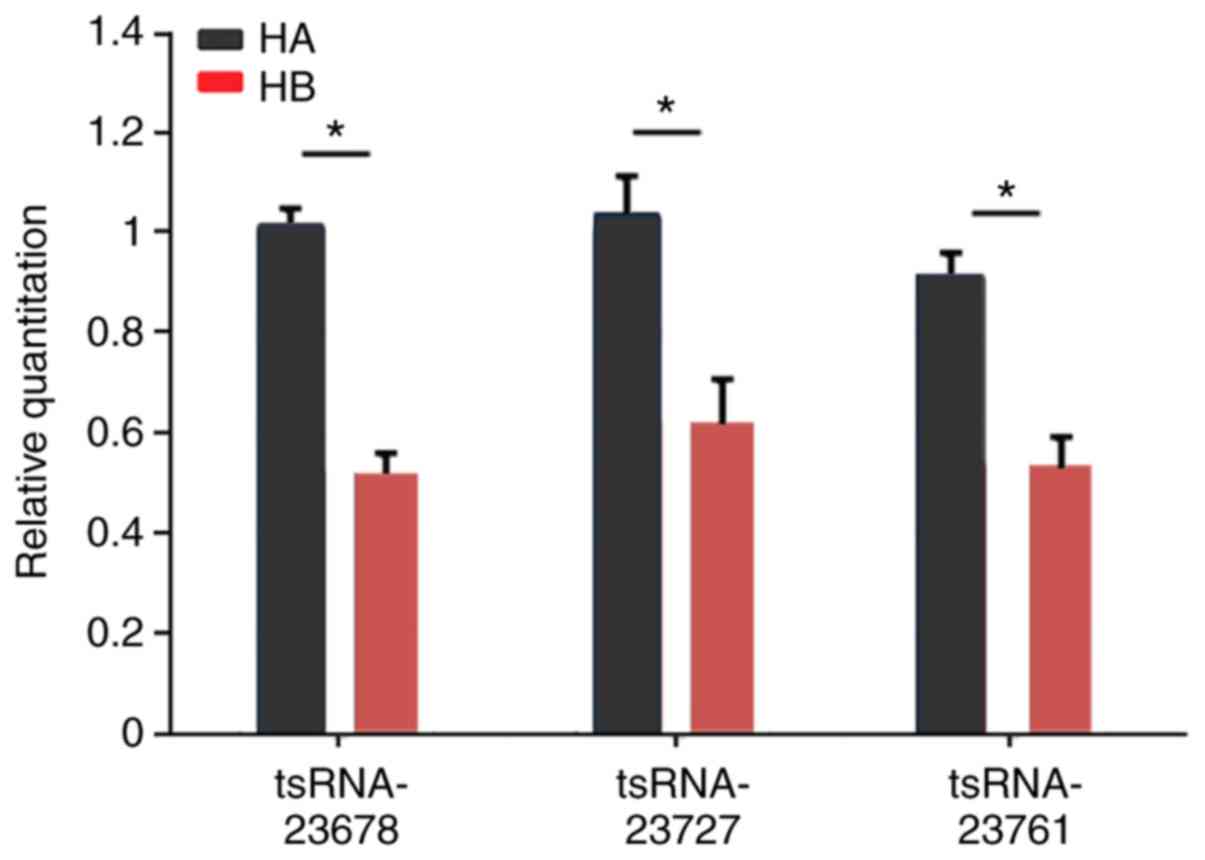
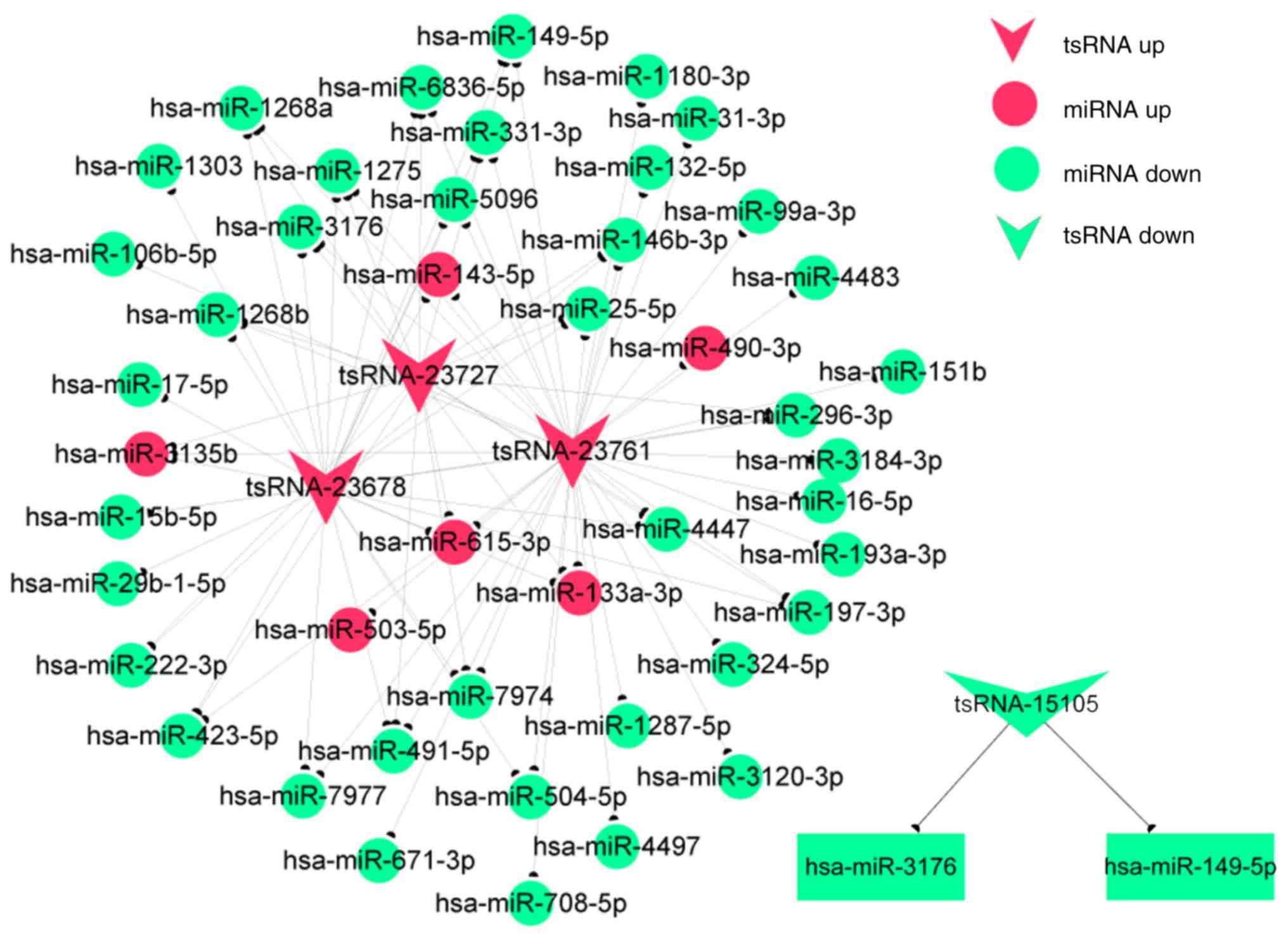
|
1
|
Bayat A, McGrouther DA and Ferguson MW:
Skin scarring. BMJ. 326:88–92. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sidgwick GP, Iqbal SA and Bayat A: Altered
expression of hyaluronan synthase and hyaluronidase mRNA may affect
hyaluronic acid distribution in keloid disease compared with normal
skin. Exp Dermatol. 22:377–379. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Syed F, Ahmadi E, Iqbal SA, Singh S,
McGrouther DA and Bayat A: Fibroblasts from the growing margin of
keloid scars produce higher levels of collagen I and III compared
with intral-esional and extralesional sites: Clinical implications
for lesional site-directed therapy. Br J Dermatol. 164:83–96. 2011.
View Article : Google Scholar
|
|
4
|
Wolfram D, Tzankov A, Pulzl P and
Piza-Katzer H: Hypertrophic scars and keloids-a review of their
pathophysiology, risk factors, and therapeutic management. Dermatol
Surg. 35:171–181. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Qi J, Liu Y, Hu K, Zhang Y, Wu Y and Zhang
X: MicroRNA-26a inhibits hyperplastic scar formation by targeting
Smad2. Exp Ther Med. 15:4332–4338. 2018.PubMed/NCBI
|
|
6
|
Zhu Z, Ding J, Shankowsky HA and Tredget
EE: The molecular mechanism of hypertrophic scar. J Cell Commun
Signal. 7:239–252. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
van der Veer WM, Bloemen MC, Ulrich MM,
Molema G, van Zuijlen PP, Middelkoop E and Miessen FB: Potential
cellular and molecular causes of hypertrophic scar formation.
Burns. 35:15–29. 2009. View Article : Google Scholar
|
|
8
|
Rumsey N, Clarke A and White P: Exploring
the psychosocial concerns of outpatients with disfiguring
conditions. J Wound Care. 12:247–252. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Leblebici B, Adam M, Bagis S, Tarim AM,
Noyan T, Akman MN and Haberal MA: Quality of life after burn
injury: The impact of joint contracture. J Burn Care Res.
27:864–868. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zuccaro J, Ziolkowski N and Fish J: A
systematic review of the effectiveness of laser therapy for
hypertrophic burn scars. Clin Plast Surg. 44:767–779. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li S, Xu Z and Sheng J: tRNA-Derived small
RNA: A novel regulatory small non-coding RNA. Genes (Basel).
9:E2462018. View Article : Google Scholar
|
|
12
|
Anderson P and Ivanov P: tRNA fragments in
human health and diease. FEBS Lett. 588:4297–4304. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pliatsika V, Loher P, Telonis AG and
Rigoutsos I: MINTbase: A framework for the interactive exploration
of mitochondrial and nucleartRNA fragments. Bioinformatics.
32:2481–2489. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zheng LL, Xu WL, Liu S, Sun WJ, Li JH, Wu
J, Yang JH and Qu LH: tRFCancer: A web server to detect
tRNA-derived small RNA fragments (tRFs) and their expression in
multiple cancers. Nucleic Acids Res. 44:W185–W193. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lee YS, Shibata Y, Malhotra A and Dutta A:
A novel class of small RNAs: tRNA-derived RNA fragments (tRFs).
Genes Dev. 23:2639–2649. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang Q, Lee I, Ren J, Ajay SS, Lee YS and
Bao X: Identification and functional characterization of
tRNA-derived RNA fragments (tRFs) in respiratory syncytial virus
infection. Mol Ther. 21:368–379. 2013. View Article : Google Scholar :
|
|
17
|
Tomita K, Ogawa T, Uozumi T, Watanabe K
and Masaki H: A cytotoxic ribonuclease which specifically cleaves
four isoac-cepting arginine tRNAs at their anticodon loops. Proc
Natl Acad Sci USA. 97:8278–8283. 2000. View Article : Google Scholar
|
|
18
|
Deng J, Ptashkin RN, Wang Q, Liu G, Zhang
G, Lee I, Lee YS and Bao X: Human metapneumovirus infection induces
significant changes in small noncoding RNA expression in airway
epithelial cells. Mol Ther Nucleic Acids. 3:e1632014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gonzale N and Goleberg DJ: Update on the
treatment of scars. J Drugs Dermatol. 18:5502019.
|
|
20
|
Ault P, Plaza A and Paratz J: Scar massage
for hypertrophic burns scarring-A systematic review. Burns.
44:24–38. 2018. View Article : Google Scholar
|
|
21
|
Tu L, Huang Q, Fu S and Liu D: Aberrantly
expressed long noncoding RNAs in hypertrophic scar fibroblast in
vitro: A microarray study. Int J Mol Med. 41:1917–1930.
2018.PubMed/NCBI
|
|
22
|
Liu Y, Zhong L, Liu D, Ye H, Mao Y and Hu
Y: Differential miRNA expression profiles in human keratinocytes in
response to protein kinase C inhibitor. Mol Med Rep. 16:6608–6619.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen MH, Chen YJ, Ye YQ, Su Y and Li CY:
The factors affecting analysis of gene expression level by semi
qRT-PCR technique. Guzhou Agricultural Sciences. 29:28–30.
332009.
|
|
24
|
Lowe TM and Eddy SR: tRNAscan-SE: A
program for improved detection of transfer RNA genes in genomic
sequence. Nucleic Acids Res. 25:955–964. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kumar P, Mudunuri SB, Anaya J and Dutta A:
tRFdb: A database for transfer RNA fragments. Nucleic Acids Res.
43:D114–D115. 2015. View Article : Google Scholar
|
|
26
|
Pliatsika V, Loher P, Magee R, Telonis AG,
Londin E, Shigematsu M, Kirino Y and Rigoutsos I: MINTbase v2.0: A
comprehensive database for tRNA-derived fragments that includes
nuclear and mitochondrial fragments from all The Cancer Genome
Atlas projects. Nucleic Acids Res. 46:D152–D159. 2018. View Article : Google Scholar :
|
|
27
|
Conesa A, Madrigal P, Tarazona S,
Gomez-Cabrero D, Cervera A, McPherson A, Szcześniak MW, Gaffney DJ,
Elo LL, Zhang X and Mortazavi A: A survey of best practices for
RNA-seq data analysis. Genome Biol. 17:132016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang L, Feng Z, Wang X and Zhang X:
DEGseq: An R package for identifying differentially expressed genes
for RAN-seq data. Bioinformatics. 26:136–138. 2009. View Article : Google Scholar
|
|
29
|
Adnan M, Morton G and Hadi S: Analysis of
rpoS and bolA gene expression under various 0stress-induced
environments in planktoic and biofilm phase using 2(-ΔΔCT) method.
Mol Cell Biochem. 357:275–282. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kim HK, Fuchs G, Wang S, Wei W, Zhang Y,
Park H, Roy-chaudhuri B, Li P, Xu J, Chu K, et al: A
transfer-RNA-derived small RNA regulates ribosome biogenesis.
Nature. 552:57–62. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kumar P, Anaya J, Mudunuri SB and Dutta A:
Meta-analysis of tRNA derived RNA fragments reveals that they are
analysis of tRNA derived RNA fragments reveals that they are
evolutionarily conserved and associate with AGO proteins to
recognize specific RNA targets. BMC Biol. 12:782014. View Article : Google Scholar
|
|
32
|
Keam SP and Hutvagner G: tRNA-Derived
fragments (tRFs): Emerging new roles for an ancient RNA in the
regulation of gene expression. Life (Basel). 5:1638–1651. 2015.
|
|
33
|
Balatti V, Pekarsky Y and Croce CM: Role
of the tRNA-derived Small RNAs in cancer: New potential biomarkers
and target for therapy. Adv Cancer Res. 135:173–187. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Maute RL, Schneider C, Sumazin P, Holmes
A, Califano A, Basso K and Dalla-Favera R: tRNA-derived microRNA
modulates proliferation and the DNA damage response and is
down-regulated in B cell lymphoma. Proc Natl Acad Sci USA.
110:1404–1409. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sobala A and Hutvagner G: Small RNAs
derived from the 5′ end of tRNA can inhibit protein translation in
human cells. RNA Biol. 10:553–563. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gebetsberger J, Zywicki M, Kunzi A and
Polacek N: tRNA-derived fragments target the ribosome and function
as regulatory non-coding RNA in Haloferax volcanii. Archaea.
2012:2609092012. View Article : Google Scholar
|
|
37
|
Goodarzi H, Liu X, Nguyen HC, Zhang S,
Fish L and Tavazoie SF: Endogenous tRNA-derived fragments suppress
breast cancer progression via YBX1 displacement. Cell. 161:790–802.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kumar P, Kuscu C and Dutta A: Biogenesis
and function of transfer RNA-related fragments (tRFs). Trends
Biochem Sci. 41:679–689. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Atiyeh BS: Nonsurgical management of
hypertrophic scars: Evidence-based therapies, standardpractices,
and emerging methods. Aesthetic Plast Surg. 31:468–492. 2007.
View Article : Google Scholar
|
|
40
|
Nehme A, Cerutti C, Dhaouadi N, Gustin MP,
Courand PY, Zibara K and Bricca G: Atlas of tissue
renin-angiotensin-aldo-sterone system in human: A transcriptomic
meta-analysis. Sci Rep. 5:100352015. View Article : Google Scholar
|
|
41
|
Slack C: Ras signaling in aging and
metabolic regulation. Nutr Healthy Aging. 4:195–205. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Koul HK, Pal M and Koul S: Role of p38 MAP
kinase signal transduction in solid tumors. Genes Cancer.
4:342–359. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Coelho MA, de Carne Trecesson S, Rana S,
Zecchin D, Moore C, Molina-Arcas M, East P, Spencer-Dene B, Nye E,
Barnouin K, et al: Oncogenic RAS signaling promotes tumor
immunoresistance by stabilizing PD-L1 mRNA. Immunity.
47:1083–1099.e6. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Krishna S, Yim DG, Lakshmanan V, Tirumalai
V, Koh JL, Park JE, Cheong JK, Low JL, Lim MJ, Sze SK, et al:
Dynamic expression of tRNA-derived small RNAs define cellular
states. EMBO Rep. 20:e477892019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Bi S, Cao C, Chai LL, Li SR and Yang DY:
Regulatory mechanism of miR-29 over TGF-β1 and COL1 in scar cells.
Eur Rev Med Pharmacol Sci. 21:2512–2517. 2017.PubMed/NCBI
|
|
46
|
Xiao K, Luo X, Wang X and Gao Z:
MicroRNA185 regulates transforming growth factor-β1 and collagen1
in hypertrophic scar fibroblasts. Mol Med Rep. 15:1489–1496. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zeng G, Zhong F, Li J, Luo S and Zhang P:
Resveratrol-mediated reduction of collagen by inhibiting
proliferation and producing apoptosis in human hypertrophic scar
fibroblasts. Biosci Biotechnol Biochem. 77:2389–2396. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Qi J, Liu Y, Hu K, Zhang Y, Wu Y and Zhang
X: MicroRNA-205-5p regulates extracellular matrix production in
hyperplastic scars by targeting Smad2. Exp Ther Med. 17:2284–2290.
2019.
|
|
49
|
Li Q: Inhibitory SMADs: Potential
regulators of ovarian function. Biol Reprod. 92:502015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhang Y, Alexander PB and Wang XF: TGF-β
family signaling in the control of cell proliferation and survival.
Cold Spring Harb Perspect Biol. 9:a0221452017. View Article : Google Scholar
|
|
51
|
Zhang Z, Kuang F, Liu CL, Chen B, Tang WB
and Li XJ: Effects of silencing Smad ubiquitination regulatory
factor 2 on the function of human hypertrophic scar-derived
fibroblasts. Zhonghua Shao Shang Za Zhi. 33:145–151. 2017.In
Chinese. PubMed/NCBI
|
|
52
|
Asnaghi L, White DT, Key N, Choi J, Mahale
A, Alkatan H, Edward DP, Elkhamary SM, Al-Mesfer S, Maktabi A, et
al: ACVR1C/SMAD2 signaling promotes invasion and growth in
retinoblastoma. Oncogene. 38:2056–2075. 2019. View Article : Google Scholar :
|
|
53
|
Huang C, Wu XF and Wang XL: Trichostatin
Ainhibits phenotypic transition and induces apoptosis of the
TAF-treated normal colonic epithelial cells through regulation of
TGF-β pathway. Int J Biochem Cell Biol. 114:1055652019. View Article : Google Scholar
|
|
54
|
Yamazaki S and Nakauchi H: Bone marrow
Schwann cells induce hematopoietic stem cell hibernation. Int J
Hematol. 99:695–698. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
LIU N, Li Y, SUN L, JIANG J and ZHANG J:
Expressions of Smad2 and Smad4 proteins in breast carcinoma tissue
and significances. J Jilin University (Medicine Edition).
42:763–767. 2016.
|
|
56
|
Li J, Cen B, Chen S and He Y: MicroRNA-29b
inhibits TGF-β1-induced fibrosis via regulation of the TGF-β1/Smad
pathway in primary human endometrial stromal cells. Mol Med Rep.
13:4229–4237. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Gallant-Behm CL, Piper J, Lynch JM, Seto
AG, Hong SJ, Mustoe TA, Maari C, Pestano LA, Dalby CM, Jackson AL,
et al: A MicroRNA-29 mimic (Remlarsen) represses extracellular
matrix expression and fibroplasia in the skin. J Invest Dermatol.
139:1073–1081. 2019. View Article : Google Scholar
|
|
58
|
Wang H, Wang B, Zhang A, Hassounah F, Seow
Y, Wood M, Ma F, Klein JD, Price SR and Wang XH: Exosome-mediated
miR-29 transfer reduces muscle atrophy and kidney fibrosis in mice.
Mol Ther. 27:571–583. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Luo S, He F, Luo J, Dou S, Wang Y, Guo A
and Lu J: Drosophila tsRNAs preferentially suppress general
translation machinery via antisense pairing and participate in
cellular starvation response. Nucleic Acids Res. 46:5250–5268.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Schopman NC, Heynen S, Haasnoot J and
Berkhout B: A miRNA-tRNA mix-up: TRNA origin of proposed miRNA. RNA
Biol. 7:573–576. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Li M, Liu DW and Lei W: Advances in the
research of effects of competing endogenous RNAs and their
regulatory networks in pathological scars of skin. Zhonghua Shao
Shang Za Zhi. 35:701–704. 2019.In Chinese. PubMed/NCBI
|
|
62
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Zhang YJ, Yang XS, Wu PS, Li X, Zhang XF,
Chen XQ and YU ZX: Effects of angiotensin II and losartan on the
growth and proliferation of hepatic stellate cells. Di Yi Jun Yi Da
Xue Xue Bao. 23:219–221. 2003.PubMed/NCBI
|
|
64
|
Wang R, Chen J, Zhang Z and Cen Y: Role of
chymase in the local renin-angiotensin system in keloids:
Inhibition of chymase may be an effective therapeutic approach to
treat keloids. Drug Des Devel Ther. 9:4979–4988. 2015.PubMed/NCBI
|
|
65
|
Demir CY, Ersoz ME, Erten R, Kocak OF,
Sultanoglu Y and Basbugan Y: Comparison of enalapril, candesartan
and intralesional triamcinolone in reducing hypertrophic scar
development: An experimental study. Aesthetic Plast Surg.
42:352–361. 2018. View Article : Google Scholar : PubMed/NCBI
|