Introduction
In recent years, there has been an increase in the
prevalence of and financial burden associated with allergic
disorders, including bronchial asthma, allergic rhinitis and atopic
dermatitis (1). Remarkably, more
than half of diagnosed allergic diseases are attributed to house
dust mite (HDM) allergens (2).
The HDM species Dermatophagoides pteronyssinus (Der p) and
Dermatophagoides farinae (Der f) are the main indoor sources
of inhaled allergens triggering IgE-mediated anaphylactic
reactions. HDM is the most common cause of respiratory allergy
worldwide. (2). The rate of HDM
allergy is high in developed countries; previous clinical studies
showed that HDM allergic sensitization is >20% in Europe
(3) and ≤40% in North America
(4). In-depth analysis of HDM
allergens is required to elucidate the mechanism of HDM allergen
sensitization and to inform the development of diagnosis and
treatment for HDM allergies (5).
Of the 39 HDM allergen groups that have been
identified (6), Group I and Group
II HDM allergen reactivity are the most prevalent, followed by
Group 23 and Group 24 (7-9). Hence, Group I and II HDM allergens
are considered to be the major allergens of HDM. Cysteine protease
activity of the Group I HDM allergen Der p 1 has been shown to
enhance the immunoglobulin E (IgE) antibody response selectively
(10), and Der p 1 proteolytic
activity leads to augmented IgE reactivity to Der p 1 itself as
well as to other allergens in the microenvironment (11). Furthermore, Der p 2 has been shown
to induce inflammatory allergenic effects via binding of Toll-like
receptor 4 (TLR4) (12,13). Thus, Group 1 and 2 HDM allergens
exhibit IgE-binding activity with sera from most HDM-allergic
patients (14) and have been
shown to induce T helper type 2 (Th2) immune responses through
cysteine protease functions (15)
and facilitation of TLR4 signaling (12), respectively. However, the
mechanisms mediating the allergic reactions triggered by most HDM
allergens, which may vary substantially from group to group, have
not yet been resolved.
IgE interaction with exogenous allergens promotes
mast cell degranulation, which induces inflammation (16). The domain of each allergen bound
by serum IgEs are known as B cell IgE-binding epitopes (17). Analysis of B cell IgE epitopes can
provide good indicators for allergy diagnosis, prediction of the
clinical severity of allergic diseases and monitoring of the
development of allergen tolerance (18). B cell epitopes for Group 1, 2, 3,
7, 11, 13 and 33 HDM allergens have been identified with
peptide-displaying phage and artificial synthetic peptide scanning
technologies (19-23). In our previously study, the Group
24 HDM allergen, Der f 24, was found to be a major HDM allergen and
to function as a ubiquinol cytochrome c reductase binding protein
(UQCRB) homolog (9). In a
multitude of species, UQCRB proteins play an important role in the
maintenance of mitochondrial complex III for electron transport and
cellular oxygen sensing (24).
The only UQCRB proteins that have been reported to exhibit
allergenic activity are those from Der f and Der p (25). The Der f and Der p mite species
contribute differently to HDM induced allergic disease, potentially
due to their differing geographical distributions (26) or inherent characteristic
differences between them (18,27). Importantly, it has been shown
previously that Der p 24 exhibits strong IgE-binding activity via
an immuno-dominant IgE epitope in its N-terminal 32-residue region
(25); however, the dominant IgE
epitope of Der f 24 may not be the same as that of its homolog Der
p 24, particularly given the differing protein sequences of the two
allergens. It remains to be determined how the allergenic
properties of the UQCRB protein in Der f differs from and/or
resembles the Der p UQCRB protein.
Detailed knowledge about the epitopes responsible
for IgE-binding of allergenic peptides can help to inform allergy
diagnosis and prognosis, and may facilitate the rational design of
hypoallergenic candidate immunotherapeutic vaccines. The Der f 24
UQCRB protein homolog shows strong IgE reactivity with serum from
HDM-allergic patients in vitro and in vivo,
indicating that Der f 24 may play a key role in HDM sensitization,
and thus, may have potential for the improvement of diagnosis and
treatment (9).
The mechanism by which UQCRB protein homologs affect
allergic reactions has not been resolved, and an analysis of the
immunodominant IgE epitope of Der f 24 has yet to be reported.
Therefore, the aim of the present study was to identify and
characterize the IgE-binding epitope of Der f 24 using IgE-western
blotting, enzyme-linked immunosorbent assays (ELISAs) and IgE-dot
blotting. The IgE epitope immuno-dominance of Der f 24 was explored
with IgE-binding activity assays with Der f-human UQCRB hybrid
proteins. Finally, homology modeling was used to predict the
structural position of the IgE-binding site within Der f 24.
Materials and methods
Serum samples
Serum samples from 30 individuals with HDM allergy
(13 males and 17 females; age range, 18-55 years) and 24
non-allergic individuals (10 males and 14 females; age range, 18-55
years) were obtained from the First Affiliated Hospital of
Guangzhou Medical College (Guangdong, China). The allergic patient
sample consisted entirely of individuals who had experienced
anaphylaxis caused by HDM exposure and who had IgE levels >3,
measured using a clinical ImmunoCAP allergen detection system
(Phadia; Thermo Fisher Scientific, Inc.). Patient
clinicopathological information, including sex, age, ImmunoCAP
HDM-specific IgE levels and previous diagnoses are presented in
Table I. Ethics approval for the
current study was obtained from the First Affiliated Hospital of
Guangzhou Medical College (approval no. 2012-51). Informed consent
was obtained from all individual participants included in the
study. All procedures involving human participants were in
accordance with the ethical standards of the committee. Sera pooled
from 10 patients with HDM allergy were used to detect the ability
of IgE-binding to recombinant Der (rDer) f 24 and hybrid
proteins.
 | Table IClinicopathological characteristics
of patients with HDM allergy. |
Table I
Clinicopathological characteristics
of patients with HDM allergy.
| Patient ID | Sex | Age, years | ImmunoCAP
HDM-specific IgE testa | Clinical
history |
|---|
| 1 | Female | 23 | 3 | - |
| 2 | Female | 18 | 3 | BA |
| 3 | Male | 35 | 3 | AR |
| 4 | Female | 51 | 4 | BA |
| 5 | Male | 29 | 4 | AR |
| 6 | Female | 36 | 4 | AR |
| 7 | Male | 52 | 4 | AR |
| 8 | Male | 19 | 5 | BA |
| 9 | Female | 47 | 5 | AR+BA |
| 10 | Male | 28 | 5 | - |
| 11 | Male | 34 | 5 | AR+BA |
| 12 | Female | 53 | 6 | AR |
| 13 | Female | 21 | 6 | AR+BA |
| 14 | Male | 43 | 6 | BA |
| 15 | Female | 55 | 6 | AR+BA |
Animals
Female BALB/c mice (4-5 weeks old) were purchased
from Guangdong Medical Laboratory Animal Center (Foshan, China),
and housed in a specific pathogen-free environment with a
relatively stable temperature (24 ± 1°C) and humidity (55 ± 10%)
for >1 week before experimentation. All studies involving mice
were approved by the Animal Ethical and Welfare Committee of the
School of Medicine of Shenzhen University (Guangdong, China).
Analysis of sequence homology
The amino acid (aa) sequences of Der f 24 (GenBank
no. AJK91617.1), Der p 24 (GenBank no. KP893174.1), human UQCRB
(GenBank no. EAW91749) and mouse UQCRB (GenBank no. NM_026219) were
obtained from the National Center for Biotechnology Information
library (NCBI; https://www.ncbi.nlm.nih.gov/protein/) and imported
into DNAMAN 8 software (version 8; Lynnon LLC) for homology
comparison. FASTA-format sequences were obtained for further
analysis.
Construction of vectors and expression
and purification of hybrid proteins
Human UQCRB has not been reported to have IgE
epitopes that induce an autoimmune response. Hybrid Der f and human
UQCRB protein sequences (Table
II) were designed and the resultant cDNA sequences were
synthesized by GenScript (Nanjing) Co. Ltd. The following synthetic
genes were registered in the GenBank database: Der f 24-Hyb1 [mite,
1-59 aa; human, 60-111 aa (accession no. KP939229)]; Der f 24-Hyb2
[human, 1-59 aa; mite, 60-118 aa (accession no. KP939230)]; Der f
24-Hyb3 [mite, 1-78 aa; human, 79-111 aa (accession no. KP939231)];
and Der f 24-Hyb4 [mite, 1-39 aa; human, 40-111 aa (accession no.
KP939232)]. These cDNAs were cloned into pET-His vectors (Miaoling
Bioscience and Technology Co., Ltd.). The recombinant plasmids were
confirmed by DNA sequencing, and for protein expression, 100 ng DNA
was transformed into Escherichia coli BL21 (DE3) pLysS
competent cells (Novagen; Merck KGaA) by heat shock. The expression
and purification of recombinant protein was performed as described
previously (28). The rDer f 24
protein and the hybrid proteins were isolated in the form of
inclusions. They were purified by Ni-NTA gel affinity
chromatography (GE Healthcare) and subjected to IgE-western
blotting or IgE-dot blotting. rDer f 24 protein quality was
evaluated by determining the activity of IgE-binding with
HDM-allergic sera, and required a positive rate of ~50% using 10
individual HDM-allergic sera in IgE-ELISA (9). The recombinant protein
concentrations were determined using the Bradford method (Bio-Rad
Laboratories, Inc.). The recombinant proteins (40 μg/well)
were analyzed using sodium dodecyl sulfate-polyacrylamide gel
electrophoresis (SDS-PAGE) and the gel was stained with Coomassie
Brilliant Blue R-250 Staining Solution (Bio-Rad Laboratories,
Inc.).
 | Table IISummary of aa sequence constitution
of hybrid proteins. |
Table II
Summary of aa sequence constitution
of hybrid proteins.
| Name | Aa sequence | No. aa |
|---|
| Der f 24-Hyb1 |
MVHLTKTLRFINNPGFRKFYYGLQGYNKYGLYYDDFYDYT
DAAHLEAV RRLPPDLYDQHMFRIKRALDLNLKHQILPKE
QWTKYEEENFYLEPYLKEVIRERKEREEWAKK | 111 aa (mite,1-59
aa; human, 60-111 aa) |
| Der f 24-Hyb2 |
MAGKQAVSASGKWLDGIRKWYYNAAGFNKLGLMRDDTI
YEDEDVKEAIRRLPENLYNDRTYRL VRASQLEITKQFLPKE
QWPSYEEDMDKGRFLTPYLDEVMKEKKEKEEWINFLSKD | 118 aa (human, 1-59
aa; mite, 60-118 aa) |
| Der f 24-Hyb3 |
MVHLTKTLRFINNPGFRKFYYGLQGYNKYGLYYDDFYD
YTDAAHLEAVRRLPPDLYDQHTYR LVRASQLEITKQFLPK
EQWTKYEEENFYLEPYLKEVIRERKEREEWAKK | 111 aa (mite, 1-78
aa; human, 79-111 aa) |
| Der f 24-Hyb4 |
MVHLTKTLRFINNPGFRKFYYGLQGYNKYGLYYDDFYDYE
DEDVKEAIRRLPENLYNDRMFRIKRALDLNLKHQILPKEQ
WTKYEEENFYLEPYLKEVIRERKEREEWAKK | 111 aa (mite, 1-39
aa; human, 40-111 aa) |
Synthesis of overlapping
polypeptides
Five overlapping Der f 24 polypeptides (31-33 aa)
were designed and synthesized by Shanghai Qiang Yao Biotechnology
Co. Ltd. The overlapping regions were 10 aa long. The polypeptides
were coupled with bovine serum albumin (BSA; Amresco, LLC) or
ovalbumin (OVA; Sigma-Aldrich; Merck KGaA) at the cysteine residue
of the N- or C-terminus using a 3-maleimidobenzoic acid
N-hydroxysuccinimide method, which was performed by Shanghai Qiang
Yao Biotechnology Co. Ltd.
IgE western blotting and IgE dot
blotting
rDer f 24 and hybrid proteins were separated by 12%
SDS-PAGE and transferred to polyvinylidene difluoride (PVDF)
membranes (EMD Millipore) for western blotting. For dot blotting, 2
μl rDer f 24 (concentration 1 μg/μl) and
synthetic polypeptides (BSA-1-32 aa, BSA-23-54 aa, BSA-45-76 aa,
BSA-67-98 aa and BSA-89-118 aa) were dotted onto PVDF membranes.
The membranes were blocked with 5% Difco™ skim milk (DSM; BD
Biosciences) diluted in Tris buffered saline containing 0.05%
Tween-20 at 4°C overnight. Western blot membranes were incubated
with a mixture of allergic sera from different 10 patients with HDM
allergy (1:10 dilution) for 2 h at 37°C. For dot blotting, a mix of
10 serum samples from HDM-allergic patients with high IgE-binding
to rDer f 24 and a mix of 3 non-allergic sera samples (1:10
dilution) were incubated for 2 h at 37°C. Following washing, the
membranes were incubated with horse radish peroxidase-conjugated
mouse anti-human IgE antibody (1:2,000 dilution; cat. no. 9160-05;
SouthernBiotech) for 1.5 h at 37°C. Antigen-antibody complexes on
membranes were visualized using a Pierce™ 3'-diaminobenzidine
substrate kit (Thermo Fisher Scientific, Inc.).
IgE-ELISA
rDer f 24 protein and conjugated polypeptides (200
ng/well; diluted in carbonate buffer; pH 9.6) were coated onto
96-well plates, and incubated at 4°C overnight. The plates were
blocked with 5% DSM in phosphate-buffered saline containing 0.05%
Tween-20 (PBST) for 3 h at 37°C. Washed plates were incubated with
sera (1:10 dilution) from 15 HDM-allergic patients and 15
non-allergic individuals for 2 h at 37°C. Serum IgEs were then
detected by incubating with a mouse anti-human IgE horseradish
peroxidase-conjugated antibody (1:2,000 dilution; SouthernBiotech)
for 1.5 h at 37°C. The plates were washed with PBST buffer. The
reaction was detected by adding 100 μl
3,3,5,5′-tetramethylbenzidine substrate (1 mM; Invitrogen; Thermo
Fisher Scientific, Inc.), and absorbance was measured at 450 nm
using a microplate reader (Bio-Rad Laboratories, Inc.). After
screening, the 15 HDM allergic sera all displayed positive reaction
to allergen Der f 24.
Sensitization of mice
Female BALB/c mice (6-8 weeks of age; 17-20 g in
weight) were purchased from Guangzhou Experimental Animal Center
(Guangdong, China). The mice (n=3) were sensitized by
intraperitoneal injection of 50 μg OVA or OVA-1-32 aa
conjugate with Imject™ Alum adjuvant (2:1 v/v in total volume 200
μl; Thermo Fisher Scientific, Inc.). On day 21 after
injection, the mice were euthanized by cervical dislocation under
deep isoflurane anesthesia and the sera were collected. Serum
levels of specific IgE and IgG targeting the BSA-1-32 conjugate
were measured by ELISA as described previously (29). The health status of the
experimental mice was monitored twice daily and humane endpoints
were used to determine if the mice met criteria to be euthanized.
These criteria included weight loss >10-15%, lethargy, inability
to stand and anorexia. Mice that met the designated criteria were
euthanized by cervical dislocation under deep isoflurane
anesthesia.
RBL-2H3 degranulation assay
The rat mast cell leukemia cell line, RBL-2H3
(Guangzhou Cellcook Biotech Co., Ltd.) was cultured in complete
Dulbecco's modified eagle medium with 4.0 mM L-glutamine, sodium
pyruvate penicillin (100 U/ml), 100 μg/ml streptomycin,
non-essential amino acids and 10% fetal bovine serum (Gibco; Thermo
Fisher Scientific, Inc.) in a humidified incubator at 37°C and 5%
CO2. A cell degranulation assay was performed as
previously described (30). The
RBL-2H3 mast cell cells were seeded in 96-well plates for 18 h.
After washing three times using Tyrode's buffer (Beijing Solarbio
Science & Technology, Co., Ltd.), cells were exposed to sera of
sensitized mice for 2 h and then stimulated with 10 μg/ml
BSA-1-32aa conjugate for 45 min. Degranulation was assessed by
measurement of β-hexosaminidase release into the supernatant and of
unreleased enzyme in the respective cell lysate.
Der f 24 3D structural modeling
The structure of cytochrome b-c1 complex subunit 7
(QCR7, also known as UQCRB) template from Bos Taurus was
obtained from the RCSB Protein Data Bank (http://www.rcsb.org/; PDB code, 5klv.1), and was used
as template for 3D modeling of the Der f 24 protein. The 3D
structure of Der f 24 was modeled in SWISS-MODEL online software
(http://swissmodel.expasy.org/) based on
the bovine QCR7 template. Ramachandran plots and root mean square
deviation (RMSD) structure assessment methods were used to assess
the reliability of the predicted 3D model using the PDBsum database
(http://www.ebi.ac.uk/pdbsum/) which
specializes in structural analyses and pictorial representations of
models. The loci of 5 synthetic polypeptides in the Der f 24
protein were indicated with color-coding for IgE-binding activity
analysis. The key residues in the 1-32 aa domain region of Der f 24
were evaluated using the Immune epitope database (IEDB) prediction
online tool (https://www.iedb.org/) (31). Amino acid property scales for
predicting antigenic determinants were developed using the BepiPred
Linear Epitope Prediction tool (version 1.0; http://www.cbs.dtu.dk/services/bepipred/).
Statistical analysis
Experimental data are presented as mean ± SD.
Statistical significance was calculated using GraphPad Prism 7
(GraphPad Software, Inc.). Group differences were determined by
analysis of variance followed by Dunnett's test for multiple
comparisons. P<0.05 were considered to indicate a statistically
significant difference.
Results
Low sequence homology of UQCRB protein
between Der f and Homo sapiens
The Der f 24 gene encodes a protein sequence of 118
aa and is registered in the NCBI library. As presented in Fig. 1, a homology comparison in DNAMAN 8
showed Der f 24 was highly homologous to Der p 24 (96.61%). Human
UQCRB protein was highly homologous to mouse UQCRB (86.61%).
However, low aa homology was observed between Der f 24 and the
human UQCRB protein (39.34%).
IgE-binding of Der f 24 and hybrid
proteins
Pooled samples of serum from patients with HDM
allergy were found to have improved IgE specificity compared to
individual samples in preliminary experiments. Therefore, pooled
serum samples were used for screening the immunodominant IgE
epitope of the allergen. The Der f and human components of purified
rDer f 24, hybrid Der f proteins and human UQCRB protein are
presented in Fig. 2A. IgE-western
blotting was performed on these proteins by probing with sera
pooled from HDM-allergic patients. All of the recombinant proteins
except for Der f 24-Hyb2, which lacked the N-terminal 59-residue
region of Der f 24, showed positive IgE-binding (Fig. 2B and C).
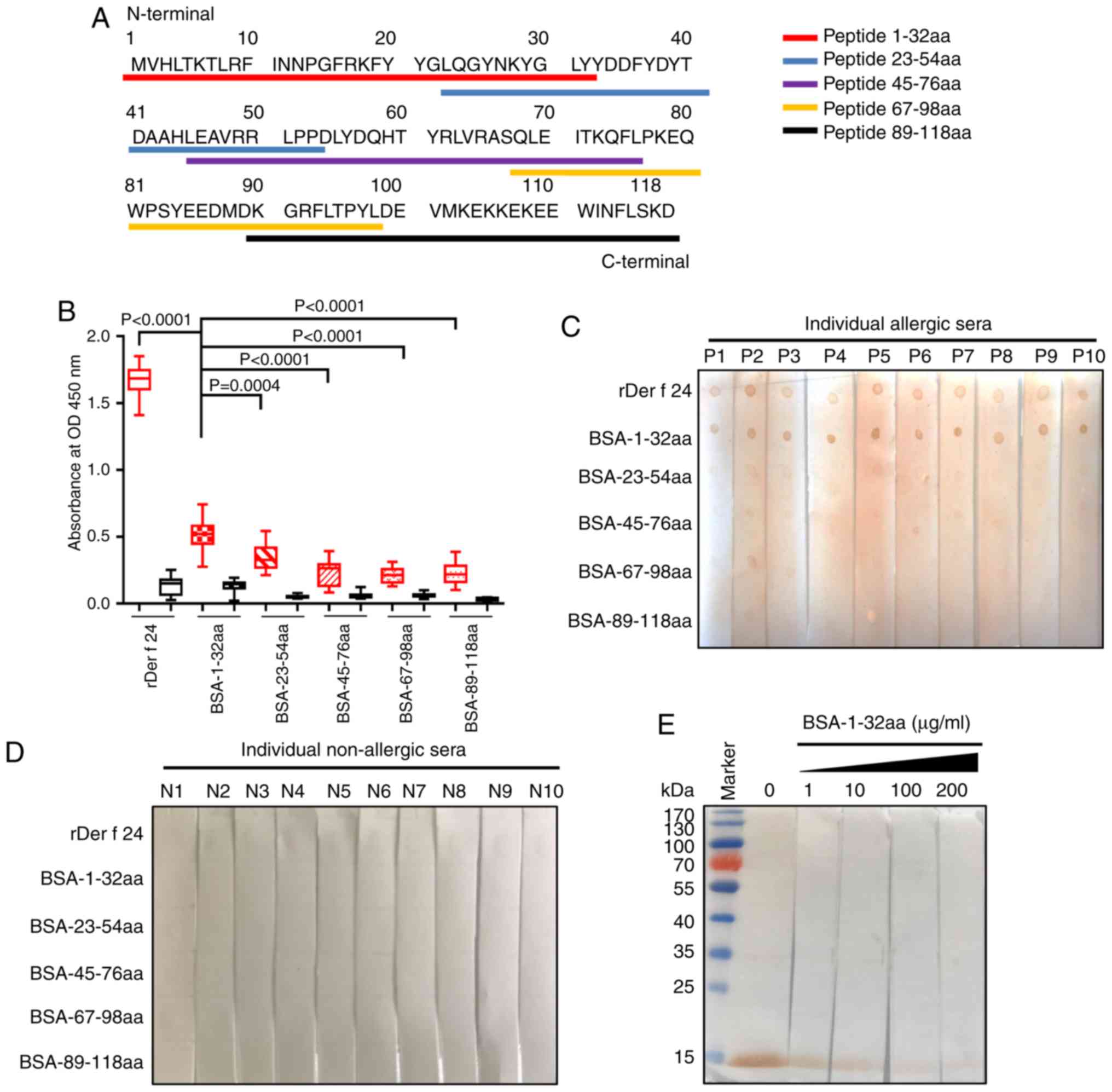
Identification of the Der f 24 B cell
epitope in the N-terminal 32-residue region
The IgE epitopes of Der f 24 were further
investigated using overlapping synthetic polypeptides coupled with
BSA at the N- or C-terminus (Fig.
3A; Table II). IgE-ELISA
results revealed that the 1-32 aa peptide group had a stronger
binding to HDM allergic-serum IgE compared with that for the other
four groups (Fig. 3B). Likewise,
IgE-dot blotting showed that the BSA-1-32 aa group had a robust
positive reaction to pooled sera from 10 allergic individuals,
while the other groups did not have a strong positive reaction
(Fig. 3C and D). The rDer f 24
protein was subjected to SDS-PAGE and transferred to PVDF membrane
for competitive IgE western blotting. With the addition of
increasing concentrations of the BSA-1-32aa conjugate, the capacity
of rDer f 24 binding to IgE from HDM allergic sera became weaker
(Fig. 3E). These results
suggested that the B cell epitope of Der f 24 may be located mainly
in the N-terminal 32-residue region.
Peptide 1-32 aa domain of Der f 24
produces high anti-body titers in sensitized mice
To further explore B cell epitope immunogenicity,
the titers of IgGs or IgEs targeting the BSA-1-32 aa conjugate were
detected mice sensitized with OVA- or OVA-1-32aa conjugate, as OVA
induces a Th2 response in vivo. At 3 weeks
post-sensitization, the OVA-1-32 aa group produced high specific
IgG and IgE antibody titers against the BSA-1-32 aa (Fig. 4A and B), confirming a role for the
N-terminal 1-32 region of Der f 24 in IgE-binding activity.
Importantly, serum from mice immunized with OVA-1-32aa, but not
OVA-injected mice, promoted significant release of β-hexosaminidase
from the mast cell line, RBL-2H3 with the addition of BSA-1-32 aa
conjugate (Fig. 4C).
Additionally, a competitive IgE-ELISA showed that pooled serum
samples of the OVA-1-32 aa-sensitized mice reduced the ability of
IgE in serum pooled from 10 HDM allergic patients to bind to the
BSA-1-32 aa antigen (Fig. 4D).
These data indicated that the 1-32 aa domain of Der f 24 contains
the immunodominant IgE epitopes of Der f 24.
Structural location analysis of IgE
epitope of Der f 24
Superimposing the 3D structural model of Der f 24 on
a bovine QCR7 template revealed sequence identity between the
template and Der f 24 of 46.60%. Ramachandran plots and RMSD
assessment showed that none of the residues were present in the
disallowed region, 88 residues were present in the most favored
regions, and the overall G-factor of the Der f 24 structure model
was -0.01 (Fig. S1). In
addition, structural analysis of homology modelling using the
SWISS-MODEL system showed that the 3D structure of Der f 24 was
highly similar to that of the QCR7 template (Fig. S2). The RMSD assessment indicated
that there were 118 aligned residues in the superimposition (RMSD
value, 0.207). These results indicate that the backbone dihedral
angles, phi and psi, of the 3D model were within the acceptable
range.
Homology modeling in the SWISS-MODEL software
revealed that the secondary structure of Der f 24 consisted of
three α-helices and a free loop (Fig.
5). The structure was positioned on the outside of whole folded
protein. This external location may facilitate IgE-binding.
According to the results of IEDB prediction, lysine (Lys) 28 and
tyrosine (Tyr) 29 residues were located on the α-helices and were
important for antigen-binding (Table III; Fig. S3). As the peptide 1-32 aa region
(red domain in Fig. 5) of Der f
24 was located in a protruding region of the structure, the
N-terminal 32-residue region may be more easily accessible for
binding by specific IgEs.
 | Table IIIBepiPred Linear Epitope Prediction
scores of each residue in the N-terminus peptide of Der f 24
(residues 1-32). |
Table III
BepiPred Linear Epitope Prediction
scores of each residue in the N-terminus peptide of Der f 24
(residues 1-32).
| Position | Amino acid | Score |
|---|
| 1 | M | −1.518 |
| 2 | V | −1.238 |
| 3 | H | −0.99 |
| 4 | L | −1.215 |
| 5 | T | −1.172 |
| 6 | K | −1.325 |
| 7 | T | −1.327 |
| 8 | L | −1.042 |
| 9 | R | −0.495 |
| 10 | F | −0.401 |
| 11 | I | −0.221 |
| 12 | N | −0.383 |
| 13 | N | −0.063 |
| 14 | P | 0.007 |
| 15 | G | 0.076 |
| 16 | F | 0.234 |
| 17 | R | −0.095 |
| 18 | K | −0.281 |
| 19 | F | −0.628 |
| 20 | Y | −0.694 |
| 21 | Y | −0.489 |
| 22 | G | −0.488 |
| 23 | L | −0.308 |
| 24 | Q | 0.104 |
| 25 | G | 0.131 |
| 26 | Y | 0.347 |
| 27 | N | 0.024 |
| 28 | K | 0.129 |
| 29 | Y | −0.004 |
| 30 | G | −0.131 |
| 31 | L | −0.169 |
| 32 | Y | −0.487 |
Discussion
In the present study, the immunodominant IgE epitope
of Der f 24 was identified using IgE-binding activity assays with
Der f-human UQCRB hybrid proteins. The N-terminal region of Der f
24 displayed strong IgE-binding activity and induced high titers of
specific IgGs and IgEs in sensitized mice. Additionally, the
N-terminal region of Der f 24 induced IgE production in
vivo. Homology modeling revealed the structural orientation of
this B cell epitope of Der f 24. The identification of the
immunodominant IgE epitope of Der f 24 has important implications
for HDM allergy diagnosis and treatment.
As allergen-specific IgE antibodies play a crucial
role in original immune responses, IgE epitopes of allergens have
been investigated (32). The B
cell epitope of the allergen can be defined in a linear or
conformational form (33,34). Although an allergen may have
multiple types of epitopes, IgE-binding of an allergen is highly
dependent on its immunodominant epitope (35). For example, the Der p 5 allergen
has the linear sequence 90-DRLMQRKDLDIFEQYNLEM-108 and consists
conformationally of a bundle of three anti-parallel α-helices
(36). Although the Group 11
allergenic Blomia tropicalis paramyosin protein contains
multiple IgE- and IgG-immunoreactive regions scattered throughout
the molecule, its dominant IgE- and IgG-binding regions have been
mapped to the aa positions 336-557 and 698-875, respectively
(37). Based on the present
results, it is reasonable to conclude that Der f 24 also has
multiple IgE-binding epitopes, of which the immunodominant IgE
epitope would be important in HDM allergy development, and
therefore was investigated in this study.
The ubiquitous presence of UQCRB homologs across
diverse species, including HDMs and humans, suggest a highly
conserved biological functionality (38). Der f 24 was found to be highly
homologous to Der p 24, but to share a relatively low sequence
homology with human UQCRB, which was expected based on the
selective allergenicity of the HDM UQCRB homologs. Conversely,
human and mouse UQCRB were found to be highly homologous. Among the
hybrid proteins tested for IgE-binding in the current study, only
the Der f 24-Hyb2 protein that lacked the N-terminal 59-residue
region of Der f 24 was not immunoreactive. All of the other
synthetic hybrid protein variants were immunoreactive, suggesting
strongly that the B cell epitope of Der f 24 is located, at least
mostly, in the N-terminal 59-residue region, where a 37.29%
sequence homology (22 out of 59 aa) was found between Der f and
human UQCRB. This sequence divergence is likely to underlie, at
least to some extent, the IgE-binding activity of Der f 24. In the
present study, activity assays using the rDer f 24 protein as a
UQCRB-like protein were hard to perform due to quality control of
the recombinant protein. The rDer f 24 protein required a positive
rate of ~50% using 10 individual HDM allergic serum samples for
IgE-ELISA according to our previous study (9).
The identification of IgE immunodominant epitopes of
HDM allergens can elucidate the mechanisms of allergen-induced
sensitization and provide useful information for the prevention and
treatment of allergic diseases. A study by Greene and Thomas
(39) found five Der p 1 linear
IgE epitopes (aa 15-33, 60-80, 81-94, 101-111 and 155-187).
Furthermore, a study by Van't Hof et al (40) identified an immunodominant IgE
epitope in Der p 2 that was located near the N terminus (aa 65-78),
similar to the IgE epitope in Der f 24 identified in the present
study. Previously, peptide-displaying phage technology and
artificial synthesized peptide scanning technology have been used
to identify immunodominant epitopes (37,40). Additionally, our previous study
showed that the immunodominant IgE epitopes of Der p 24 located in
the N-terminal 32-residue region triggered IgE production in
vivo (25). In the present
study, artificial hybrid proteins and synthesized polypeptides were
used to identify the N-terminal region of Der f 24 as a major
mediator of the IgE-binding activity of Der f 24.
OVA can be used as a carrier protein, like BSA, to
enhance immune response activation. Unlike BSA, OVA generates a
strong Th2 response in vivo (41). The N-terminal 1-32 aa
region of the Der f 24-OVA conjugates produced specific IgE
responses in vivo in the current study. Notably, structural
modeling showed that the N-terminal 32 residues of Der f 24 form a
structural protrusion that may be conducive to specific
IgE-binding. Mutation experiments are required to investigate the
importance of the Lys 28 and Tyr 29 residues of Der f 24 for
IgE-binding. The results of the present study were similar to that
of homolog Der p 24, indicating that the major allergen Group 24 in
HDM may share a similar dominant IgE epitope and produce similar
clinical significance (25).
At present, allergen immunotherapy is an effective
way of treating IgE-mediated allergies (42). As a single epitope often may not
perform well for allergic diagnosis or therapy, HDM whole protein
extract is the preferred choice during clinical application
(7). Identification of more
immunodominant IgE epitopes of HDM allergens is required for
further refinement. Hence, elucidation of the IgE epitopes of
allergens can serve to facilitate the rational design of
hypoallergenic candidate vaccine immunotherapies for allergies
(43). Natural HDM allergen
extracts and vaccines based on these complex molecular extracts are
known to produce adverse secondary effects (44,45). Genetic recombinant hybrid proteins
have emerged as another approach to immunotherapy development
(46). Good outcomes of such
immunotherapy approaches are dependent upon clarification of
immunodominant epitopes of allergens (45). Carrier-bound HDM Group 1
allergen-derived peptides have been produced for HDM allergen
immunotherapy development (47).
A previous study demonstrated that recombinant hybrid proteins
consisting of reassembled Der p 1 and Der p 2 fragments can be used
as a safe hypoallergenic molecule for achieving HDM allergen
tolerance and vaccination (2).
Thus, the identification of the immunodominant epitope of Der f 24
contributes to the theoretical foundation for developing a
hypoallergenic vaccine for HDM allergies.
In conclusion, the results of the present study
suggest that the immunodominant IgE epitope of Der f 24 is located
mainly in the N-terminal 32 aa region. These findings may help to
elucidate the mechanisms of HDM allergy sensitization. Based on the
finding of the current study and previously published studies, HDM
allergens have the potential to induce allergic reactions by way of
protein functions or by interaction with IgE B cell epitopes.
Supplementary Data
Acknowledgments
We thank the other members of Kunmei Ji's lab for
their critical comments and, we thank the Department of Allergy and
Clinical Immunology of the First Affiliated Hospital of Guangzhou
Medical College (Guangdong, China) for supporting serum sample
collection.
Abbreviations:
|
HDM
|
house dust mite
|
|
Der f 24
|
group 24 allergen of
Dermatophagoides farina
|
|
UQCRB
|
ubiquinol-cytochrome c reductase
binding protein
|
|
IgE
|
immunoglobulin E
|
|
ELISA
|
enzyme-linked immunosorbent assay
|
|
IEDB
|
immune epitope database
|
Funding
The present study was supported in part by research
funding from the National Natural Science Foundation of China
(grant. no. 81571570), Guangdong Province (grants. no.
2016A020215176, 2016A030313039, 2017A010105014 and 2018A050506083),
and Shenzhen City 2016 Biochemistry Discipline Construction.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
CJJ and JKM conceived and designed the study. ZLC
and JJC analyzed the data. WLL, ZLC, ZZ, YBH and YSH performed the
studies. JJC and KJ contributed reagents materials and analysis
tools. JJC, ZZ and KJ wrote the manuscript.
Ethics approval and consent to
participate
Permission to conduct this study was obtained from
the Ethics Committee of the First Affiliated Hospital of Guangzhou
Medical College (no. 2012-51). Informed consent was obtained from
all individual participants included in the study. All procedures
involving human participants were in accordance with the ethical
standards of the committee.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
D'Amato G, Liccardi G, D'Amato M and
Holgate S: Environmental risk factors and allergic bronchial
asthma. Clin Exp Allergy. 35:1113–1124. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ciprandi G, Puccinelli P, Incorvaia C and
Passalacqua G: Italian Cometa Study Group: The relevance of house
dust mites allergy in clinical practice: The epidemiological impact
on allergen immunotherapy. Immunotherapy. 9:1219–1224. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bousquet PJ, Chinn S, Janson C, Kogevinas
M, Burney P and Jarvis D: European Community Respiratory Health
Survey I: Geographical variation in the prevalence of positive skin
tests to environmental aeroallergens in the European community
respiratory health survey I. Allergy. 62:301–309. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chew GL, Reardon AM, Correa JC, Young M,
Acosta L, Mellins R, Chew FT and Perzanowski MS: Mite sensitization
among Latina women in New York, where dust-mite allergen levels are
typically low. Indoor Air. 19:193–197. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang L and Zhu R: Immunotherapy of house
dust mite allergy. Hum Vaccin Immunother. 13:2390–2396. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pomés A, Davies JM, Gadermaier G, Hilger
C, Holzhaouser T, Lidholm J, Lopata AL, Mueller GA, Nandy A,
Radauer C, et al: WHO/IUIS allergen nomenclature: Providing a
common language. Mol Immunol. 100:3–13. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Thomas WR, Smith WA, Hales BJ, Mills KL
and O'Brien RM: Characterization and immunobiology of house dust
mite allergens. Int Arch Allergy Immunol. 129:1–18. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Weghofer M, Grote M, Resch Y, Casset A,
Kneidinger M, Kopec J, Thomas WR, Fernández-Caldas E, Kabesch M,
Ferrara R, et al: Identification of Der p 23, a peritrophin-like
protein, as a new major Dermatophagoides pteronyssinus allergen
associated with the peritrophic matrix of mite fecal pellets. J
Immunol. 190:3059–3067. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chan TF, Ji KM, Yim AK, Liu XY, Zhou JW,
Li RQ, Yang KY, Li J, Li M, Law PT, et al: The draft genome,
transcriptome, and microbiome of Dermatophagoides farinae reveal a
broad spectrum of dust mite allergens. J Allergy Clin Immunol.
135:539–548. 2015. View Article : Google Scholar
|
|
10
|
Gough L, Schulz O, Sewell HF and Shakib F:
The cysteine protease activity of the major dust mite allergen Der
p 1 selectively enhances the immunoglobulin E antibody response. J
Exp Med. 190:1897–902. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gough L, Sewell HF and Shakib F: The
proteolytic activity of the major dust mite allergen Der p 1
enhances the IgE antibody response to a bystander antigen. Clin Exp
Allergy. 31:1594–1598. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Trompette A, Divanovic S, Visintin A,
Blanchard C, Hegde RS, Madan R, Thorne PS, Wills-Karp M, Gioannini
TL, Weiss JP and Karp CL: Allergenicity resulting from functional
mimicry of a Toll-like receptor complex protein. Nature.
457:585–588. 2009. View Article : Google Scholar
|
|
13
|
Park BS, Lee NR, Kim MJ, Kim SY and Kim
IS: Interaction of Der p 2 with Toll-like receptor 4 and its effect
on cytokine secretion. Biomed Sci Letters. 21:152–159. 2015.
View Article : Google Scholar
|
|
14
|
Hales BJ, Martin AC, Pearce LJ, Laing IA,
Hayden CM, Goldblatt J, Le Souef PN and Thomas WR: IgE and IgG
anti-house dust mite specificities in allergic disease. J Allergy
Clin Immunol. 118:361–367. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jacquet A: Innate immune responses in
house dust mite allergy. ISRN Allergy. 2013.735031:2013.
|
|
16
|
Amin K: The role of mast cells in allergic
inflammation. Respir Med. 106:9–14. 2012. View Article : Google Scholar
|
|
17
|
Pomes A: Relevant B cell epitopes in
allergic disease. Int Arch Allergy Immunol. 152:1–11. 2010.
View Article : Google Scholar :
|
|
18
|
Matsuo H, Yokooji T and Taogoshi T: Common
food allergens and their IgE-binding epitopes. Allergol Int.
64:332–343. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bronnert M, Mancini J, Birnbaum J,
Agabriel C, Liabeuf V, Porri F, Cleach I, Fabre A, Deneux I,
Grandné V, et al: Component-resolved diagnosis with commercially
available D. pteronyssinus Der p 1 Der p 2 and Der p 10: Relevant
markers for house dust mite allergy. Clin Exp Allergy.
42:1406–1415. 2012. View Article : Google Scholar
|
|
20
|
Chou H, Tam MF, Lee SS, Tang RB, Lin TH,
Tai HY, Chen YS and Shen HD: Asp159 is a critical core amino acid
of an IgE-binding and cross-reactive epitope of a dust mite
allergen Der f 7. Mol Immunol. 48:2130–2134. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chruszcz M, Chapman MD, Vailes LD, Stura
EA, Saint-Remy JM, Minor W and Pomes A: Crystal structures of mite
allergens Der f 1 and Der p 1 reveal differences in surface-exposed
residues that may influence antibody binding. J Mol Biol.
386:520–530. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
de Halleux S, Stura E, VanderElst L,
Carlier V, Jacquemin M and Saint-Remy JM: Three-dimensional
structure and IgE-binding properties of mature fully active Der p
1, a clinically relevant major allergen. J Allergy Clin Immunol.
117:571–576. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Takai T, Kato T, Yasueda H, Okumura K and
Ogawa H: Analysis of the structure and allergenicity of recombinant
pro- and mature Der p 1 and Der f 1: Major conformational IgE
epitopes blocked by prodomains. J Allergy Clin Immunol.
115:555–563. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cho YS, Jung HJ, Seok SH, Payumo AY, Chen
JK and Kwon HJ: Functional inhibition of UQCRB suppresses
angiogenesis in zebrafish. Biochem Biophys Res Commun. 433:396–400.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cai ZL, Chen JJ, Zhang Z, Hou YB, He YS,
Sun JL and Ji K: Identification of immunodominant IgE binding
epitopes of Der p 24, a major allergen of Dermatophagoides
pteronyssinus. Clin Transl Allergy. 9:282019. View Article : Google Scholar :
|
|
26
|
Chen ZG, Li YT, Wang WH, Tan KS, Zheng R,
Yang LF, Guan WJ, Hong HY and Yang QT: Distribution and
determinants of dermatophagoides mites sensitization of allergic
rhinitis and allergic asthma in China. Int Arch Allergy Immunol.
180:17–27. 2019. View Article : Google Scholar
|
|
27
|
Thomas WR: Geography of house dust mite
allergens. Asian Pac J Allergy Immunol. 28:211–224. 2010.
|
|
28
|
Liu XY, Ji KM, Gao B and Liu ZG:
Expression, purification and identification of the recombinant
allergen Der p2 from Dermatophagoides pteronyssinus and
investigation on its immunological activities. Chin J Zoonoses.
009:929–935. 2009.
|
|
29
|
Lauber B, Molitor V, Meury S, Doherr MG,
Favrot C, Tengvall K, Bergvall K, Leeb T, Roosje P and Marti E:
Total IgE and allergen-specific IgE and IgG antibody levels in sera
of atopic dermatitis affected and non-affected Labrador- and Golden
retrievers. Vet Immunol Immunopathol. 149:112–118. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Taylor JA, Karas JL, Ram MK, Green OM and
Seidel-Dugan C: Activation of the high-affinity immunoglobulin E
receptor Fc epsilon RI in RBL-2H3 cells is inhibited by Syk SH2
domains. Mol Cell Biol. 15:4149–4157. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang Q, Wang P, Kim Y, Haste-Andersen P,
Beaver J, Bourne PE, Bui HH, Buus S, Frankild S, Greenbaum J, et
al: Immune epitope database analysis resource (IEDB-AR). Nucleic
Acids Res. 36:Web Server Issue. pp. W513–W518. 2008, View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Costa MA, Duro G, Izzo V, Colombo P,
Mirisola MG, Locorotondo G, Cocchiara R and Geraci D: The
IgE-binding epitopes of rPar j 2, a major allergen of Parietaria
judaica pollen, are heterogeneously recognized among allergic
subjects. Allergy. 55:246–250. 2000. View Article : Google Scholar
|
|
33
|
Banerjee B, Greenberger PA, Fink JN and
Kurup VP: Conformational and linear B-cell epitopes of Asp f 2, a
major allergen of Aspergillus fumigatus, bind differently to
immuno-globulin E antibody in the sera of allergic bronchopulmonary
aspergillosis patients. Infect Immun. 67:2284–2291. 1999.PubMed/NCBI
|
|
34
|
Szalai K, Fuhrmann J, Pavkov T, Scheidl M,
Wallmann J, Brämswig KH, Vrtala S, Scheiner O, Keller W, Saint-Remy
JM, et al: Mimotopes identify conformational B-cell epitopes on the
two major house dust mite allergens Der p 1 and Der p 2. Mol
Immunol. 45:1308–1317. 2008. View Article : Google Scholar
|
|
35
|
Stanley JS, King N, Burks AW, Huang SK,
Sampson H, Cockrell G, Helm RM, West CM and Bannon GA:
Identification and mutational analysis of the immunodominant IgE
binding epitopes of the major peanut allergen Ara h 2. Arch Biochem
Biophys. 34:244–253. 1997. View Article : Google Scholar
|
|
36
|
Laskowski RA, MacArthur MW, Moss DS and
Thornton JM: PROCHECK-a program to check the stereochemical quality
of protein structures. J App Cryst. 26:283–291. 1993. View Article : Google Scholar
|
|
37
|
Ramos JD, Cheong N, Lee BW and Chua KY:
Peptide mapping of immunoglobulin E and immunoglobulin G
immunodominant epitopes of an allergenic Blomia tropicalis
paramyosin, Blo t 11. Clin Exp Allergy. 33:511–517. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Jung HJ, Kim KH, Kim ND, Han G and Kwon
HJ: Identification of a novel small molecule targeting UQCRB of
mitochondrial complex III and its anti-angiogenic activity. Bioorg
Med Chem Lett. 21:1052–1056. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Greene WK and Thomas WR: IgE binding
structures of the major house dust mite allergen Der p I. Mol
Immunol. 29:257–262. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Van't Hof W, Driedijk PC, van den Berg M,
Beck-Sickinger AG, Jung G and Aalberse RC: Epitope mapping of the
Dermatophagoides pteronyssinus house dust mite major allergen Der p
II using overlapping synthetic peptides. Mol Immunol. 28:1225–1232.
1991. View Article : Google Scholar
|
|
41
|
Herrick CA, MacLeod H, Glusac E, Tigelaar
RE and Bottomly K: Th2 responses induced by epicutaneous or
inhalational protein exposure are differentially dependent on IL-4.
J Clin Invest. 105:765–775. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Moingeon P and Mascarell L: Novel routes
for allergen immuno-therapy: Safety, efficacy and mode of action.
Immunotherapy. 4:201–212. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chen KW, Blatt K, Thomas WR, Swoboda I,
Valent P, Valenta R and Vrtala S: Hypoallergenic Der p 1/Der p 2
combination vaccines for immunotherapy of house dust mite allergy.
J Allergy Clin Immunol. 130:435–443. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Winther L, Arnved J, Malling HJ, Nolte H
and Mosbech H: Side-effects of allergen-specific immunotherapy: A
prospective multi-centre study. Clin Exp Allergy. 36:254–260. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Nishiyama C, Fukada M, Usui Y, Iwamoto N,
Yuuki T, Okumura Y and Okudaira H: Analysis of the IgE-epitope of
Der f 2, a major mite allergen, by in vitro mutagenesis. Mol
Immunol. 32:1021–1029. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ferreira F, Wallner M, Breiteneder H,
Hartl A, Thalhamer J and Ebner C: Genetic engineering of allergens:
Future therapeutic products. Int Arch Allergy Immunol. 128:171–178.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Fanuel S, Tabesh S, Mokhtarian K,
Saroddiny E, Fazlollahi MR, Pourpak Z, Falak R and Kardar GA:
Construction of a recombinant B-cell epitope vaccine based on a Der
p1-derived hypoallergen: A bioinformatics approach. Immunotherapy.
10:537–553. 2018. View Article : Google Scholar : PubMed/NCBI
|