlation
between miR-199a-5p and SOX4 expression in 40 tumor tissues
(r=-0.7441, P<0.01; Fig. 3C).
Furthermore, overexpression of miR-199a-5p markedly inhibited SOX4
(P<0.01; Fig. 3D), whereas
inhibition of miR-199a-5p significantly upregulated SOX4 expression
in SCC-4 and HSC-2 cells (P<0.01; Fig. 3E). Luciferase reporter assay
revealed that miR-199a-5p markedly inhibited luciferase activity
when compared with mimics NC, whereas miR-199a-5p inhibitor
significantly enhanced luciferase activity (P<0.01; Fig. 3F). However, miR-199a-5p
mimic/inhibitor did not affect luciferase activity in cells
transfected with psi-CHECK-2-SOX4-mut-3′-UTR (Fig. 3F). Collectively, these results
indicated that miR-199a-5p may suppress SOX4 expression by
targeting its 3′-UTR in OSCC.
Knockdown of SOX4 inhibits OSCC cell
proliferation, invasion and migration
To explore the role of SOX4 in OSCC cells, SCC-4 and
HSC-2 cells were transfected with si-SOX4 or si-Scramble, and cell
proliferation, invasion and migration were evaluated using CCK-8,
Transwell invasion and wound-healing assays, respectively. Firstly,
the effects of si-SOX4 (si-SOX4-1, si-SOX4-2 and si-SOX4-3) on SOX4
expression in OSCC cells were analyzed. As shown in Fig. 4A and B, all three siRNAs
significantly inhibited SOX4 expression in SCC-4 and HSC-2 cells
compared with si-Scramble (P<0.01); si-SOX4-1 exhibited the
strongest effect and was selected for subsequent experiments. As
shown in Fig. 4C and D, knockdown
of SOX4 markedly suppressed proliferation of SCC-4 and HSC-2 cells
compared with si-Scramble (P<0.05). Furthermore, cell invasion
and migration were significantly inhibited in SCC-4 and HSC-2 cells
transfected with si-SOX4 (P<0.01; Fig. 4E-G). These results indicated that
SOX4 may act as an oncogene in OSCC via modulating cell
proliferation, migration and invasion.
 | Figure 4Knockdown of SOX4 suppresses OSCC
cell proliferation, migration and invasion. (A and B) SCC-4 and
HSC-2 cells were transfected with si-SOX4 (si-SOX4-1, si-SOX4-2 and
si-SOX4-3) or si-Scramble, and the effects of si-SOX4 on SOX4
expression were evaluated by reverse transcription-quantitative
polymerase chain reaction. (C and D) Cell proliferation was
evaluated by Cell Counting Kit-8 assay in SCC-4 and HSC-2 cells
transfected with si-SOX4 or si-Scramble. (E and F) Cell invasion
was evaluated using Transwell invasion assay in SCC-4 and HSC-2
cells transfected with si-SOX4 or si-Scramble (magnification,
×200). (G and H) SCC-4 and HSC-2 cells were transfected with
si-SOX4 or si-Scramble, and cell migration was evaluated by
wound-healing assay (magnification, ×200). *P<0.05,
**P<0.01 vs. si-Scramble. Data are presented as the
means ± standard deviation of three independent experiments. OD,
optical density; si, small interfering RNA; SOX4, SRY-box 4. |
Overexpression of SOX4 rescues the
suppressive effects of miR-199a-5p on OSCC cells
To further investigate whether SOX4 is a functional
target of miR-199a-5p in OSCC cells, SCC-4 and HSC-2 cells were
transfected with miR-199a-5p mimics/mimics NC or were
co-transfected with miR-199a-5p mimics and pcDNA-SOX4/pcDNA-vector
plasmids, and cell invasion and migration were determined using
Transwell invasion and wound-healing assays, respectively. Firstly,
the transfection efficiency of pcDNA-SOX4 was evaluated in SCC-4
and HSC-2 cells; SOX4 was significantly upregulated compared with
in cells transfected with the pcDNA-vector (P<0.01; Fig. 5A). Subsequently, the results
demonstrated that overexpression of miR-199a-5p markedly suppressed
cell migration; however, upregulation of SOX4 significantly
increased miR-199a-5p-mediated migration of SCC-4 and HSC-2 cells
(P<0.01; Fig. 5B and C).
Furthermore, the results confirmed that the suppressive effect of
miR-199a-5p on cell invasion was rescued by SOX4 in SCC-4 and HSC-2
cells (P<0.01; Fig. 5D and E).
These data suggested that the suppressive effects of miR-199a-5p on
cell migration and invasion may be rescued by SOX4 overexpression
in OSCC cells.
Discussion
Previous studies have demonstrated that miR-199a-5p
functions as a tumor suppressor gene or oncogene in numerous types
of cancer, including human colorectal cancer and gastric cancer
(22,33). However, the roles of miR-199a-5p
in OSCC remain elusive. In this study, the results demonstrated
that miR-199a-5p was markedly downregulated in human OSCC tissues
and cell lines, and low miR-199a-5p expression was associated with
clinicopathological features and predicted poor overall survival in
patients with OSCC. Furthermore, overexpression of miR-199a-5p
suppressed the invasion and migration of OSCC cells through
inhibiting the EMT process. In addition, SOX4 was identified as a
target gene of miR-199a-5p in OSCC cells, and the suppressive
effects of miR-199a-5p on cell migration and invasion were rescued
by overexpression of SOX4 in OSCC cells.
It has been reported that miR-199a dysregulation is
associated with the progression and pathogenesis of cancer, and
previous studies have proposed conflicting roles for miR-199a in
tumor progression and carcinogenesis. For example, a previous study
demonstrated that miR-199a may act as a tumor suppressor in renal
cancer (34), whereas another
study documented that miR-199a may serve an oncogenic role in small
cell carcinoma of the cervix (35). In addition, a previous study
reported that miR-199a-5p attenuates proliferation and metastasis,
and stem cell-like characteristics in breast cancer (36). However, little is currently known
about the functional role of miR-199a-5p in OSCC. The present study
performed RT-qPCR to investigate miR-199a-5p expression in OSCC
tissues and adjacent normal oral epithelial tissues. The results
revealed that miR-199a-5p was significantly downregulated in OSCC
tissues compared with in normal oral epithelial tissues.
Furthermore, miR-199a-5p expression in OSCC tissues with lymph node
metastasis was lower than that in tissues without lymph node
metastasis, thus suggesting that miR-199a-5p may serve an important
role in the progression of OSCC. In addition, low expression of
miR-199a-5p in patients with OSCC was associated with
clinicopathological features and predicted a poor prognosis.
Downregulation of miR-199a-5p was also confirmed in OSCC cell
lines. These data suggested that miR-199a-5p may act as a tumor
suppressor in OSCC tumorigenesis and may serve as a biomarker in
OSCC diagnosis. This study also revealed that overexpression of
miR-199a-5p significantly inhibited the migration and invasion of
OSCC cells; however, the potential molecular mechanism requires
further investigation.
EMT, which is characterized by the acquisition of a
mesenchymal phenotype and loss of epithelial characteristics,
serves a critical role in tumor metastasis and is associated with
tumor recurrence (14,37). EMT can be evaluated by measuring
alterations in the expression levels of molecular markers,
including N-cadherin, E-cadherin and vimentin (38,39). Previous studies reported that
miR-199a-5p modulates metastasis via regulating the EMT process in
various types of cancer, such as human gastric cancer and
colorectal cancer (40,41). In the present study, it was
demonstrated that overexpression of miR-199a-5p upregulated
E-cadherin, and downregulated N-cadherin, vimentin and fibronectin
in OSCC cell lines. These data indicated that miR-199a-5p may
inhibit invasion and migration via suppressing EMT.
The transcription factor SOX4 has been demonstrated
to induce EMT through modulating expression of enhancer of zeste 2
polycomb repressive complex 2 subunit, which acts as an epigenetic
regulator (12,13). SOX4 is an EMT-related gene that
governs traditional EMT transcription factors, including ZEB, Snail
and Twist family members (13).
Notably, SOX4 is upregulated in numerous types of human cancer,
including esophageal cancer (42), endometrial cancer (43), gastric cancer (44), ovarian cancer (45) and osteosarcoma (46), thus suggesting that SOX4 may be a
key oncogene influencing the progression and metastasis of tumors.
Previous studies have reported that some miRNAs suppress metastasis
in various types of cancer by targeting the EMT regulator SOX4
(5,47). Additionally, numerous miRNAs have
been revealed to regulate cell proliferation, invasion and
metastasis by targeting SOX4 in human cancer, including miR-138,
miR-129-2 and miR-204 (44,45). In this study, the results
indicated that SOX4 may be a target gene of miR-199a-5p in OSCC
cells. Correlation analysis also revealed a negative correlation
between miR-199a-5p and SOX4 expression in tumor tissues.
Furthermore, knockdown of SOX4 by siRNA suppressed OSCC cell
proliferation, migration and invasion; SOX4 knockdown had similar
effects to miR-199a-5p over-expression. Additionally, restoration
of SOX4 reversed the inhibitory effects of miR-199a-5p on OSCC
cells, suggesting that miR-199a-5p may act as a tumor suppressor
through repressing the oncogene SOX4. Notably, there may be more
targets for miR-199a-5p. In future studies, we aim to transfect
OSCC cells with miR-199a-5p mimics or inhibitor and use a DNA chip
to identify genes with altered expression and a miR-199a-5p binding
sequence in the 3′-UTR. Furthermore, associated signaling pathways
will also be investigated in the future.
In conclusion, the present findings demonstrated
that miR-199a-5p was significantly downregulated in human OSCC
tissues and cell lines. Overexpression of miR-199a-5p suppressed
invasion and migration of OSCC cells through blocking the SOX4/EMT
pathway. The identified miR-199a-5p/SOX4 axis may provide novel
insights into the molecular mechanisms underlying the progression
and metastasis of cancer, and targeting miR-199a-5p may be a
potential therapeutic strategy for the treatment of OSCC in the
future.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
DW, WW, BS, YZ, XY, GL and JY performed the
experiments, contributed to data analysis and wrote the paper. DW,
WW, BS, YZ, XY, GL and JY analyzed the data. YS designed the study,
and contributed to data analysis and experimental materials. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
All individuals provided written informed consent
for the use of human specimens for clinical research. The present
study was approved by The First Affiliated Hospital of Xinxiang
Medical University Ethics Committees.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Andersen CL, Christensen LL, Thorsen K,
Schepeler T, Sørensen FB, Verspaget HW, Simon R, Kruhøffer M,
Aaltonen LA, Laurberg S and Ørntoft TF: Dysregulation of the
transcription factors SOX4, CBFB and SMARCC1 correlates with
outcome of colorectal cancer. Br J Cancer. 100:511–523. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Min R, Zun Z, Siyi L, Wenjun Y, Lizheng W
and Chenping Z: Increased expression of Toll-like receptor-9 has
close relation with tumour cell proliferation in oral squamous cell
carcinoma. Arch Oral Biol. 56:877–884. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang H, Xia J, Wang K and Zhang J: Serum
autoantibodies in the early detection of esophageal cancer: A
systematic review. Tumour Biol. 36:95–109. 2015. View Article : Google Scholar
|
|
4
|
Genden EM, Ferlito A, Bradley PJ, Rinaldo
A and Scully C: Neck disease and distant metastases. Oral Oncol.
39:207–212. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hu F, Min J, Cao X, Liu L, Ge Z, Hu J and
Li X: MiR-363-3p inhibits the epithelial-to-mesenchymal transition
and suppresses metastasis in colorectal cancer by targeting Sox4.
Biochem Biophys Res Commun. 474:35–42. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Penzo-Méndez AI: Critical roles for SoxC
transcription factors in development and cancer. Int J Biochem Cell
Biol. 42:425–428. 2010. View Article : Google Scholar :
|
|
7
|
Lee CJ, Appleby VJ, Orme AT, Chan WI and
Scotting PJ: Differential expression of SOX4 and SOX11 in
medulloblastoma. J Neurooncol. 57:201–214. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Castillo SD, Matheu A, Mariani N,
Carretero J, Lopez-Rios F, Lovell-Badge R and Sanchez-Cespedes M:
Novel transcriptional targets of the SRY-HMG box transcription
factor SOX4 link its expression to the development of small cell
lung cancer. Cancer Res. 72:176–186. 2012. View Article : Google Scholar
|
|
9
|
Liu P, Ramachandran S, Ali Seyed M,
Scharer CD, Laycock N, Dalton WB, Williams H, Karanam S, Datta MW,
Jaye DL and Moreno CS: Sex-determining region Y box 4 is a
transforming oncogene in human prostate cancer cells. Cancer Res.
66:4011–4019. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liao YL, Sun YM, Chau GY, Chau YP, Lai TC,
Wang JL, Horng JT, Hsiao M and Tsou AP: Identification of SOX4
target genes using phylogenetic footprinting-based prediction from
expression microarrays suggests that overexpression of SOX4
potentiates metastasis in hepatocellular carcinoma. Oncogene.
27:5578–5589. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Medina PP, Castillo SD, Blanco S,
Sanz-Garcia M, Largo C, Alvarez S, Yokota J, Gonzalez-Neira A,
Benitez J, Clevers HC, et al: The SRY-HMG box gene, SOX4, is a
target of gene amplification at chromosome 6p in lung cancer. Hum
Mol Genet. 18:1343–1352. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Koumangoye RB, Andl T, Taubenslag KJ,
Zilberman ST, Taylor CJ, Loomans HA and Andl CD: SOX4 interacts
with EZH2 and HDAC3 to suppress microRNA-31 in invasive esophageal
cancer cells. Mol Cancer. 14:242015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tiwari N, Tiwari VK, Waldmeier L, Balwierz
PJ, Arnold P, Pachkov M, Meyer-Schaller N, Schübeler D, van
Nimwegen E and Christofori G: Sox4 is a master regulator of
epithelial-mesenchymal transition by controlling Ezh2 expression
and epigenetic reprogramming. Cancer Cell. 23:768–783. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kang Y and Massagué J:
Epithelial-mesenchymal transitions: Twist in development and
metastasis. Cell. 118:277–279. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wu X, Zeng Y, Wu S, Zhong J, Wang Y and Xu
J: MiR-204, down-regulated in retinoblastoma, regulates
proliferation and invasion of human retinoblastoma cells by
targeting CyclinD2 and MMP-9. FEBS Lett. 589:645–650. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Voorhoeve PM: MicroRNAs: Oncogenes, tumor
suppressors or master regulators of cancer heterogeneity? Biochim
Biophys Acta. 1805:72–86. 2010.
|
|
19
|
Wu BH, Luo M, Zhang DH and Zhou X:
Deformation control and chatter suppression in 5-Axis milling of
thin-walled blade. Adv Mater Res. 188:314–318. 2011. View Article : Google Scholar
|
|
20
|
Shen Q, Cicinnati VR, Zhang X, Iacob S,
Weber F, Sotiropoulos GC, Radtke A, Lu M, Paul A, Gerken G and
Beckebaum S: Role of microRNA-199a-5p and discoidin domain receptor
1 in human hepatocellular carcinoma invasion. Mol Cancer.
9:2272010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nam EJ, Yoon H, Kim SW, Kim H, Kim YT, Kim
JH, Kim JW and Kim S: MicroRNA expression profiles in serous
ovarian carcinoma. Clin Cancer Res. 14:2690–2695. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
He XJ, Ma YY, Yu S, Jiang XT, Lu YD, Tao
L, Wang HP, Hu ZM and Tao HQ: Up-regulated miR-199a-5p in gastric
cancer functions as an oncogene and targets klotho. BMC Cancer.
14:2182014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Worley LA, Long MD, Onken MD and Harbour
JW: Micro-RNAs associated with metastasis in uveal melanoma
identified by multiplexed microarray profiling. Melanoma Res.
18:184–190. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhong J, Huang R, Su Z, Zhang M, Xu M,
Gong J, Chen N, Zeng H, Chen X and Zhou Q: Downregulation of
miR-199a-5p promotes prostate adenocarcinoma progression through
loss of its inhibition of HIF-1α. Oncotarget. 8:83523–83538. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Koshizuka K, Hanazawa T, Kikkawa N, Arai
T, Okato A, Kurozumi A, Kato M, Katada K, Okamoto Y and Seki N:
Regulation of ITGA3 by the anti-tumor miR-199 family inhibits
cancer cell migration and invasion in head and neck cancer. Cancer
Sci. 108:1681–1692. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
27
|
Kim SY, Nam SY, Choi SH, Cho KJ and Roh
JL: Prognostic value of lymph node density in node-positive
patients with oral squamous cell carcinoma. Ann Surg Oncol.
18:2310–2317. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bergers G, Brekken R, McMahon G, Vu TH,
Itoh T, Tamaki K, Tanzawa K, Thorpe P, Itohara S, Werb Z and
Hanahan D: Matrix metalloproteinase-9 triggers the angiogenic
switch during carcinogenesis. Nat Cell Biol. 2:737–744. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Natalwala A, Spychal R and Tselepis C:
Epithelial-mesenchymal transition mediated tumourigenesis in the
gastrointestinal tract. World J Gastroenterol. 14:3792–3797. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Dasgupta P, Kulkarni P, Majid S, Shahryari
V, Hashimoto Y, Bhat NS, Shiina M, Deng G, Saini S, Tabatabai ZL,
et al: MicroRNA-203 inhibits long noncoding RNA HOTAIR and
regulates tumorigenesis through epithelial-to-mesenchymal
transition pathway in renal cell carcinoma. Mol Cancer Ther.
17:1061–1069. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Imani S, Wei C, Cheng J, Khan MA, Fu S,
Yang L, Tania M, Zhang X, Xiao X, Zhang X and Fu J: MicroRNA-34a
targets epithelial to mesenchymal transition-inducing transcription
factors (EMT-TFs) and inhibits breast cancer cell migration and
invasion. Oncotarget. 8:21362–21379. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ko YH, Kim EJ, Sun DS, Won HS and Ahn YH:
Abstract 1466: QKI, a miRNA-200 target gene, suppresses
epithelial-to-mesenchymal transition in oral squamous cell
carcinoma cells. Cancer Res. 77:14662017.
|
|
33
|
Kim BK, Yoo HI, Kim I, Park J and Kim Yoon
S: FZD6 expression is negatively regulated by miR-199a-5p in human
colorectal cancer. BMB Rep. 48:360–366. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tsukigi M, Bilim V, Yuuki K, Ugolkov A,
Naito S, Nagaoka A, Kato T, Motoyama T and Tomita Y: Re-expression
of miR-199a suppresses renal cancer cell proliferation and survival
by targeting GSK-3β. Cancer Lett. 315:189–197. 2012. View Article : Google Scholar
|
|
35
|
Huang L, Lin JX, Yu YH, Zhang MY, Wang HY
and Zheng M: Downregulation of six MicroRNAs is associated with
advanced stage, lymph node metastasis and poor prognosis in small
cell carcinoma of the cervix. PLoS One. 7:e337622012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen J, Shin VY, Siu MT, Ho JC, Cheuk I
and Kwong A: miR-199a-5p confers tumor-suppressive role in
triple-negative breast cancer. BMC Cancer. 16:8872016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Nauseef JT and Henry MD:
Epithelial-to-mesenchymal transition in prostate cancer: Paradigm
or puzzle? Nat Rev Urol. 8:428–439. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yang J and Weinberg RA:
Epithelial-mesenchymal transition: At the crossroads of development
and tumor metastasis. Dev Cell. 14:818–829. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zeisberg M and Neilson EG: Biomarkers for
epithelial-mesenchymal transitions. J Clin Invest. 119:1429–1437.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhao X, He L, Li T, Lu Y, Miao Y, Liang S,
Guo H, Bai M, Xie H, Luo G, et al: SRF expedites metastasis and
modulates the epithelial to mesenchymal transition by regulating
miR-199a-5p expression in human gastric cancer. Cell Death Differ.
21:1900–1913. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hu Y, Liu J, Jiang B, Chen J, Fu Z, Bai F,
Jiang J and Tang Z: MiR-199a-5p loss up-regulated DDR1 aggravated
colorectal cancer by activating epithelial-to-mesenchymal
transition related signaling. Dig Dis Sci. 59:2163–2172. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kang M, Li Y, Liu W, Wang R, Tang A, Hao
H, Liu Z and Ou H: miR-129-2 suppresses proliferation and migration
of esophageal carcinoma cells through downregulation of SOX4
expression. Int J Mol Med. 32:51–58. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Huang YW, Liu JC, Deatherage DE, Luo J,
Mutch DG, Goodfellow PJ, Miller DS and Huang TH: Epigenetic
repression of microRNA-129-2 leads to overexpression of SOX4
oncogene in endometrial cancer. Cancer Res. 69:9038–9046. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhou X, Li L, Su J and Zhang G: Decreased
miR-204 in H. pylori-associated gastric cancer promotes cancer cell
proliferation and invasion by targeting SOX4. PLoS One.
9:e1014572014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yeh YM, Chuang CM, Chao KC and Wang LH:
MicroRNA-138 suppresses ovarian cancer cell invasion and metastasis
by targeting SOX4 and HIF-1α. Int J Cancer. 133:867–878. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wang J, Xu G, Shen F and Kang Y: miR-132
targeting cyclin E1 suppresses cell proliferation in osteosarcoma
cells. Tumour Biol. 35:4859–4865. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Li S, Qin X, Li Y, Zhang X, Niu R, Zhang
H, Cui A, An W and Wang X: MiR-133a suppresses the migration and
invasion of esophageal cancer cells by targeting the EMT regulator
SOX4. Am J Transl Res. 7:1390–1403. 2015.PubMed/NCBI
|