Introduction
Breast cancer is a malignant tumor that occurs in
the epithelial tissue of the breast glands and is the most
frequently occurring cancer in women all over the world (1). The global incidence of breast cancer
has been on the rise since the late 1970s and breast cancer has
become a major public health problem in current society. Despite
the improved prognosis of patients with breast cancer resulting
from early diagnosis, radical surgery, chemotherapy, radiotherapy,
hormonal therapy and the development of targeted therapy, breast
cancer remains the leading cause of cancer mortality in women
(2,3). The high mortality rates in breast
cancer patients are associated with recurrence, drug resistance and
the high frequency of metastasis (4). In most cases, patients with
metastasis are not eligible for surgery and chemotherapy or
radiotherapy, which do not contribute significantly to cure them
(5-7). Metastasis remains a major obstacle
in achieving a radical cure and preventing death of breast cancer
patients. Therefore, it is essential to understand the molecular
pathways involved in causing metastasis of breast cancer.
Tumor metastatic invasiveness is linked with the
increased migration capacity of epithelial cells. This process is
known as the epithelial-mesenchymal transition (EMT) (8). Epithelial cancer cells in the
lactiferous duct having undergone EMT are capable of undergoing
metastasis and are more invasive (9-11).
Transforming growth factor (TGF)-β1 has been reported to induce EMT
(12). The loss of epithelial
(E)-cadherin, an adherens junction cell surface protein expressed
in epithelial cells is the principal characteristic of EMT.
Vimentin, which is the major cytoskeletal component of mesenchymal
cells was increased significantly by the treatment of TGF-β1
(13). In breast cancer, EMT both
elevates the migratory capacity and invasive potential of tumor
cells and initiates pro-tumorigenic alterations in the tumor
microenvironment (14). CORM-A1
and DETA/NO combination therapy showed antimetastatic action and
resulted in normalization of endothelial metabolism, diminution of
platelet activation, and inhibition of EMT progression (12). Coenzyme Q0 attributed
to the phosphatidyl inositol 3 kinase (PI3K)/protein kinase B
(AKT)/nuclear factor (NF)κB/matrix metalloproteinase-9 signaling
pathway through reactive oxygen species-mediated apoptosis can
inhibit the progression of metastasis as well as EMT (in
vitro and in vivo) (15). Metastasis of breast cancer remains
challenging for the current clinical treatments and more than half
of the patients suffer from a disease relapse due to distant
metastases (16). Therefore,
further research into the under-lying molecular mechanism and
development of more targets of EMT and metastasis of breast cancer
is urgent.
Circular RNA (circRNA) is a novel type of endogenous
non-coding RNA without a 5′ cap and 3′ poly (A) end. They are
widely expressed in the central nervous system and enrich
biological functions as demonstrated by some reports such as acting
as miRNA sponges, regulating transcription or binding to the miRNA
binding proteins (17,18). Previously, growing evidence has
confirmed that circRNAs are associated with multiform cancerous
biological progression (19,20), and an increasing number of
functional circRNAs are discovered in breast cancer. For instance,
targeting the circBMPR2/microRNA (miR)-553/USP4 axis can be used as
a potential therapeutic approach for breast cancer; circ_0103552
predicts dismal prognosis and promotes breast cancer cell
proliferation and invasion by sponging miR-1236 (21,22); hsa_ circ_001569 can facilitate
metastasis through the modulation of the PI3K-AKT pathway, which is
an unfavorable prognostic factor of breast cancer (23). However, little is known about the
global change of circRNA profiles during EMT, which is an important
step in tumor metastasis.
In this study, expression profiles of circRNAs in
breast cancer cells with TGF-β1 induction were analyzed to confirm
the EMT related circRNAs, suggesting that the different expression
levels of circRNAs may be closely related to breast cancer
metastasis. This study validated circSCYL2, which could suppress
breast cancer cell migration and invasion, may serve as biomarkers
during tumor metastasis. The present study aimed to explore the
functions of circRNAs and furnish novel therapeutic targets for
tumor metastasis.
Materials and methods
Cell culture and establishment of EMT
model
The human breast cancer cell lines MCF-7 and
MDA-MB-231 used in this research were obtained from Yingbio
Technology. MCF-7 cells were incubated in Dulbecco's modified
Eagle's medium (10-013-CVR; Corning, Inc.) with 10% (v/v)
heat-inactivated fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific, Inc.) and 1% penicillin-streptomycin (P/S, E607011;
Sangon Biotech Co., Ltd.) and were cultured in a humidified 37°C
incubator with 5% CO2. While MDA-MB-231 cells were
incubated in Leibovitz's 15 medium (L15, E600016-0500; BBI
Solutions) with 10% (v/v) FBS and 1% P/S and were cultured in a
humidified 37°C incubator with 100% air. These cells, which were
incubated in medium with 0.5% FBS, were treated with 20 ng/ml
TGF-β1 (100-21-10; Peprotech, Inc.) for 48 h (24,25). The control cells were incubated in
medium with 0.5% FBS and treated with DMSO. Then, cell pictures
were captured by an inverted phase-contrast microscope (Shanghai
Optical Sixth Factory). The cells treated by TGF-β1 were named as
cell-EMT and the control cells were named as cell-blank.
Western blotting
The total protein of cells (MCF-7-EMT, MCF-7-blank,
MDA-MB-231-EMT and MDA-MB-231-blank) was extracted using RIPA Lysis
Buffer (Thermo Fisher Scientific, Inc.) and the concentration was
measured using a bicinchoninic acid assay kit. Total 150 µg
protein samples were separated via 10% SDS-PAGE and transferred to
a polyvinylidene fluoride membrane (EMD Millipore). After blocking
in 5% milk at room temperature for 3 h, the membranes were
incubated with primary antibodies against E-cadherin (cat. no.
sc-52327; 1:500; Santa Cruz Biotechnology, Inc.), Vimentin (cat.
no. ab8978; 1:500; Abcam) and GAPDH (cat. no. 60004-1-Ig; 1:1,000;
Proteintech, Inc.) overnight at 4°C. This was followed by
incubation with goat anti-mouse IgG-horseradish peroxidase antibody
(cat. no. A0216; 1:1,000; Beyotime Institute of Biotechnology) at
room temperature for 2 h and the blots were detected using the
enhanced chemiluminescence reagent (Thermo Fisher Scientific,
Inc.). Images were captured by a ChemiDoc Gel Imaging system
(Thermo Fisher Scientific, Inc.) and optical density was analyzed
by Image J software (version 1.8.0; National Institute of Health).
Results represent the means of three independent experiments
performed in triplicates. P-values were determined by using t-tests
and P<0.05 was considered to indicate a statistically
significant difference.
RNA isolation, RNA library construction
and sequencing
Total RNA from two paired samples (MC-7 EMT and
MCF-7 blank, MDA-MB-231 EMT and MDA-MB-231 blank) was extracted
using the TRIZOL reagent (Invitrogen; Thermo Fisher Scientific,
Inc.), followed by an assessment for quantity and purity using the
microspectrophotometer (TGem Tiangen Biotech Co., Ltd.). The RNA
integrity number was analyzed using a UV Spectrophotometer
(UV-2000; Tanon Science and Technology Co., Ltd.). Thereafter, 1
µg total RNA from each sample was used to prepare the
library according to the following procedures: Ribosomal RNAs
(rRNAs) were removed from total RNA using an rRNA probe and RNase
H. The ribosomal-depleted RNA of target fragment size (300 bp) was
obtained using metal ions, reverse transcribed into cDNA, connected
with adapters, followed by fragment sorting, enrichment with PCR
and then purified library products were quantified and evaluated
using the Quant-iT™ PicoGreen® dsDNA Assay kit (Beckman
Coulter, Inc.) and Agilent 2200. The process of reverse
transcription in RNA library construction was as follows:
First-strand cDNA was synthesized by the reagents of Actinomycin D
(Vazyme Biotech Co., Ltd.), first Strand Enzyme Mix (Vazyme Biotech
Co., Ltd.), first Strand Buffer, dNTPs random primer using the
following process at 25°C for 10 min, 42°C for 15 min and 75°C for
15 min. Second Strand/End Repair Enzyme Mix, second Strand Marking
Buffer and dNTPs were mixed at 16°C for 60 min to synthesize the
second-strand cDNA. All libraries were sequenced by Illumina Hiseq
X Ten (Illumina, Inc.).
Bioinformatics analysis
Quality control of raw reads was evaluated by
Fast-QC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/).
The remaining clean reads were mapped to the reference human genome
(GRCh38) using HISAT2 software (version 2.1.0; https://daehwankimlab.github.io/hisat2/)
and to predict the circRNAs, ACFS2 (https://github.com/arthuryxt/acfs) was used to
identify and quantify circRNA from these mapping splice junction
reads. The differentially expressed circRNAs between EMT and blank
were analyzed using the DE-Seq algorithm, and significant
differences were defined as absolute |log2 fold-change|>1, with
a false discovery rate (FDR) <0.05. The miRanda/RNAhybird was
used to predict the relationship between all miRNAs and
differential mRNA and differential circRNA (the intersection of the
two pieces of prediction software) was used as the final target
gene prediction result. The parameters set in miRanda were energy
miranda <−20 and score miranda >150, and in RNAhybird was
energy RNAhybird <−25. Function and pathway analysis of these
target genes of differentially expressed circRNAs were assessed
using Gene Ontology (GO) (http://www.geneontology.org) and the Kyoto
Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.jp/kegg) databases, respectively.
The significance level was assessed using the Fisher test and the
significant enrichment was obtained by considering a P<0.05.
circRNA/miRNA/mRNA interactions were subsequently investigated and
the circRNA-miRNA-mRNA network was constructed by Cytoscape
software (version 3.6.1; https://cyto-scape.org).
RT-qPCR
Total RNA was extracted and quantified and then
reverse transcribed into complementary DNA (cDNA) using a Reverse
Transcription kit (K1622; Thermo Fisher Scientific, Inc.). The
procedure of reverse transcription was 25°C for 5 min, 42°C for 60
min, 70°C for 5 min. Primers (Table
SI) for actin, E-cadherin, Vimentin, GAPDH, circRNAs were
obtained from Shanghai Sangon Pharmaceutical Co., Ltd., and the
RT-qPCR analysis was performed using Applied Biosystems; Thermo
Fisher Scientific, Inc. (ABI Q6). The kit used for qPCR was 2X SYBR
Green PCR Master Mix (Roche Diagnostics) and the thermocycling
conditions were 95°C for 10 min, 45 cycles of 95°C for 15 sec; 60°C
for 60 sec. The temperature of melt curve was 60 to 99°C. The actin
and GAPDH was used as an internal control. Relative quantification
and calculation were done through the comparative threshold (Cq)
cycle method (2−ΔΔCq) (26).
PCR and Sanger sequencing
cDNA extraction was performed as previously
described, gDNA was extracted using Genomic DNA Isolation kit
(Bio-Rad Laboratories, Inc.). Divergent primers and convergent
primers listed in Table I were
designed for the PCR reaction. The cDNA and gDNA PCR products were
observed using 2% agarose gel electrophoresis. CircRNAs of purified
PCR products were validated by Sanger sequencing and the results
were compared with the reference sequence (UCSC
chr12_100298175_100282943_+15232-SCYL2).
 | Table IPrimer sequences for circSCYL2 in
breast cancer cells. |
Table I
Primer sequences for circSCYL2 in
breast cancer cells.
| Primer | Sequences,
5′-3′ | Product expected
length/bp | Tm/°C |
|---|
| Divergent
primer-F |
CCTTCCCCTATATCTCCAGACATTA | 184 | 60 |
| Divergent primer
-R |
GACAGGATTTCCCATTACAGCA | | 60 |
| Convergent
primer-F |
CTTCTTACTGTCCAGCATCCTTTAG | 127 | 60 |
| Convergent
primer-R |
CTGGAGATATAGGGGAAGGTAGATT | | 60 |
Human breast cancer and normal
tissues
A total of 20 paired breast cancer specimens and
normal adjacent tissues (>2 cm from the edge of tumor tissue)
were obtained from patients undergoing surgical resection at the
Second Affiliated Hospital of Nanchang University, between January
2018 and July 2019). The age range of patients is 40 to 65 years
old, with a median age of 51±2.42 years old. The patients had the
inclusion criteria as follows: i) Diagnosed as breast cancer by
pathological confirmation; ii) without radiotherapy or chemotherapy
before surgery; iii) complete clinical and pathological data. The
exclusion criteria were as follows: i) Combined with other tumors
and serious diseases and ii) incomplete clinical and pathological
data. All the patients provided written consent and the study was
approved by the Ethics Committee of the Second Affiliated Hospital
of Nanchang University. Tissue fragments were immediately frozen in
liquid nitrogen at the time of surgery and stored at −80°C.
Plasmids construction and cell
transfection
Human circ-SCYL2 cDNA with BamHI (GGATCC) and
EcoRI (GAATTC) restriction sites was synthesized by Yingbio
Technology and cloned into the pcDNA circRNA vector, which
contained a front circular frame and a back circular frame. MCF-7
and MDA-MB-231 cells were seeded in 6-well plates at a
concentration of 3×105 cells/well; when the cells grew
to 80-90% confluency, the fresh culture medium was exchanged from
each well. Transfection of 2 ug vector was carried out using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. An empty vector was used
as the negative control. After transfection for 48 h, RT-qPCR was
performed to detect the expression level of circSCYL2.
Transwell assay
Transwell migration was performed using 0.8
µm 24-well transwell chambers (353097; FALCON™, Corning,
Inc.) and transwell invasion was performed using BioCoat™
Matrigel® 0.8 µm 24-well trans-well chamber
(354480, BioCoat; Thermo Fisher Scientific, Inc.). After
transfection for 48 h, the cells were digested, centrifuged with
200 × g at room temperature for 3 min and resuspended in a
serum-free medium to adjust the concentration to 1.5×105
cells/ml. A cell suspension of 500 µl was inoculated into
the upper chamber and 700 µl medium containing 10% FBS was
added to the lower chamber as a chemoattractant. After incubation
for 24 h, the transwell chamber was clipped and the invaded or
migrated cells were fixed with formaldehyde at room temperature for
30 min and stained using crystal violet at room temperature for 30
min. The cells on the upper surface of the membrane were wiped off
with cotton swabs and those on the lower surface were photographed
and counted by ImageJ software (version 1.8.0). The numbers of
invaded and migrated cells were counted in 3 randomly selected
fields.
Statistical analysis
RT-qPCR was repeated at least in three independent
experiments in every sample. Data were presented as mean ± SD of
three or more independent experiments. The significant expression
of circSCYL2 in breast cancer tissues and normal tissues was valued
by paired Student's t-test, and other significance was assessed
with unpaired Student's t-test using SPSS 24.0 software (IBM,
Corps.). P<0.05 was considered to indicate a statistically
significant difference.
Results
Establishment of EMT model in breast
cancer cells
After TGF-β1 stimulation, breast cancer cells
gradually displayed the characteristics of mesenchymal cells; the
cells became elongated and circular or polygonal progressively with
the extension of pseudopodia, and they became spindle-shaped, while
their intercellular space grew wider, and the cells loos-ened up
with an irregular distribution (Fig.
1A). To examine the establishment of EMT, the expression of
E-cadherin and Vimentin in TGF-β1 stimulated cells was detected by
RT-qPCR and western blotting analysis. It was shown that the mRNA
and protein expression levels of E-cadherin significantly
decreased, whereas the expression of Vimentin was statistically
elevated when compared to the blank (Fig. 1B and C). These results suggest
that EMT of breast cancer cells has been established successfully
by TGF-β1 induction.
Analysis of circRNA transcript data and
differentially expressed circRNAs
To investigate the roles of circRNA in EMT of the
breast cancer induced by TGF-β, the present study analyzed the
circRNA expression profiles of MCF-7 and MDA-MB-231 cells using RNA
sequencing (RNA-seq) analysis. The sequencing experimental flow
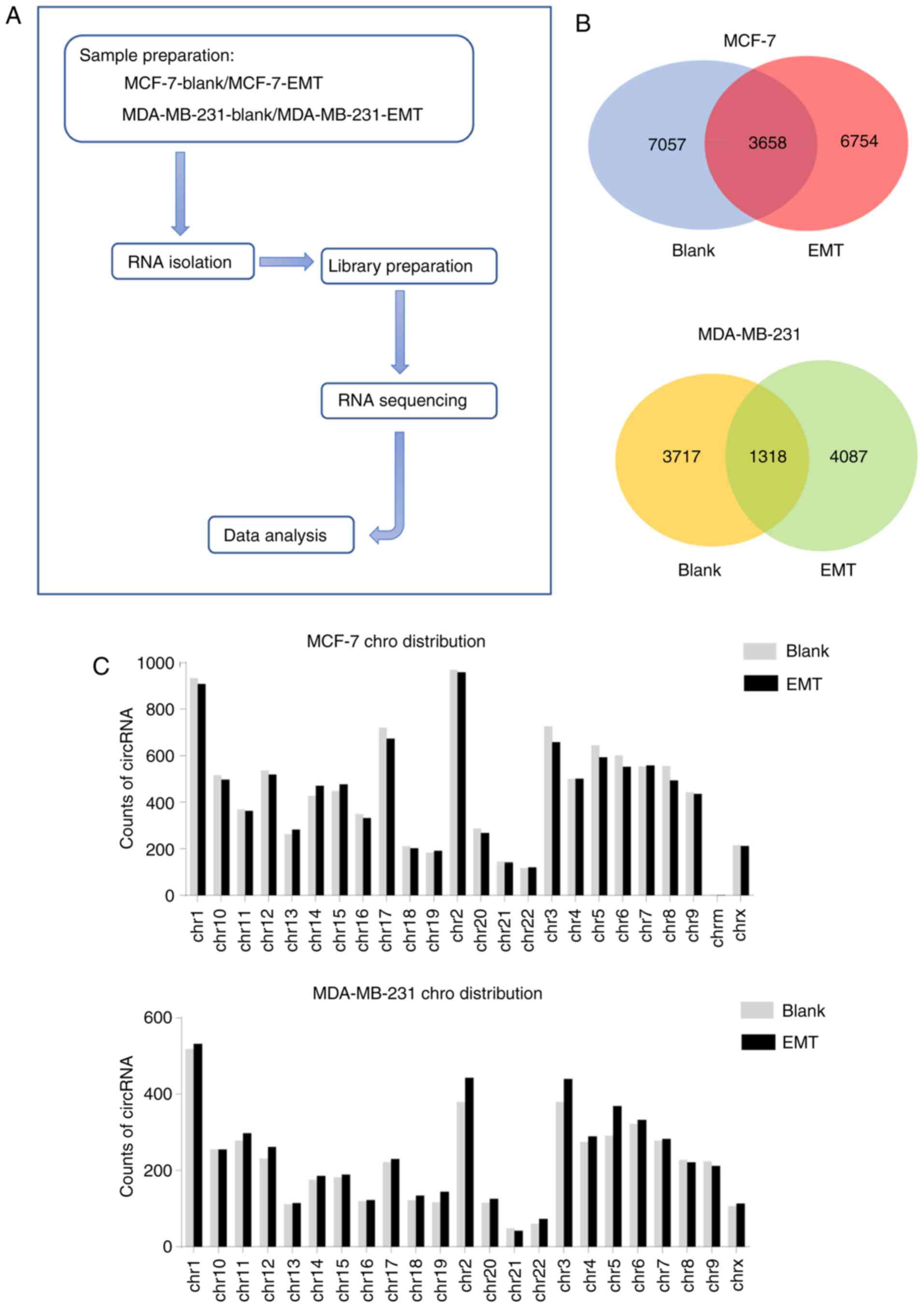
chart of this study is shown in Fig.
2A. A total of ~77 (MCF-7-blank), 105 (MCF-7-EMT), 90
(MDA-MB-231-blank) and 95 (MDA-MB-231-EMT) million clean reads were
obtained. A total of 87-97% of these reads were mapped to the
reference genome and ~18-22% of clean reads were realigned to
determine junction reads. A total of 23,347 circRNAs were predicted
from junction reads, among these, 10,715 and 10,412 circRNAs were
detected in the blank and EMT group, respectively in MCF-7 cells,
while in MDA-MB-231 cells, the data was 5,035 and 5,405,
respectively (Table II).
 | Table IISummary of the sequencing reads
alignment to the reference genome. |
Table II
Summary of the sequencing reads
alignment to the reference genome.
| Type | MCF-7-blank | MCF-7-EMT |
MDA-MB-231-blank | MDA-MB-231-EMT |
|---|
| All reads | 77697606 | 105327346 | 90555230 | 95327320 |
| Mapped reads | 75372155 (97%) | 97792240 (93%) | 79454343 (88%) | 83504290 (87%) |
| Junction Mapped
reads | 15980915 (21%) | 23008054 (22%) | 16930465 (18%) | 18926756 (20%) |
| cirRNA number
reads | 10,715 | 10,412 | 5,035 | 5,405 |
The Venn diagram in Fig. 2B reveals circRNAs distribution and
helps understanding the shared and unique circRNAs between blank
and EMT. In the current study 7,057 and 6,754 circRNAs were
detected exclusively in blank and EMT of MCF-7 cells, respectively,
and only 3,658 circRNAs were shared between the two groups. Also,
3,717 and 4,087 circRNAs were exclusively expressed in blank and
EMT of MDA-MB-231 cells respectively, only 1,318 circRNAs
over-lapped in both groups (Fig.
2B). These circRNAs of the blank and EMT in two cell lines were
widely distributed among all chromosomes; the number of circRNAs in
each chromosome showed no significant difference between the blank
and EMT groups (Fig. 2C).
Differentially expressed circRNAs were identified
between blank and EMT of the two cells to analyze the role of
circRNA in EMT. The Volcano plot exhibited circRNAs profiles that
were significantly differentially expressed between the blank and
EMT groups of two cells. There were 1,402 significantly
differentially expression circRNAs between EMT and blank of MCF-7
cells, among them, 773 circRNAs were significantly upregulated and
629 were downregulated in the EMT group. While in MDA-MB-231 cells,
322 circRNAs were significantly differentially expressed between
the blank and EMT groups, of which, 163 circRNAs were upregulated
and 159 were downregulated within blank and EMT (Fig. 2D). The Venn diagram shows the
common and unique dysregulated circRNAs in the two cells. Among all
these significantly dysregulated circRNAs, only 7 (0.4%) circRNAs
were significantly upregulated and 16 (1%) circRNAs were
significantly downregulated in both the MCF-7 EMT group and the
MDA-MB-231 EMT group, compared with the corresponding blank groups
(Fig. 2E).
GO and KEGG analysis of differently
expressed circRNAs
To explore the potential functional roles of
circRNAs, a circRNA-mediated competing endogenous RNA network in
tumor cells was constructed to predict differentially expressed
mRNAs that were targeted by the differentially expressed circRNAs.
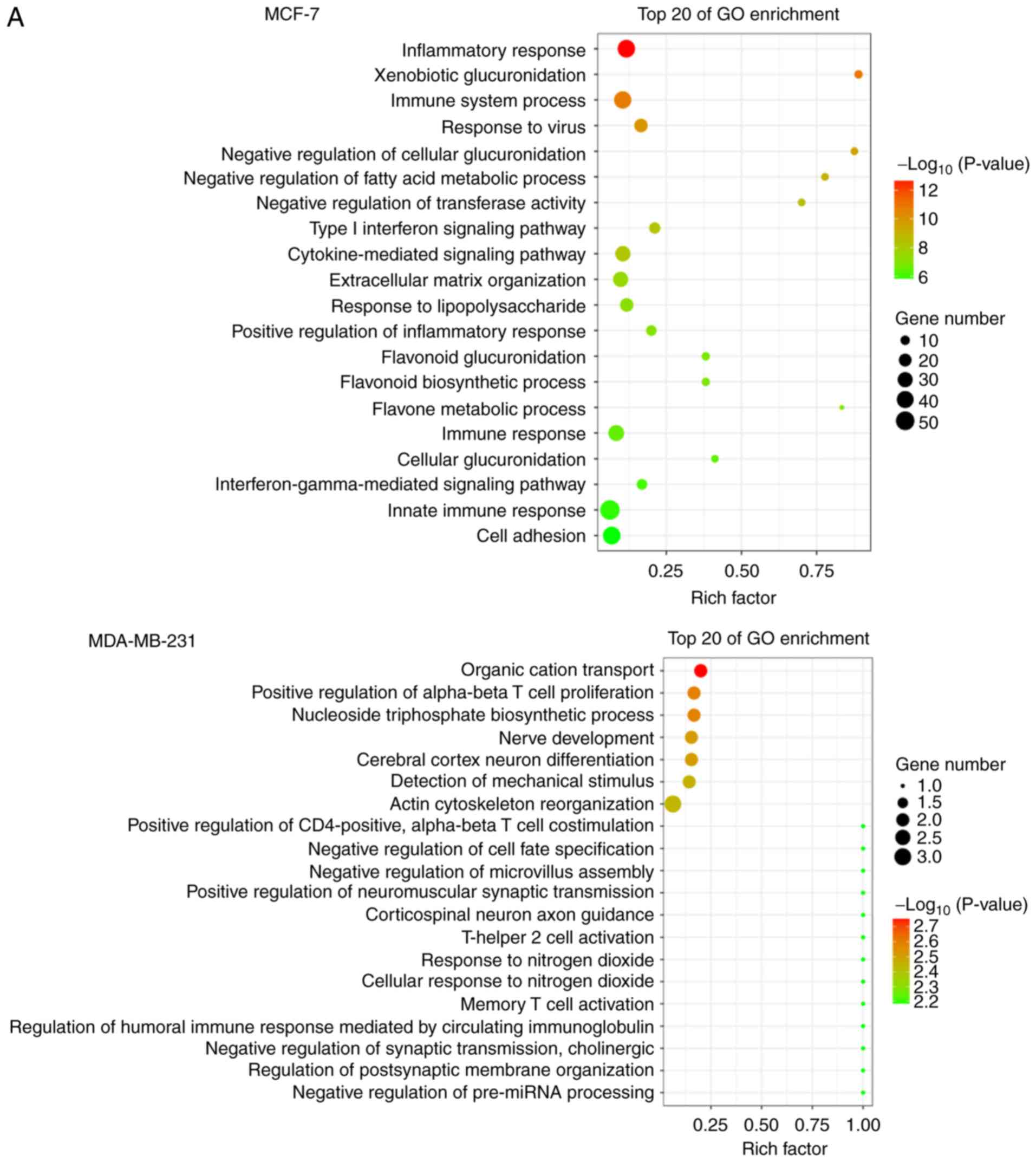
GO analysis was applied to explore the biological functions of host
mRNA. Interestingly, it was found that the most significantly
enriched functional terms were associated with the
immune/inflammatory response and cell adhesion/adherence junctions
in both MCF-7 and MDA-MB-231 cells, such as 'inflammatory re
sponse', 'immune system process', 'cell adhesion', 'positive
regulation of α-βT cell proliferation', and 'actin cytoskeleton
reorganization' (Fig. 3A).
The host genes were classified by KEGG function
annotations to analyze the pathways (Fig. 3B). In MCF-7 cells, KEGG analysis
con sistently revealed various signaling pathways involved in
regulating tumor growth, such as 'TNF signaling pathway', 'Steroid
hormone biosynthesis', 'NF-κB signaling pathway' and 'PI3K-AKT
pathway'. Similarly in MDA-MB-231 cells, pathways such as
'ECM-receptor interaction' and 'Calcium signaling pathway' were
enriched, which were reported to be associated with tumor cell
proliferation and migration (27-31).
Construction of the circRNA-miRNA-mRNA
interaction network
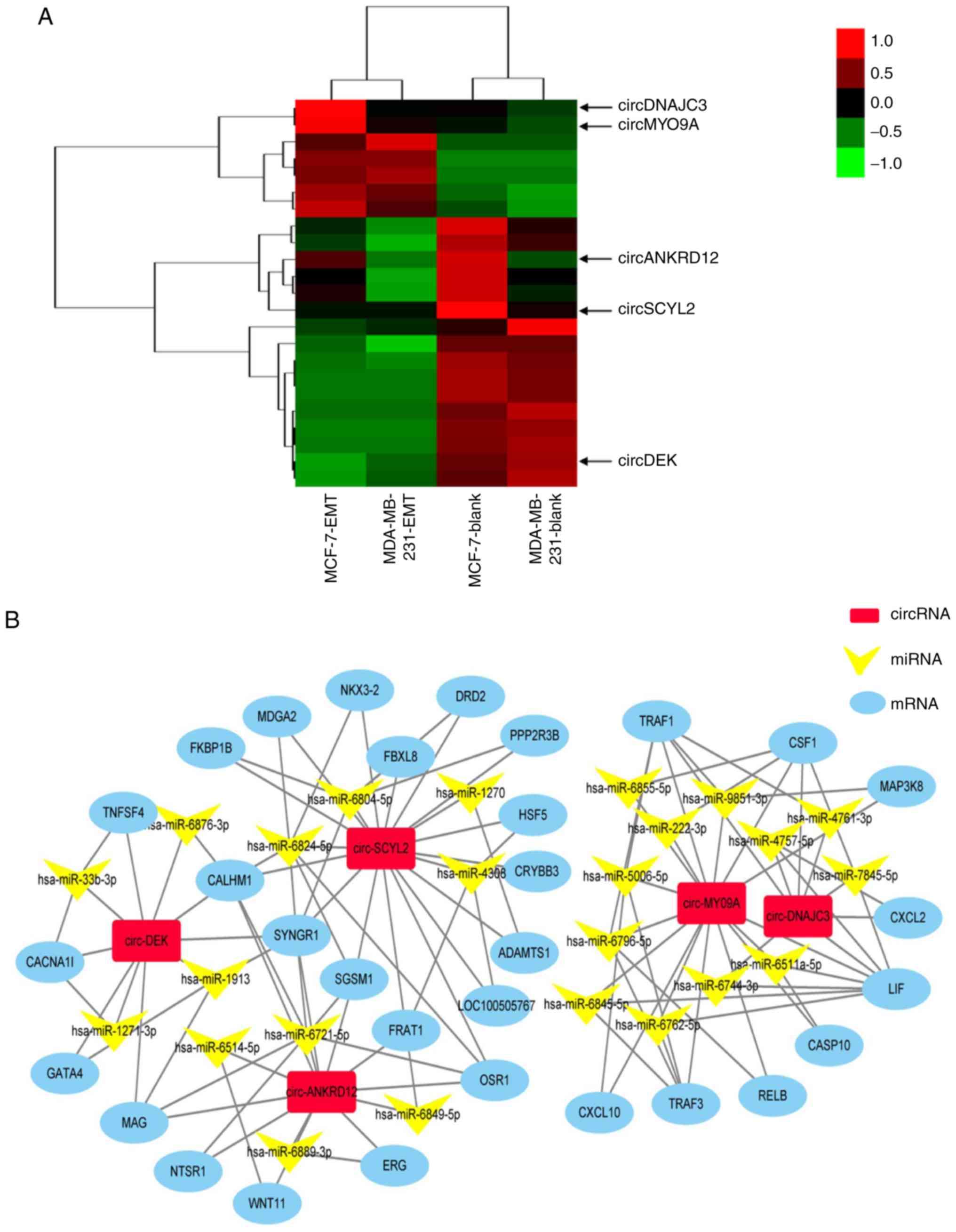
Next, to appraise the valuable circRNAs, the
expression changes of the simultaneously dysregulated circRNAs in
EMT and blank groups of two cells are shown in Fig. 4A. Although differentially
expressed circRNAs have been proposed to be closely associated with
EMT (12,15), the roles of circRNAs in breast
cancer EMT remain unclear. The present study aimed to identify
novel circRNAs that regulate EMT of breast cancer cells. A total of
five circRNAs were screened from these simultaneously dysregulated
circRNAs as the following criteria: i) Up or downregulation was
identified in EMT groups of the two cells, compared with their
blank groups, respectively; ii) higher fold-change and iii) more
abundant expression level. Heatmaps assessed and compared the
expression changes of circRNAs in EMT and blank groups of two cells
(Fig. 4A). The circRNA-miRNA-mRNA
network was constructed by predicting the differentially expressed
genes of five circRNAs and some target genes of these five
circRNAs, such as OSR1, DRD2, MAP3K8, and TNFSF4 that are related
to cancer (Fig. 4B) (32-35).
Verification of candidate circRNAs by
RT-qPCR
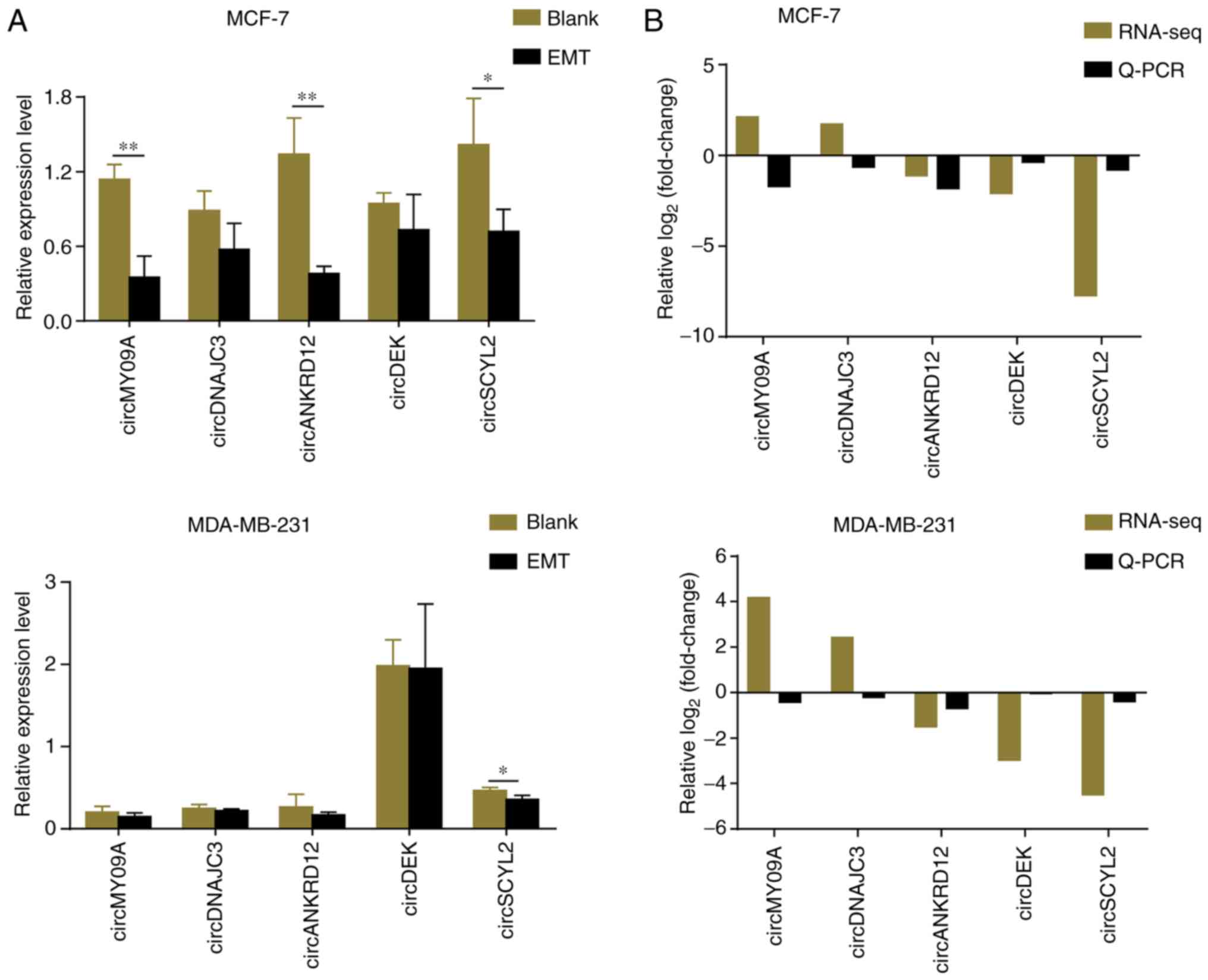
The sequencing data of five candidate circRNAs was
validated using RT-qPCR in EMT groups of MCF-7 and MDA-MB-231 cells
and matched with their blanks, respectively (Fig. 5A). On comparing RNA-seq results
with RT-qPCR information, it was found that three of the five
circRNAs were downregulated in EMT groups, which was consistent
with RNA sequencing. As the stability of circRNA sequencing is not
the same as mRNA sequencing, the results of two circRNAs between
qPCR and RNA-seq were not matched. Notably, circSCYL2 was the most
downregulated circRNA (Fig.
5B).
circSCYL2 is significantly downregulated
in breast cancer
Due to the significant downregulation of circSCYL2
in EMT groups compared with that in the blank group, the current
study next assessed the exon structure of circSCYL2. Convergent
primers were designed to amplify SCYL2 mRNA and divergent primers
to amplify circSCYL2. Using cDNA and genomic DNA (gDNA) from breast
cancer cell lines as templates, circSCYL2 was only amplified by
divergent primers in cDNA, and no amplification product was
observed in gDNA (Fig. 6A).
circSCYL2 derived from exon 2 to 4 of the SCYL2 gene with a length
of 508 nt, which was confirmed the back-spliced junction in the
RT-qPCR product of circSCYL2 with the expected size by Sanger
sequencing (Fig. 6B and C). To
investigate the expression of circSCYL2 in breast cancer, the
expression level of circSCYL2 in 20 pairs of human breast cancer
tissues and normal breast tissues were detected, and the results
demonstrated that the expression of circSCYL2 was significantly
decreased in breast cancer tissues compared with in normal breast
tissues (Fig. 6D).
Overexpression of circSCYL2 inhibits the
migration and invasion of breast cancer cells
The results of the present study demonstrated that
circSCYL2 is downregulated in breast cancer tissues and cell lines.
To investigate the function of circSCYL2 in breast cancer
progression, the circSCYL2 expression construct was transfected
into MCF-7 and MDA-MB-231 cells, and the expression of circSCYL2
was significantly increased. Meanwhile, the expression of
E-cadherin mRNA was enhanced in MCF-7 and MDA-MB-231 cells with
over-expression of circSCYL2, while the expression of Vimentin was
significantly reduced (Fig. 7A).
A matrigel-coated transwell invasion assay was used to examine the
abilities of cell migration and invasion. Transwell assays
indicated that the migration and invasion abilities of breast
cancer cell lines were also suppressed by over-expression of
circSCYL2 (Fig. 7B).
Discussion
Breast cancer metastasis is the leading cause of
mortality in patients; metastatic tumors can occur after a period
of clinical therapy anytime from months to decades, leaving the
survivors uncertain about their longer-term prognosis (36). Recent research has suggested that
circRNAs are involved in various breast cancer biological
procession (37,38), but its function in EMT of breast
cancer remains largely unclear. In this study, the whole
transcriptome RNA-Seq was used to construct circRNA expression
profiles in two breast cancer cells MCF-7 and MDA-MB-231 which were
induced by TGF-β1. A total of 7 upregulated circRNAs and 16
downregulated circRNAs were identified to be shared in the
MCF-7-EMT and MDA-MB-231-EMT groups. Functional investigations
revealed that these differently expressed circRNAs were associated
with immune/inflammatory response, cell proliferation/cell adhesion
functions and relative pathways. The present study identified and
validated that circSCYL2 was downregulated in breast cancer tissues
and cell lines; the overexpression of circSCYL2 in breast cancer
cells inhibited the abilities of cell migration and invasion.
Previous studies have revealed that circRNAs play
important roles in the regulation of multiple diseases, especially
cancers (39-42). Aberrant expression of circRNAs was
also correlated with progression, drug resistance and prognosis of
cancers (21). Increasing
evidence indicates that one of the important functions of circRNAs
is to serve as miRNA sponges to regulate target genes, therefore
the circRNA-miRNA-mRNA network was constructed to find the target
genes of differentially expressed circRNAs (43). For instance, hsa_circ_0058124
promotes papillary thyroid cancer tumorigenesis and invasiveness
through the NOTCH3/GATAD2A axis, elevated levels of hsa_circ_006100
in gastric cancer promote cell growth and metastasis via
miR-195/GPRC5A signaling (44,45). The current study identified 7
significantly upregulated circRNAs and 16 downregulated circRNAs
that were shared in both the MCF-7 EMT group and the MDA-MB-231 EMT
group which demonstrated that these circRNAs may play important
roles in breast cancer metastasis.
circSCYL2 arose from the SCYL2 gene and consisted of
the head-to-tail splicing of exon 2-4. A total of 16 differentially
expressed target genes of circSCYL2 were found in the
circRNA-miRNA-mRNA network and they were downregulated in MCF-7-EMT
and MDA-MB-231-EMT cells. Among them, the odd-skipped related
transcription factor 1 (OSR1) gene, which is located on human
chromosome 2p24.1 and encodes a zinc-finger transcription factor
was focused on. Previous studies suggested that OSR1 suppresses the
progression of gastric cancer and renal cell carcinoma (46,47). Another study also found that OSR1
was downregulated in tongue squamous cell carcinoma (TSCC) cells
and specimens and that OSR1 overexpression inhibited TSCC cell
migration and invasion, while its knockdown promoted TSCC cell
migration and invasion (48).
There was also some evidence that OSR1 is a target of TGF-β
signaling (49), which has a
predominant role and its activation can lead to the expression of
key transcription factors of EMT, such as SNAIL, ZEB and basic
helix-loop-helix transcription factors (50). During the malignant phases of
breast cancer progression, the TGF-β signaling pathway elicits
tumor promoting effects particularly by driving the EMT, which
enhances tumor cell migration, invasion and ultimately metastasis
to distant organs (51). The
current study showed circSCYL2 and its target gene OSR1 targeting
the TGF-β signaling pathway were downregulated in the EMT model of
breast cancer cells, which demonstrates that circSCYL2 may regulate
EMT through OSR1/TGF-β axis.
In conclusion, circRNAs expression profiles were
identified and differentially expressed circRNAs were screened
between EMT and the blank group of two breast cancer cells, and
these circRNAs were associated with immune/inflammatory response
and cell proliferation/cell adhesion functions by bioinformatic
analysis. The present study found circSCYL2 and its target gene
OSR1 were downregulated in breast cancer cells, and the
overexpression of circSCYL2 in breast cancer cells inhibited the
abilities of cell migration and invasion. All these suggest that
further research of circRNAs related to EMT progression may provide
a potential diagnostic and therapeutic biomarker for breast cancer.
Therefore, the relationship of circRNAs was only inferred and more
biological experiments are still needed to verify the interactions
among them; the molecular mechanism underlying the association of
circRNAs with tumor metastasis should be experimentally identified
and characterized in the future.
Supplementary Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SD was involved in the conceptualization of the
study and in funding acquisition. CY, XL, XZ and HZ performed the
experiments and were involved in the investigative aspects of the
study. CY was involved in data curation. XL was involved in the
study methodology. XZ and HZ were involved in the interpretation of
the results and in the provision of software, and also created the
figures. CY, XL and XZ were involved in the writing of the original
draft and in the revision of the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
the Second Affiliated Hospital of Nanchang University. All patients
provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhou J, Zhang WW, Peng F, Sun JY, He ZY
and Wu SG: Downregulation of hsa_circ_0011946 suppresses the
migration and invasion of the breast cancer cell line MCF-7 by
targeting RFC3. Cancer Manag Res. 10:535–544. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rodgers RJ, Reid GD, Koch J, Deans R,
Ledger WL, Friedlander M, Gilchrist RB, Walters KA and Abbott JA:
The safety and efficacy of controlled ovarian hyperstimulation for
fertility preservation in women with early breast cancer: A
systematic review. Hum Reprod. 32:1033–1045. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhou SY, Chen W, Yang SJ, Xu ZH, Hu JH,
Zhang HD, Zhong SL and Tang JH: The emerging role of circular RNAs
in breast cancer. Biosci Rep. 39:BSR201906212019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hattori M and Iwata H: Advances in
treatment and care in meta-static breast cancer (MBC): Are there
MBC patients who are curable? Chin Clin Oncol. 7:232018. View Article : Google Scholar
|
|
6
|
Andersson Y, Bergkvist L, Frisell J and de
Boniface J: Long-term breast cancer survival in relation to the
metastatic tumor burden in axillary lymph nodes. Breast Cancer Res
Treat. 171:359–369. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang Z, Katsaros D, Biglia N, Shen Y, Fu
Y, Loo LWM, Jia W, Obata Y and Yu H: High expression of long
non-coding RNA MALAT1 in breast cancer is associated with poor
relapse-free survival. Breast Cancer Res Treat. 171:261–271. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sobierajska K, Ciszewski WM, Wawro ME,
Wieczorek-Szukała K, Boncela J, Papiewska-Pajak I, Niewiarowska J
and Kowalska MA: TUBB4B downregulation is critical for increasing
migration of metastatic colon cancer cells. Cells. 8:E8102019.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Arnold JM, Gu F, Ambati CR, Rasaily U,
Ramirez-Pena E, Joseph R, Manikkam M, San Martin R, Charles C, Pan
Y, et al: UDP-glucose 6-dehydrogenase regulates hyaluronic acid
production and promotes breast cancer progression. Oncogene. July
15–2019.Epub ahead of print.
|
|
10
|
Gruenbacher G and Thurnher M: Mevalonate
metabolism in cancer stemness and trained immunity. Front Oncol.
8:3942018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bhowmik SK, Ramirez-Peña E, Arnold JM,
Putluri V, Sphyris N, Michailidis G, Putluri N, Ambs S, Sreekumar A
and Mani SA: EMT-induced metabolite signature identifies poor
clinical outcome. Oncotarget. 6:42651–42660. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Porshneva K, Papiernik D, Psurski M,
Łupicka-Słowik A, Matkowski R, Ekiert M, Nowak M, Jarosz J, Banach
J, Milczarek M, et al: Temporal inhibition of mouse mammary gland
cancer metastasis by CORM-A1 and DETA/NO combination therapy.
Theranostics. 9:3918–3939. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang HF, Wang SS, Zheng M, Dai LL, Wang K,
Gao XL, Cao MX, Yu XH, Pang X, Zhang M, et al: Hypoxia promotes
vasculogenic mimicry formation by vascular endothelial growth
factor A mediating epithelial-mesenchymal transition in salivary
adenoid cystic carcinoma. Cell Prolif. 52:e126002019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Singh S and Chakrabarti R: Consequences of
EMT-driven changes in the immune microenvironment of breast cancer
and therapeutic response of cancer cells. J Clin Med. 8:E6422019.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang HL, Thiyagarajan V, Shen PC, Mathew
DC, Lin KY, Liao JW and Hseu YC: Anti-EMT properties of CoQ0
attributed to PI3K/AKT/NFKB/MMP-9 signaling pathway through
ROS-mediated apoptosis. J Exp Clin Cancer Res. 38:1862019.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gener P, Rafael D, Seras-Franzoso J, Perez
A, Pindado LA, Casas G, Arango D, Fernández Y, Díaz-Riascos ZV,
Abasolo I and Schwartz S Jr: Pivotal role of AKT2 during dynamic
phenotypic change of breast cancer stem cells. Cancers (Basel).
11:E10582019. View Article : Google Scholar
|
|
17
|
Holdt LM, Kohlmaier A and Teupser D:
Molecular roles and function of circular RNAs in eukaryotic cells.
Cell Mol Life Sci. 75:1071–1098. 2018. View Article : Google Scholar :
|
|
18
|
Patop IL, Wüst S and Kadener S: Past,
present, and future of circRNAs. EMBO J. 38:e1008362019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Du WW, Yang W, Liu E, Yang Z, Dhaliwal P
and Yang BB: Foxo3 circular RNA retards cell cycle progression via
forming ternary complexes with p21 and CDK2. Nucleic Acids Res.
44:2846–2858. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Guarnerio J, Bezzi M, Jeong JC, Paffenholz
SV, Berry K, Naldini MM, Lo-Coco F, Tay Y, Beck AH and Pandolfi PP:
Oncogenic role of fusion-circRNAs derived from cancer-associated
chromosomal translocations. Cell. 165:289–302. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liang Y, Song X, Li Y, Ma T, Su P, Guo R,
Chen B, Zhang H, Sang Y, Liu Y, et al: Targeting the
circBMPR2/miR-553/USP4 axis as a potent therapeutic approach for
breast cancer. Mol Ther Nucleic Acids. 17:347–361. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yang L, Song C, Chen Y, Jing G and Sun J:
Circular RNA circ_0103552 forecasts dismal prognosis and promotes
breast cancer cell proliferation and invasion by sponging miR-1236.
J Cell Biochem. 120:15553–15560. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xu JH, Wang Y and Xu D: Hsa_circ_001569 is
an unfavorable prognostic factor and promotes cell proliferation
and metastasis by modulating PI3K-AKT pathway in breast cancer.
Cancer Biomark. 25:193–201. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hatta M, Miyake Y, Uchida K and Yamazaki
J: Keratin 13 gene is epigenetically suppressed during transforming
growth factor-beta1-induced epithelial-mesenchymal transition in a
human keratinocyte cell line. Biochem Biophys Res Commun.
496:381–386. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Suzuki S, Toyoma S, Tsuji T, Kawasaki Y
and Yamada T: CD147 mediates transforming growth factor-β1-induced
epithelial-mesenchymal transition and cell invasion in squamous
cell carcinoma of the tongue. Exp Ther Med. 17:2855–2860.
2019.PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
27
|
Martínez-Reza I, Díaz L and García-Becerra
R: Preclinical and clinical aspects of TNF-α and its receptors
TNFR1 and TNFR2 in breast cancer. J Biomed Sci. 24:902017.
View Article : Google Scholar
|
|
28
|
Manna PR, Ahmed AU, Yang S, Narasimhan M,
Cohen-Tannoudji J, Slominski AT and Pruitt K: Genomic profiling of
the steroidogenic acute regulatory protein in breast cancer: In
silico assessments and a mechanistic perspective. Cancers (Basel).
11:E6232019. View Article : Google Scholar
|
|
29
|
Saleh R, Taha RZ, Sasidharan Nair V,
Alajez NM and Elkord E: PD-L1 blockade by atezolizumab
downregulates signaling pathways associated with tumor growth,
metastasis, and hypoxia in human triple negative breast cancer.
Cancers (Basel). 11:E10502019. View Article : Google Scholar
|
|
30
|
Piperigkou Z and Karamanos NK: Estrogen
receptor-mediated targeting of the extracellular matrix network in
cancer. Semin Cancer Biol. July 13–2019.Epub ahead of print.
PubMed/NCBI
|
|
31
|
Sharma S, Wu SY, Jimenez H, Xing F, Zhu D,
Liu Y, Wu K, Tyagi A, Zhao D, Lo HW, et al: Ca2+ and
CACNA1H mediate targeted suppression of breast cancer brain
metastasis by AM RF EMF. EBioMedicine. 44:194–208. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gao JL, Peng K, Shen MW, Hou YH, Qian XB,
Meng XW, Ji FH, Wang LN and Yang JP: Suppression of WNK1-SPAK/OSR1
attenuates bone cancer pain by regulating NKCC1 and KCC2. J Pain.
20:1416–1428. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Prabhu VV, Madhukar NS, Gilvary C, Kline
CLB, Oster S, El-Deiry WS, Elemento O, Doherty F, VanEngelenburg A,
Durrant J, et al: Dopamine receptor D5 is a modulator of tumor
response to dopamine receptor D2 antagonism. Clin Cancer Res.
25:2305–2313. 2019. View Article : Google Scholar
|
|
34
|
Wu Q, Li J, Li Z, Sun S, Zhu S, Wang L, Wu
J, Yuan J, Zhang Y, Sun S and Wang C: Exosomes from the
tumour-adipocyte interplay stimulate beige/brown differentiation
and reprogram metabolism in stromal adipocytes to promote tumour
progression. J Exp Clin Cancer Res. 38:2232019. View Article : Google Scholar
|
|
35
|
Chu H, Hui G, Yuan L, Shi D, Wang Y, Du M,
Zhong D, Ma L, Tong N, Qin C, et al: Identification of novel piRNAs
in bladder cancer. Cancer Lett. 356:561–567. 2015. View Article : Google Scholar
|
|
36
|
Ma B, Wells A and Clark AM: The
pan-therapeutic resistance of disseminated tumor cells: Role of
phenotypic plasticity and the metastatic microenvironment. Semin
Cancer Biol. July 31–2019.Epub ahead of print. PubMed/NCBI
|
|
37
|
Tang H, Huang X, Wang J, Yang L, Kong Y,
Gao G, Zhang L, Chen ZS and Xie X: circKIF4A acts as a prognostic
factor and mediator to regulate the progression of triple-negative
breast cancer. Mol Cancer. 18:232019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ye F, Gao G, Zou Y, Zheng S, Zhang L, Ou
X, Xie X and Tang H: circFBXW7 inhibits malignant progression by
sponging miR-197-3p and encoding a 185-aa protein in
triple-negative breast cancer. Mol Ther Nucleic Acids. 18:88–98.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Garikipati VNS, Verma SK, Cheng Z, Liang
D, Truongcao MM, Cimini M, Yue Y, Huang G, Wang C, Benedict C, et
al: Circular RNA CircFndc3b modulates cardiac repair after
myocardial infarction via FUS/VEGF-A axis. Nat Commun. 10:43172019.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang Y, Huang R, Cheng M, Wang L, Chao J,
Li J, Zheng P, Xie P, Zhang Z and Yao H: Gut microbiota from
NLRP3-deficient mice ameliorates depressive-like behaviors by
regulating astrocyte dysfunction via circHIPK2. Microbiome.
7:1162019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Su Y, Feng W, Shi J, Chen L, Huang J and
Lin T: circRIP2 accelerates bladder cancer progression via
miR-1305/Tgf-β2/smad3 pathway. Mol Cancer. 19:232020. View Article : Google Scholar
|
|
42
|
Chen X, Mao R, Su W, Yang X, Geng Q, Guo
C, Wang Z, Wang J, Kresty LA, Beer DG, et al: Circular RNA
circHIPK3 modulates autophagy via MIR124-3p-STAT3-PRKAA/AMPKα
signaling in STK11 mutant lung cancer. Autophagy. 1–13. 2019.
|
|
43
|
Tian C, Tang X, Zhu X, Zhou Q, Guo Y, Zhao
R, Wang D and Gong B: Expression profiles of circRNAs and the
potential diagnostic value of serum circMARK3 in human acute
stanford type A aortic dissection. PLoS One. 14:e02190132019.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yao Y, Chen X, Yang H, Chen W, Qian Y, Yan
Z, Liao T, Yao W, Wu W, Yu T, et al: Hsa_circ_0058124 promotes
papillary thyroid cancer tumorigenesis and invasiveness through the
NOTCH3/GATAD2A axis. J Exp Clin Cancer Res. 38:3182019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Liang M, Huang G, Liu Z, Wang Q, Yu Z, Liu
Z, Lin H, Li M, Zhou X and Zheng Y: Elevated levels of
hsa_circ_006100 in gastric cancer promote cell growth and
metastasis via miR-195/GPRC5A signalling. Cell Prolif.
52:e126612019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhang Y, Yuan Y, Liang P, Guo X, Ying Y,
Shu XS, Gao M Jr and Cheng Y: OSR1 is a novel epigenetic silenced
tumor suppressor regulating invasion and proliferation in renal
cell carcinoma. Oncotarget. 8:30008–30018. 2017.PubMed/NCBI
|
|
47
|
Otani K, Dong Y, Li X, Lu J, Zhang N, Xu
L, Go MY, Ng EK, Arakawa T, Chan FK, et al: Odd-skipped related 1
is a novel tumour suppressor gene and a potential prognostic
biomarker in gastric cancer. J Pathol. 234:302–315. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Chen W, Wu K, Zhang H, Fu X, Yao F and
Yang A: Odd-skipped related transcription factor 1 (OSR1)
suppresses tongue squamous cell carcinoma migration and invasion
through inhibiting NF-κB pathway. Eur J Pharmacol. 839:33–39. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Gallolu Kankanamalage S, Karra AS and Cobb
MH: WNK pathways in cancer signaling networks. Cell Commun Signal.
16:722018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Liao JY, Wu J, Wang YJ, He JH, Deng WX, Hu
K, Zhang YC, Zhang Y, Yan H, Wang DL, et al: Deep sequencing
reveals a global reprogramming of lncRNA transcriptome during EMT.
Biochim Biophys Acta Mol Cell Res. 1864:1703–1713. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Suriyamurthy S, Baker D, Ten Dijke P and
Iyengar PV: Epigenetic reprogramming of TGF-β signaling in breast
cancer. Cancers (Basel). 11:E7262019. View Article : Google Scholar
|