Introduction
The Fos proto-oncogene, activator protein-1 (AP-1)
transcription factor subunit (c-fos) gene is a
proto-oncogene belonging to the immediate early gene family
(1). c-Fos and c-Jun (a member
of the Jun family of transcription factors) form a heterodimer
through their leucine zipper plus basic domain (2), resulting in the formation of the
AP-1 complex, which recognizes and binds AP-1 response elements in
the promoter and enhancer regions of target genes, thus converting
extracellular signals into changes in gene expression (3). c-Fos is involved in a number of
important cellular events, including cell proliferation,
differentiation and survival (4), and is activated by the MAPK-ERK1/2
signaling pathway, which also positively regulates enterovirus 71
(EV71) replication (5-8).
EV71 is a member of the Picornaviridae
family, which are small non-enveloped, positive-strand RNA viruses
with a genome size of ~7,400 nt. EV71 is the major etiological
agent of hand, foot and mouth disease, which endangers global
public health security (8).
Additionally, this virus causes neurological disease and even
death, particularly in young children (9,10). Picornavirus infection profoundly
affects host cell mRNA translation (8). The 2A and 3C proteases of EV71
cleave eukaryotic translation initiation factor 4 (eIF4) G and
poly(A) binding protein (PABP), respectively (11). The eIF4G protein is a component
of the eIF4F cap-binding complex that is crucial for cap-dependent
translation (11). PABP is an
important factor for cellular mRNA translation that interacts with
several translation initiation factors (11,12). Cleavage of eIF4G and PABP is
considered to contribute to the shutdown of cellular protein
synthesis (12).
Control of protein translation is crucial for cell
proliferation, differentiation, mitosis and programmed apoptosis.
Furthermore, abnormal regulation of translation initiation is often
an important cause of tumors and disease (13-16). The translation initiation of most
eukaryotic proteins relies on the mRNA 5′-m7G cap
structure (12). However,
certain RNA viruses, including EV71, can utilize internal ribosome
entry site (IRES) sequences, which are located in mRNA
5′-untranslated regions (UTRs), to recruit the 40S ribosome
directly to the vicinity of the initiation codon, independently of
the cap structure (17,18).
Previous studies have demonstrated that numerous
cellular genes also harbor IRES elements in the 5′UTR of their mRNA
(19,20). Cap-dependent initiation is com
promised during mitosis, viral infection, hypoxia or apoptosis
(19). A number of
IRES-containing mRNAs use IRES-mediated translation to protect
cells from stress conditions or to induce programmed cell death
(19,21). Therefore, it was hypothesized
that cellular IRES-mediated translation serves an important role in
cellular processes under various conditions (22). For example, the IRES of the
oncogene c-myc (23), the
tumor suppressor gene p53 (24) and the cellular transcription
factor c-jun (25) can
all contribute to the regulation of cellular activities. The latter
pairs with c-Fos to form AP-1 (2).
The most obvious features of cellular IRES sequences
are their GC-rich nature and length (>150 bases) (26). IRES sequences require IRES
trans-acting factors (ITAFs) to recruit the 40S ribosomal subunit,
and cellular IRES sequences promote the selective synthesis of
certain proteins during situations when cap-dependent translation
is compromised (27).
In our previous study, ribosome profiling revealed
that the translation efficiency of c-fos was upregulated
when cellular protein synthesis was stopped by EV71 infection,
which may be regulated by IRES-mediated translation (28). The translation efficiency was
defined as the relative increase in mRNA fragments protected by
ribosomes (28). The present
study investigated the mechanism by which the translation
efficiency of c-fos mRNA is upregulated when cellular
protein synthesis is shut off, and demonstrated the presence of an
IRES element in the 5′UTR of the c-fos mRNA. Further
analysis revealed that nucleotides 31-205 nt of the c-fos
5′UTR are essential for IRES-mediated translation. Furthermore, two
well-known ITAFs, poly(C)-binding protein 2 (PCBP2) and La
autoantigen (La), were found to regulate the activity of the
c-fos IRES and to bind to the 5′UTR of the c-fos
mRNA. In addition, EV71 infection activated the IRES in the
c-fos 5′UTR and may contribute to the increase in c-Fos
levels.
Materials and methods
Plasmid construction
The bicistronic reporter vector pR-F and phpR-F
constructs were gifts from Professor Anne E. Willis (MRC Toxicology
Unit, University of Cambridge, Cambridge, UK) (29). The EV71 infectious clone,
pSVA-EV71, was a gift from Professor Zhiyong Lou (Tsinghua
University, Beijing, China). The empty Flag-tagged plasmid
pCE-puro-3xFlag was a gift from Professor Akio Kihara (Faculty of
Pharmaceutical Sciences, Hokkaido University, Sapporo, Japan)
(30). Plasmid dl-mouse mammary
tumour virus (MMTV) IRES, which contains the MMTV IRES sequence,
was a gift from Professor Marcelo López-Lastra (Escuela de
Medicina, Pontificia Universidad Católica de Chile, Santiago,
Chile) (31). The aforementioned
individuals are the original producers of the plasmids.
PCR was performed using DNA polymerase Ex
Taq® (Takara Biotechnology Co., Ltd.) according to the
manufacturer's protocol. The thermocycling conditions were as
follows: 95°C for 5 min; 95°C for 30 sec, the annealing temperature
was determined according to the primer Tm value and reacted for 30
sec, the extension time was determined according to the length of
the target fragment and the response was at 72°C; 95°C for 7 min;
4°C for preservation. A 2% agarose gel was used for
electrophoresis, and ethidium bromide staining was used to detect
the target fragments. HeLa cells were used to create a cDNA library
[for detailed methods, please refer to reverse
transcription-quantitative (RT-qPCR) analysis] as the PCR template.
The pSVA-EV71 plasmid was extracted by alkaline lysis method and
used as the PCR template. The gapdh fragment and the coding
regions of PTB, PCBP2, La, hnRNP K and P97 were PCR amplified from
a cDNA library. The 5′UTR of c-fos was PCR amplified from
cDNA library and the 5′UTR of EV71 was PCR amplified from
pSVA-EV71. The full-length 5′UTR cDNA of the c-fos mRNA, the
gapdh cDNA fragment (951-1,250 bp), the EV71 5′UTR and
serial truncations of the c-fos 5′UTR were produced using
PCR amplification with forward and reverse primers containing
EcoRI and NcoI endonuclease restriction sites,
respectively. All products were inserted separately into the dual
luciferase vector pR-F between the EcoRI and NcoI
sites, and the c-fos 5′UTR was also inserted into phpR-F
between the EcoRI and NcoI sites. The c-fos
5′UTR was inserted upstream of the translation start codon of the
firefly luciferase gene in the promoter-less pGL3-basic vector
between the MluI and BglII sites. pGL3-basic,
pGL3-SV40 and β-galactosidase (β-gal) were purchased from Promega
Corporation. The coding regions of polypyrimidine tract-binding
protein (PTB), PCBP2 and La were produced by PCR amplification
using forward and reverse primers containing BamHI and
NotI endonuclease restriction sites, while primers for the
heterogeneous nuclear ribonucleoprotein (hnRNP) K and
death-associated protein 5 (P97) coding regions contained
SalI and NotI sites. These coding regions were
separately inserted into the empty Flag-tagged plasmid
pCE-puro-3xFlag between the EcoRI and NcoI sites or
the SalI and NotI sites, respectively. All plasmids
were verified using DNA sequencing (Sanger sequencing; performed by
Sangon Biotech Co., Ltd.). The sequences of all forward and reverse
primers were as follows: c-fos 5′UTR forward, 5′-GCG GAA TTC
ATT CAT AAA ACG CTT GTT ATA AAA GCA GTG GCT GCG G-3′ and reverse,
5′-AAT TAT CCA TGG CGT GGC GGT TAG GCA AAG CCG GG-3′; gapdh
forward, 5′-CCG GAA TTC TAT GAT GAC ATC AAG AAG GTG GTG AAG C-3′
and reverse, 5′-TAT CCA TGG TGA GGG TCT CTC TCT TCC TCT TG-3′;
EV71 5′UTR forward, 5′-CAA GAA TTC TTA AAA CAG CCT GTG GGT
TGC ACC CAC TC-3′ and reverse, 5′-GCC CCA TGG TGT TTG ACT GTA TTG
AGA GTT AAT ATA AAG TTG AGG GTG-3′; c-fos 5′UTR 1-30
forward, 5′-AAT TCA TTC ATA AAA CGC TTG TTA TAA AAG CAG TGC-3′ and
reverse, 5′-CAT GGC ACT GCT TTT ATA ACA AGC GTT TTA TGA ATG-3′;
c-fos 5′UTR 31-113 forward, 5′-ATA GAA TTC GCT GCG GCG CCT
CGT ACT CCA AC-3′ (also used as the forward for c-fos 5′UTR
31-137, c-fos 5′UTR 31-164 and c-fos 5′UTR 31-185)
and reverse, 5′-TAT CCA TGG GTT CGC TGC GCC GCG GCC GCC GGC TCA GTC
TTG-3′; c-fos 5′UTR 114-205 forward, 5′-ATA GAA TTC GAG CAG
TGA CCG TGC TCC TAC CCA GC-3′ and reverse, 5′-ATA CCA TGG CGT GGC
GGT TAG GCA AAG CCG GG-3′ (also used as the reverse for
c-fos 5′UTR 45-205, c-fos 5′UTR 60-205, c-fos
5′UTR 75-205 and c-fos 5′UTR 93-205); c-fos 5′UTR
31-137 reverse, 5′-AAT ACC ATG GTG GGT AGG AGC ACG GCC ACT G-3′;
c-fos 5′UTR 31-164 reverse, 5′-TAT TCC ATG GAG ACA GGT GGG
CGC TGT GAA G-3′; c-fos 5′UTR 31-185 reverse, 5′-TTA TAC CAT
GGG GGC GAG GGG CCG AGG GGC GGA GAC-3′; c-fos 5′UTR 45-205
forward, 5′-GTC GAA TTC TACTCC AAC CGC ATC TGC AGC GAG CAA C-3′;
c-fos 5′UTR 60-205 forward, 5′-ATC GAA TTC TGC AGC GAG CAA
CTG AGA AGC CAA GAC-3′; c-fos 5′UTR 75-205 forward, 5′-TAT
GAA TTC AGA AGC CAA GAC TGA GCC GGC GGC CGC GGC GCA GCG AAC-3′;
c-fos 5′UTR 93-205 forward, 5′-ATT AGA ATT CGG CGG CCG CGG
CGC AGC GAA CGA GCAG-3′; PTB forward, 5′-TAT GGA TCC ATG GAC GGC
ATT GTC CCA GAT ATA GCC-3′ and reverse, 5′-TAA TGC GGC CGC CTA GAT
GGT GGA CTT GGA GAA GGA GAC-3′; PCBP2 forward, 5′-GAG GGA TCC ATG
GAC ACC GGT GTG ATT GAA GG-3′ and reverse, 5′-ATT AGC GGC CGC CTA
GCT GCT CCC CAT GCC ACC CGT CTC-3′; La forward, 5′-GCG GGA TCC ATG
GCT GAA AAT GGT GAT AAT GAA AAG ATG GC-3′ and reverse, 5′-ATT AGC
GGC CGC CTA CTG GTC TCC AGC ACC ATT TTC TGT TTT CTG-3′; hnRNP K
forward, 5′-TCA AGT CGA CAT GGA AAC TGA ACA GCC AG-3′ and reverse,
5′-GCA TGC GGC CGC TTA GAA AAA CTT TCC AGA AT-3′; and P97 forward,
5′-AAT AGT CGA CAT GGC TTC TGG AGC CGA TTC-3′ and reverse, 5′-TAT
TGC GGC CGC TTA GCC ATA CAG GTC ATC AT-3′.
Cell culture and DNA transfection
Human rhabdomyosarcoma (RD), HeLa and 293T cell
lines (The Cell Bank of Type Culture Collection of The Chinese
Academy of Sciences) were cultured in DMEM (Gibco; Thermo Fisher
Scientific, Inc.) with 10% (v/v) FBS (HyClone; Cytiva), penicillin
(100 U/ml) and streptomycin (100 µg/ml). All cells were
cultured at 37°C in a humidified atmosphere with 5%
CO2.
For transient DNA transfection, cells
(1.2×105 cells/well) were seeded into 12-well plates at
24 h before transfection. DNA was transfected into cells using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol.
Opti-MEM (Invitrogen; Thermo Fisher Scientific, Inc.) mixed with
Lipofectamine® 2000 was used for the transfection of
plasmids at room temperature for 20 min. Cell medium was replaced
with complete culture medium at 6 h post-transfection, and after 48
h, the cells were rinsed twice with PBS, and cell extracts were
prepared using 5X Passive Lysis Buffer (Promega Corporation).
The transfection amounts of plasmids were as
follows: 200 ng for pR-F, pR-gapdh-F, pR-c-fos
5′UTR-F, pR-EV71 5′UTR-F, phpR-c-fos 5′UTR-F,
pGL3-basic, pGL3-c-fos 5′UTR, pGL3-SV40, pR-c-fos
5′UTR-F truncations, and dl-MMTV IRES; 50 ng for β-gal; and 250 or
500 ng for pCE-puro-3xFlag, pCE-puro-3xFlag-PTB, pCE -pu ro -3x F
lag-PCBP 2, p CE -pu ro -3x F lag-La, pCE-puro-3xFlag-hnRNP K and
pCE-puro-3xFlag-P97. Among them, pR-F, pR-gapdh-F,
pGL3-basic and pCE-puro-3xFlag were used as negative controls.
In vitro transcription
A MEGAscript T7 High Yield Transcription Kit (Thermo
Fisher Scientific, Inc.) was used to perform in vitro
transcription. The templates were first linearized using
BamHI and then reacted according to the manufacturer's
protocol. According to the supplier's instructions, ~80% of the
product RNA had an m7G cap (New England BioLabs, Inc.)
at the 5′end. An RNeasy Mini Kit (Qiagen GmbH) was used to purify
the RNA. RNA quantified by UV spectrophotometry was transfected
into HeLa or RD cells using Lipofectamine® 3000
(Invitrogen; Thermo Fisher Scientific, Inc.) as aforementioned.
Opti-MEM mixed with Lipofectamine® 3000 was used for the
transfection of 200 ng RNA at room temperature for 20 min. The
following plasmids were used for transcription in vitro:
pR-F, pR-gapdh-F, pR-c-fos 5′UTR-F and pR-EV71
5′UTR-F.
EV71 infection
pSVA-EV71 was linearized using SalI,
transcribed into RNA in vitro, and then transfected into RD
cells using Lipofectamine® 3000 as mentioned in the
in vitro transcription subsection. After cell death, the
culture supernatant was harvested and centrifuged at 13,000 × g for
1 min at room temperature to obtain EV71 particles. EV71 was
expanded in RD cells for three generations. The titers of viruses
were measured using a 50% tissue culture infective dose assay.
Transfected or untransfected cells were infected with EV71 at an
MOI of 5 with maintenance medium containing 2% (v/v) FBS.
Luciferase assay
Renilla luciferase (RL) and firefly
luciferase (FL) dual-luciferase activities were measured using a
Dual-Luciferase Reporter Assay System (Promega Corporation) and FL
single-luciferase activity was measured using a Luciferase Assay
System (Promega Corporation) according to the manufacturer's
protocol, with the exception that only 100 µl of each
reagent was used. For the pR-F series, FL activity was normalized
to Renilla activity. For the pGL3 series, FL activity was
normalized to β-gal activity. Signals were measured using a
luminometer.
The plasmids used to measure dual luciferase
activity were as follows: pR-F, pR-gapdh-F, pR-c-fos
5′UTR-F, pR-EV71 5′UTR-F, phpR-c-fos 5′UTR-F, dl-MMTV
IRES and pR-c-fos 5′UTR-F truncations. The plasmids for
transcription in vitro used to measure dual luciferase
activity were as follows: pR-F, pR-gapdh-F, pR-c-fos
5′UTR-F and pR-EV71 5′UTR-F. The plasmids used to measure FL
single-luciferase activity were as follows: pGL3-basic,
pGL3-c-fos 5′UTR, pGL3-SV40 and β-gal.
Lipofectamine® 2000 or Lipofectamine® 3000
(Invitrogen; Thermo Fisher Scientific, Inc.) was used for
transfection as mentioned in the in vitro transcription and
cell culture and DNA transfection subsections. At 48 h after
transfection, the cell extracts were prepared using 5X Passive
Lysis Buffer (Promega Corporation). For infection experiments,
in vitro-transcribed pR-F, pR-gapdh-F,
pR-c-fos 5′UTR-F and pR-EV71 5′UTR-F mRNA was
transfected into RD cells. After 1 h, RD cells were infected EV71
for 6, 8 and 10 h, and then harvested for FL and RL activity
measurements.
Protein analysis
Cells were washed once with ice-cold 1X PBS and
lysed in buffer containing 50 mM TRIS (pH 7.4), 150 mM NaCl, 2 mM
EDTA, 3% glycerol, 1% NP-40 and protease inhibitor cocktail
(complete, EDTA-free; Roche Diagnostics) for 30 min on ice.
Following centrifugation at 13,000 × g for 10 min at 4°C, the
supernatant was mixed with 5X SDS loading buffer containing 250 mM
TRIS (pH 6.8), 10% SDS (w/v), 50% glycerol (v/v), 5%
2-mercaptoethanol (v/v) and 0.5% Bromophenol Blue (w/v), and boiled
for 10 min at 100°C. The concentration of the protein samples was
determined using the BCA protein quantification kit (Beijing
Dingguo Changsheng Biotechnology Co., Ltd.), and the loading amount
was 20 µg per lane. Samples were resolved using 10% SDS-PAGE
and transferred onto a PVDF membrane (GE Healthcare). Membranes
were blocked with 5% non-fat milk (in 1X PBS) for 45 min at room
temperature, and then probed with the primary antibodies for 90 min
at room temperature. The primary antibodies against c-Fos (cat. no.
YM3469; 1:1,000) and Flag-tag (cat. no. YM3025; 1:5,000) were
purchased from ImmunoWay Biotechnology Company. The primary
antibody against Tubulin (cat. no. ab44928; 1:10,000) and the
secondary goat anti-mouse IgG H&L (HRP) antibody (cat. no.
ab6789; 1:10,000) were purchased from Abcam. Following incubation
with the secondary antibody for 45 min at room temperature, the
membranes were treated with Immobilon Western Chemiluminescent HRP
Substrate (EMD Millipore) and protein signals were detected using
X-ray film.
RNA-protein immunoprecipitation
(RIP)
Cells were co-transfected with 1 µg
pR-c-fos 5′UTR-F and 6 µg of various ITAFs, as
mentioned in the Cell culture and DNA transfection subsection.
Lysates (900 µl per IP reaction) were centrifuged at 1,000 ×
g at 4°C for 15 min and supernatants were aliquoted corresponding
to 3×106 cells. A 6-mg anti-Flag antibodies (cat. no.
YM3025; ImmunoWay Biotechnology Company) was crosslinked with 40
µl magnetic Protein G Dynabeads (EMD Millipore). The
crosslinked beads were incubated with aliquots of pre-cleared cell
lysates at 4°C. After washing, immunoprecipitated RNA-protein
complexes were eluted at 95°C. Eluates were treated for 10 min at
room temperature with proteinase K (Beijing Dingguo Changsheng
Biotechnology Co., Ltd.) and RNA was extracted using an RNeasy Mini
Kit (Qiagen GmbH) and treated with DNaseI (Takara Biotechnology
Co., Ltd.). cDNA synthesis and subsequent RT-PCR were performed as
described subsequently. Western blotting was performed as
aforementioned to detect protein expression. The buffer for cell
lysis and immunoprecipitation consisted of 30 mM HEPES pH 7.3, 160
mM KCl, 2.5 mM MgCl2, 1 mM dithiothreitol (DTT), 0.1%
NP-40, 0.5% Triton X-100 and 10% glycerol. The washing buffer
contained PBS pH 7.4 and 0.02% Tween, and the elution buffer
contained 100 mM TRIS-HCl pH 7.4, 5% SDS, 70 mM β-mercaptoethanol
and 5 mM DTT.
RT-qPCR analysis
RD cells were lysed using TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). Total RNA was
extracted using an RNeasy Mini Kit (Qiagen GmbH). Reverse
transcription was performed with the oligoT primer (Shanghai Sangon
Biotech Co., Ltd.), dNTPs (Takara Biotechnology Co., Ltd.) and
M-MLV Reverse Transcriptase with 5X buffer (Promega Corporation).
cDNA was synthesized at 42°C for 60 min. Thereafter, qPCR was
carried out using 2X SYBR Green Mix (Roche Diagnostics). The
sequences of the primers used were as follows: GAPDH (reference
gene) forward, 5′-AAC AGC GAC ACC CAC TCC TC-3′ and reverse, 5′-CAT
ACC AGG AAA TGA GCT TGA CAA-3′; cFOS forward, 5′-GGG GCA AGG TGG
AAC AGT TAT-3′ and reverse, 5′-CCG CTT GGA GTG TAT CAG TCA-3′. The
following thermocycling conditions were used: Initial denaturation
at 95°C for 5 min, denaturing at 95°C for 15 sec and annealing and
extension together at 60°C for 60 sec for 35 cycles. All oligoT
primers and RT-PCR primers were synthesized by Sangon Biotech Co.,
Ltd. The final data were analyzed using the 2−ΔΔCq
method (32).
RNA structure analysis
The secondary structure of the c-fos 5′UTR
was predicted by version 7.1 of the Geneious software (https://www.geneious.com/).
Statistical analysis
All experimental data were analyzed using GraphPad
Prism 5.0 (GraphPad Software, Inc.) and presented as the mean ±
standard error of the mean. Each independent experiment was
repeated at least twice. One-way ANOVA with Dunnett's multiple
comparison test was used to analyze the statistical differences.
P<0.05 was considered to indicate a statistically significant
difference.
Results
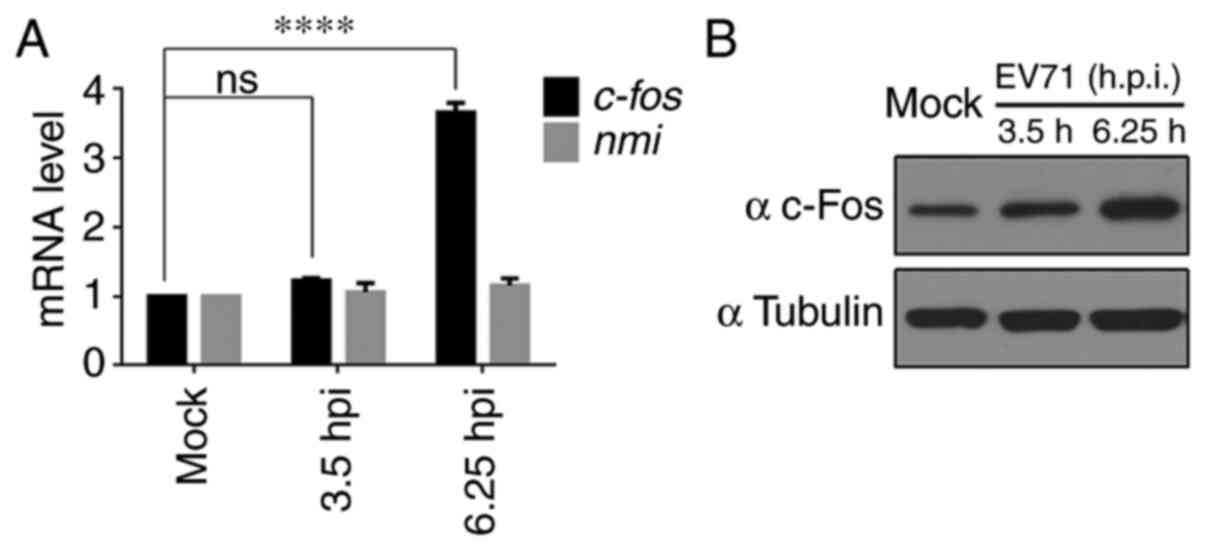
c-Fos expression is upregulated in
EV71-infected RD cells
Previously, ribosome profiling technology was used
to analyze host gene expression in EV71-infected RD cells and it
was demonstrated that the mRNA and translation efficiency of c-Fos
were upregulated (28).
c-fos mRNA expression was assessed using RT-qPCR, and
3.65-fold upregulation was observed at 6.25 h post infection, while
the expression levels of the negative control (N-Myc interactor)
remained unchanged, as expected (28) (Fig. 1A). c-Fos protein expression was
also measured, and a marked increase in c-Fos expression was
observed in EV71-infected RD cells at 6.25 h compared with mock
cells (Fig. 1B).
Identification and verification of an
IRES element in the 5′UTR of c-fos mRNA
To examine whether the high expression levels of
c-Fos were regulated by an IRES-mediated mechanism, the
c-fos 5′UTR was inserted into a bicistronic vector (pR-F),
which contained RL and FL in the first and second cistrons,
respectively. RL and FL were on the same mRNA, so RL was suitable
for system calibration. FL/RL is usually used to show IRES activity
(33,34). Additionally, two negative
controls were included, comprising an empty vector containing the
multiple cloning site in the intercistronic region (pR-F) and a
vector containing a segment from the coding region of human
gapdh (pR-gapdh-F). A plasmid containing the EV71
virus IRES (pR-EV71 5′UTR-F) was used as a positive control
(Fig. 2A). The constructs were
transfected into HeLa cells, and the luciferase activity was
measured. The results suggested that the c-fos 5′UTR
contained a potent IRES element that could direct a marked increase
in the expression of the downstream cistron (Fig. 2B). However, other
non-IRES-dependent causes, such as activation of a splicing event,
readthrough or cryptic promoter activity, had to be eliminated.
 | Figure 2Identification of IRES activity in
the c-fos 5′UTR. (A) Schematic representation of the
expression cassette in the dual-luciferase bicistronic constructs
pR-F, pR-gapdh-F, pR-c-fos 5′UTR-F and pR-EV71
5′UTR-F. EV71 5′UTR was used as the positive control. (B) Plasmids
pR-F, pR-gapdh-F, pR-c-fos 5′UTR-F and pR-EV71
5′UTR-F were transfected into HeLa cells. IRES activity was
expressed as the ratio of downstream cistron expression to upstream
cistron expression (FL/RL). pR-F was the control group. (C)
Schematic representation of constructs phpR-F and phpR-c-fos
5′UTR-F, showing the introduction of a hairpin structure at the
transcription start site of pR-F to rule out aberrant splicing
events and ribosome readthrough. (D) pR-c-fos 5′UTR-F and
phpR-c-fos 5′UTR-F were transfected into HeLa cells, and the
RL and FL activities were measured. RL and FL of pR-c-fos
5′UTR-F were the control group. (E) To examine the cryptic promoter
activity of the c-fos 5′UTR, the sequence was cloned into
the promoterless pGL3-basic vector. The pGL3-SV40 was a positive
control containing a promoter. (F) pGL3-basic, pGL3-c-fos
5′UTR and pGL3-SV40 were co-transfected with the β-gal plasmid into
293T cells. FL activity was measured and normalized against β-gal
activity. pGL3-basic was the control group. (G) The same method as
in (B) was performed in 293T cells. pR-F was the control group. (H)
To exclude the influence of transcription levels and cryptic
promoter, plasmids pR-F, pR-gapdh-F, pR-c-fos 5′UTR-F
and pR-EV71 5′UTR-F were transcribed in vitro. The
purified mRNA was transfected into HeLa cells to directly express
luciferase. pR-F was the control group. Each experiment was
independently repeated at least two times. *P<0.05,
**P<0.01, ***P<0.001 and
****P<0.0001 vs. the respective control. β-gal,
β-galactosidase; c-fos, Fos proto-oncogene, AP-1
transcription factor subunit; EV71, enterovirus 71; FL, firefly
luciferase; IRES, internal ribosome entry site; ns, not
significant; RL, Renilla luciferase; UTR, untranslated
region; phpR-F, bicistronic reporter vector containing hairpin;
pR-F, bicistronic reporter vector; pGL3, reporter vector without
promoter; SV40, simian virus 40 promoter. |
To rule out an aberrant splicing event and ribosome
readthrough, a hairpin structure was introduced at the
transcription start site of the bicistronic reporter (Fig. 2C). The values of FL and RL were
displayed separately, compared with bicistronics without hairpin,
the hairpin only inhibited the expression of the upstream RL
cistron, and in the presence of c-fos 5′UTR fragments, the
expression of FL did not decrease but increased (Fig. 2D). This result suggested that an
aberrant splicing event or ribosome readthrough could be eliminated
as possible mechanisms (35).
To rule out cryptic promoter activities exerting an
effect on the c-fos 5′UTR (36), the c-fos 5′UTR was
inserted into the pGL3-basic vector without a promoter (Fig. 2E). Compared with the pGL3-SV40
positive control, which contained an SV40-promoter,
pGL3-c-fos did not exhibit significant promoter activity
(Fig. 2F). Therefore, it was
unlikely that the c-fos 5′UTR had cryptic promoter
activity.
The activity of an IRES sequences may vary in
different cell lines, and thus, it was examined whether the
c-fos IRES activity would be different in 293T cells.
Therefore, the pR-F series constructs were transfected into 293T
cells. Compared with that in HeLa cells (Fig. 2B), the results revealed that the
IRES activity of c-fos in 293T cells was reduced to 60% of
that in HeLa cells; however, the IRES activity of EV71 in 293T
cells was increased by 1.6-fold (Fig. 2G). This suggested that
c-fos and EV71 had different IRES activities in different
cell lines.
To confirm that c-fos IRES regulated
translation but not transcription, and to exclude the effects of
transcription and cryptic promoters, the pR-F series plasmids were
transcribed in vitro, and the purified mRNA was subsequently
transfected into HeLa cells. The cells were harvested at 8 h after
transfection. Compared with the results of direct transfection of
plasmids (Fig. 2B), the
luciferase activity of the transfected mRNA was similar (Fig. 2H). This further demonstrated that
there was an IRES element in the 5′UTR of the c-fos mRNA
that regulated translation.
Mapping the c-fos IRES element
To further identify the core regions that promote
the internal initiation of translation, serial truncations of the
c-fos 5′UTR were created based on the secondary structure
predicted by the Geneious software (Fig. 3A). Subsequently, different
truncations were inserted into pR-F, and FL/RL was measured. The
results revealed that the construct comprising nucleotides 31-205
maintained most of the IRES activity (Fig. 3B). Therefore, truncations of the
31-205 nt region from the 3′-terminus were created; however, it was
observed that all truncations reduced the IRES activity (Fig. 3C). Subsequently, truncations of
the c-fos 5′UTR 31-205 nt from the 5′-terminus were created,
and as the fragments got shorter, the IRES activity gradually
decreased (Fig. 3D). These
results demonstrated that nucleotides 31-205 of the c-fos
5′UTR contributed to the maximal IRES activity, and further
truncations would have a deleterious effect on the IRES
activity.
PCBP2 and La affect c-fos IRES activity
by binding to the c-fos 5′UTR
PTB, PCBP2, La, hnRNP K and P97 are
well-characterized ITAFs that bind to the IRES region of mRNA and
induce conformational changes to facilitate recruitment of the
ribosome (31,37-40). Therefore, the present study aimed
to evaluate the effect of overexpressing each of these five
proteins on translation driven by the c-fos 5′UTR.
Therefore, the pR-c-fos 5′UTR-F plasmid was co-transfected
with different concentrations of plasmids expressing a Flag-tagged
version of PTB, PCBP2, La, hnRNP K or P97 in 293T cells. PTB has
been reported to be an ITAF of the MMTV IRES (31). Therefore, the dl-MMTV IRES and
Flag-tagged PTB were co-transfected as a positive control. The
results revealed that overexpression of PTB upregulated the IRES
activity of MMTV (Fig. 4A),
whereas PTB had little effect on c-fos-IRES activity
(Fig. 4B). PCBP2 decreased
c-fos-IRES activity (Fig.
4C), and the other three proteins enhanced c-fos-IRES
activity (Fig. 4D-F). PCBP2, La
and P97 were selected for further study to assess whether binding
to the c-fos 5′UTR influenced the activity of the
c-fos IRES.
 | Figure 4PCBP2 and La influence c-fos
IRES activity by binding to the c-fos 5′UTR. (A) dl-MMTV
IRES and 250 or 500 ng pCE-puro-3xFlag-PTB were co-transfected into
293T cells. Flag-PTB expression was detected by western blotting
simultaneously. (B) Plasmid pR-c-fos 5′UTR-F and 250 or 500
ng pCE-puro-3xFlag-PTB were co-transfected into 293T cells, as in
(A), to detect luciferase activity, followed by western blotting.
(C) Plasmid pR-c-fos 5′UTR-F and 250 or 500 ng
pCE-puro-3xFlag-PCBP2 were co-transfected. (D) Plasmid
pR-c-fos 5′UTR-F and 250 or 500 ng pCE-puro-3xFlag-La were
co-transfected. (E) Plasmid pR-c-fos 5′UTR-F and 250 or 500
ng pCE-puro-3xFlag-hnRNP K were co-transfected. (F) Plasmid
pR-c-fos 5′UTR-F and 250 or 500 ng pCE-puro-3xFlag-P97 were
co-transfected. (G) Identification of ITAFs interacting with the
c-fos 5′UTR. There was a non-specific amplification band
<100 bp at the bottom of the figure. Each experiment was
independently repeated two times. *P<0.05,
**P<0.01, ***P<0.001 and
****P<0.0001 vs. not transfected with ITAFs.
c-fos, Fos proto-oncogene, AP-1 transcription factor
subunit; EV71, enterovirus 71; FL, firefly luciferase; hnRNP K,
heterogeneous nuclear ribonucleoprotein k; IRES, internal ribosome
entry site; ITAFs, IRES trans-acting factors; La, La autoantigen;
ns, not significant; P97, death-associated protein 5; PCBP2,
poly(C)-binding protein 2; PTB, polypyrimidine tract-binding
protein; RL, Renilla luciferase; UTR, untranslated region;
dl-MMTV, bicistronic reporter vector containing MMTV IRES; IP,
immunoprecipitation; M, marker; pR-F, bicistronic reporter
vector. |
Therefore, anti-Flag RIP was performed in 293T cells
(Fig. 4G). Lysates were
generated from 293T cells after co-transfection for 48 h. The hnRNP
K [reported to interact with EV71 IRES (37)] and EV71 5′UTR were used as a
positive control, and the 3xFlag vector served as a negative
control. The results demonstrated that detectable products could be
amplified from PCBP2 and La, but not P97, immunoprecipitated mRNA,
indicating the physical interaction of PCBP2 and La with the
c-fos 5′UTR. Overall, these results suggested that PCBP2 and
La influenced the IRES activity of c-fos by binding to the
c-fos 5′UTR.
c-fos 5′UTR-mediated translation is
activated during EV71 infection of RD cells
To investigate whether the upregulation of c-Fos in
EV71-infected RD cells was caused by translation initiated by the
c-fos 5′UTR, an infection confirmation experiment was
performed. The pR-F series plasmids in Fig. 2A, which also contained a T7
promoter, were transcribed in vitro and an m7G
cap was added. In the same mRNA, the expression of RL depended on
the cap, while the expression of FL depended on the inserted
fragments (Fig. 5A). The mRNA
was transfected into RD cells, and cells were infected with 5 MOI
of EV71 for 6, 8 and 10 h at 1 h after transfection. Finally, the
luciferase activity was detected. The results demonstrated that the
IRES activities of c-fos 5′UTR and EV71 were both
significantly upregulated upon EV71 infection at 8 and 10 h
(Fig. 5B). The IRES activity of
the c-fos 5′UTR was activated during EV71 infection and the
IRES of the c-fos 5′UTR, as a translational regulatory
element on mRNA, at least partially contributed to the expression
of c-Fos protein during infection. This provided an explanation for
the mechanism by which EV71 infection upregulated c-Fos expression
at the translation level.
 | Figure 5c-fos 5′UTR-mediated
translation is activated in RD cells during EV71 infection. (A)
Schematic diagram of the in vitro transcription construct.
(B) In vitro-transcribed pR-F, pR-gapdh-F,
pR-c-fos 5′UTR-F and pR-EV71 5′UTR-F mRNA was
transfected into RD cells. After 1 h, RD cells were infected with
EV71 for 6, 8 and 10 h, and then harvested for FL and RL activity
measurements. EV71 5′UTR was used as the positive control. Each
experiment was independently repeated two times.
*P<0.05, **P<0.01,
***P<0.001 and ****P<0.0001 vs. mock.
(C) Schematic diagram of the effect of EV71 infection on host gene
mRNA translation. The image of EV71 was taken from the study by
Plevka et al (41).
c-fos, Fos proto-oncogene, AP-1 transcription factor
subunit; eIF4G, eukaryotic translation initiation factor 4G; EV71,
enterovirus 71; FL, firefly luciferase; IRES, internal ribosome
entry site; La, La autoantigen; ns, not significant; PCBP2,
poly(C)-binding protein 2; RD, rhabdomyosarcoma; RL, Renilla
luciferase; UTR, untranslated region; cap, mRNA 5′-m7G cap
structure; CDS, coding sequence; pR-F, bicistronic reporter
vector. |
A model was provided to summarize the present
findings (Fig. 5C). After EV71
infects the host cell, the viral protease 2A cleaves eIF4G,
shutting down cap-dependent translation of general host genes,
resulting in stagnating of expression (12). At this time, the IRES activity of
c-fos 5′UTR is activated by EV71 infection. The image of
EV71 was taken from the study by Plevka et al (41).
Discussion
When cells are under stress, such as starvation,
hypoxia or apoptosis, overall cellular cap-dependent translation
tends to be turned off (19,22). However, under external stress,
certain key mRNAs in cells initiate translation in an
IRES-dependent manner (19,20). A variety of cellular genes have
been reported to contain IRES elements, including p53,
c-myc and c-jun, which forms a heterodimer with
c-fos (24,25,42). A number of viruses can shut off
the host cell gene expression system to gain a competitive
advantage or for immune evasion (43-47). Therefore, studying the mechanisms
by which certain genes are upregulated during infection will help
understand the crucial proteins and signaling pathways that are
active in host-virus interactions. The present study identified an
IRES element in the c-fos 5′UTR, determined its core region
and identified two ITAFs of the IRES. Furthermore, it was observed
that EV71 infection upregulated the IRES activity of c-fos.
To the best of our knowledge, the present study was the first to
report the identification of the IRES activity in the 5′UTR of the
c-fos mRNA.
EV71 can induce host cap-dependent translation
shutoff (48,49). The expression levels of numerous
genes of the MAPK signaling pathway, including c-fos, are
upregulated in EV71-infected RD cells (50). In our previous study,
RNA-sequencing and ribosome profiling, two high-throughput
techniques, were used to analyze gene expression in RD cells. When
the general cap-dependent translation of host cell was turned off
by EV71, c-fos translation efficiency was markedly
upregulated (28). In the
present study, the sequencing results were first verified and it
was revealed that both the mRNA and protein levels of c-fos
were increased during EV71 infection. Considering that the
c-fos promoter was not markedly affected by EV71 (data not
shown), it is possible that c-fos mRNA continues to be
translated in a cap-independent manner, such as via an IRES.
In contrast to regular viral IRESs, cellular IRESs
cannot be classified because they do not exhibit sequence or
secondary structure similarities (20). Researchers have tried to use
data-base analysis to predict cellular IRESs, such as analyses
using IRSS, IRESfinder, IRESpy and IRESPred (51-54). However, the bicistronic test for
IRES element verification remains the gold standard (51). The present study used the classic
bicistronic reporter system to identify that the c-fos 5′UTR
had strong IRES activity, comparable to that of the EV71 5′UTR, and
ruled out non-IRES-mediated causes. Subsequently, two databases,
IRESpy (53) and IRESPred
(54), were used to make
predictions. The results revealed that the c-fos 5′UTR had
no typical IRES structures (data not shown). On the one hand, this
further demonstrated that the true general characteristics of
cellular IRESs have not yet been identified. On the other hand, it
suggested that the c-fos IRES has a relatively special IRES
structure, which is difficult to predict using the current IRES
library. It was considered that the identification of the
c-fos IRES adds novel information regarding the cellular
IRES library.
The activity and function of an IRES is linked to
its structure (55).
Traditionally, 5′UTRs with high GC content may have IRES activity
(26). The GC percentage of
c-fos 5′UTR fragments was counted. The full-length
c-fos 5′UTR GC content is 64%. The GC content of nucleotides
31-205, which retained the maximum IRES activity was 70%,
corresponding to the middle large stem loop and a series of small
hairpins at the 3′end. These analyses indicated that the GC-rich
regions were favorable factors for the activity of the c-fos
5′UTR, and that the IRES activity was dependent on certain
secondary structures, such as a complete stem-loop or hairpin
combination. Further studies are required to determine the tertiary
and 3D-folded structures of the c-fos 5′UTR. Thus, the
present study provided a foundation for structural research in the
future.
The majority of IRESs, particularly cellular IRESs,
require ITAFs for their function (20). It has been reported that ~50
proteins have the ability to specifically regulate cellular IRESs
(20). In the present study,
PCBP2 downregulated and La upregulated the IRES activity of the
c-fos 5′UTR dose-dependently and both of them interacted
with c-fos 5′UTR mRNA. PCBP2 is involved in
post-transcriptional and translational regulation by interacting
with single-stranded poly(C) motifs in target mRNAs (38). Two adjacent CCCC sites in the
c-fos 5′UTR that may interact with PCBP2 were observed. The
La protein has been demonstrated to interact with a variety of
cellular and viral RNAs and is involved in numerous cellular
processes. Kumar et al (39) revealed that La interacts with the
GCAC motif of the hepatitis C virus IRES to enhance viral RNA
replication. There are six GCA sites in the c-fos 5′UTR
representing candidate La interaction motifs. The specific
interaction mechanisms require further experimental verification.
It was attempted to knock down PCBP2 and La; however, no obvious
impact on c-fos IRES activity was observed (data not shown),
suggesting that there may be other functionally redundant ITAFs
involved in c-fos IRES regulation. Overall, the present
study identified PCBP2 and La as ITAFs for the c-fos
IRES.
Picornavirus exerts complex regulatory effects of
cellular IRESs (40,56). Polypyrimidine tract binding
protein-associated splicing factor (PSF) protein levels are
upregulated in Coxsackievirus B3 (CVB3) infection, and the IRES
element in the psf 5′UTR is activated during CVB3 infection
(40). The EV71 3C protease
cleaves the inhibitor protein hnRNP A1 of the apoptotic peptidase
activating factor 1 (apaf-1) IRES, enabling IRES-dependent
APAF-1 synthesis (56). The
present study demonstrated experimentally that the IRES activity of
pR-c-fos 5′UTR-F mRNA transcribed in vitro exhibited
a trend of gradual upregulation during EV71 infection, providing a
mechanism that explains how the virus upregulates c-Fos expression
at the protein level. However, the molecular details of how EV71
activates the c-fos IRES, and the effects of c-Fos on EV71,
require further exploration.
In summary, previously, the regulation of c-Fos at
the translational level was poorly understood; however, the present
results demonstrated that EV71 upregulated c-fos IRES
activity and c-Fos protein expression. The present results
demonstrated the presence of IRES activity in the c-fos
5′UTR, which is likely to be a special cellular IRES structure. The
present study provided a novel target that enriches the cellular
IRES library.
Availability of data and materials
The datasets used and/or analyzed during the
current study are available from the corresponding author on
reasonable request.
Authors' contributions
HL, YC, JZ and YL performed experiments. HL and YC
collated the data, drafted the manuscript and confirmed the
authenticity of the data. ZY provided guidance on the design of
experiments and made suggestions for the analysis of the results.
JT supervised the experiments and provided general guidance and
interpretation of the results. WQ designed the present study and
provided feasible experimental proposals. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
The authors would like to thank Professor Anne E.
Willis (MRC Toxicology Unit, University of Cambridge, Cambridge,
UK), Professor Zhiyong Lou (Tsinghua University, Beijing, China),
Professor Marcelo López-Lastra (Escuela de Medicina, Pontificia
Universidad Católica de Chile, Santiago, Chile) and Professor Akio
Kihara (Faculty of Pharmaceutical Sciences, Hokkaido University,
Sapporo, Japan) for providing plasmids.
References
|
1
|
van Dam H and Castellazzi M: Distinct
roles of Jun: Fos and Jun: ATF dimers in oncogenesis. Oncogene.
20:2453–2464. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shaulian E and Karin M: AP-1 as a
regulator of cell life and death. Nat Cell Biol. 4:E131–E136. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chiu R, Boyle WJ, Meek J, Smeal T, Hunter
T and Karin M: The c-fos protein interacts with c-JunAP-1 to
stimulate transcription of AP-1 responsive genes. Cell. 54:541–552.
1988. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tulchinsky E: Fos family members:
Regulation, structure and role in oncogenic transformation. Histol
Histopathol. 15:921–928. 2000.PubMed/NCBI
|
|
5
|
Lim H and Kim HP: Matrix
metalloproteinase-13 expression in IL-1beta-treated chondrocytes by
activation of the p38 MAPK/c-Fos/AP-1 and JAK/STAT pathways. Arch
Pharm Res. 34:109–117. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hong S, Skaist AM, Wheelan SJ and Friedman
AD: AP-1 protein induction during monopoiesis favors C/EBP: AP-1
heterodimers over C/EBP homodimerization and stimulates FosB
transcription. J Leukoc Biol. 90:643–651. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Duan H, Zhu M, Xiong Q, Wang Y, Xu C, Sun
J, Wang C, Zhang H, Xu P and Peng Y: Regulation of enterovirus 2A
protease-associated viral IRES activities by the cell's ERK
signaling cascade: Implicating ERK as an efficiently antiviral
target. Antiviral Res. 143:13–21. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shi W, Hou X, Peng H, Zhang L, Li Y, Gu Z,
Jiang Q, Shi M, Ji Y and Jiang J: MEK/ERK signaling pathway is
required for enterovirus 71 replication in immature dendritic
cells. Virol J. 11:2272014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kehle J, Roth B, Metzger C, Pfitzner A and
Enders G: Molecular characterization of an Enterovirus 71 causing
neurological disease in Germany. J Neurovirol. 9:126–128. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Schmidt NJ, Lennette EH and Ho HH: An
apparently new enterovirus isolated from patients with disease of
the central nervous system. J Infect Dis. 129:304–309. 1974.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen LL, Kung YA, Weng KF, Lin JY, Horng
JT and Shih SR: Enterovirus 71 infection cleaves a negative
regulator for viral internal ribosomal entry site-driven
translation. J Virol. 87:3828–3838. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Thompson SR and Sarnow P: Enterovirus 71
contains a type I IRES element that functions when eukaryotic
initiation factor eIF4G is cleaved. Virology. 315:259–266. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Schmeing TM and Ramakrishnan V: What
recent ribosome structures have revealed about the mechanism of
translation. Nature. 461:1234–1242. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rodnina MV and Wintermeyer W: Recent
mechanistic insights into eukaryotic ribosomes. Curr Opin Cell
Biol. 21:435–443. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Silvera D, Formenti SC and Schneider RJ:
Translational control in cancer. Nat Rev Cancer. 10:254–266. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Le Quesne JP, Spriggs KA, Bushell M and
Willis AE: Dysregulation of protein synthesis and disease. J
Pathol. 220:140–151. 2010. View Article : Google Scholar
|
|
17
|
Hellen CU and Sarnow P: Internal ribosome
entry sites in eukaryotic mRNA molecules. Genes Dev. 15:1593–1612.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Balvay L, Soto Rifo R, Ricci EP, Decimo D
and Ohlmann T: Structural and functional diversity of viral IRESes.
Biochim Biophys Acta. 1789:542–557. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Komar AA and Hatzoglou M: Internal
ribosome entry sites in cellular mRNAs: Mystery of their existence.
J Biol Chem. 280:23425–23428. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Godet AC, David F, Hantelys F, Tatin F,
Lacazette E, Garmy-Susini B and Prats AC: IRES trans-acting
factors, key actors of the stress response. Int J Mol Sci. 20:2019.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sherrill KW, Byrd MP, Van Eden ME and
Lloyd RE: BCL-2 translation is mediated via internal ribosome entry
during cell stress. J Biol Chem. 279:29066–29074. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Komar AA and Hatzoglou M: Cellular
IRES-mediated translation: The war of ITAFs in pathophysiological
states. Cell Cycle. 10:229–240. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Stoneley M, Paulin FE, Le Quesne JP,
Chappell SA and Willis AE: C-Myc 5′ untranslated region contains an
internal ribosome entry segment. Oncogene. 16:423–428. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yang DQ, Halaby MJ and Zhang Y: The
identification of an internal ribosomal entry site in the
5′-untranslated region of p53 mRNA provides a novel mechanism for
the regulation of its translation following DNA damage. Oncogene.
25:4613–4619. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Blau L, Knirsh R, Ben-Dror I, Oren S,
Kuphal S, Hau P, Proescholdt M, Bosserhoff AK and Vardimon L:
Aberrant expression of c-Jun in glioblastoma by internal ribosome
entry site (IRES)-mediated translational activation. Proc Natl Acad
Sci USA. 109:E2875–E2884. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sagliocco FA, Vega Laso MR, Zhu D, Tuite
MF, McCarthy JE and Brown AJ: The influence of 5′-secondary
structures upon ribosome binding to mRNA during translation in
yeast. J Biol Chem. 268:26522–26530. 1994. View Article : Google Scholar
|
|
27
|
Li Q, Gao WQ, Dai WY, Yu C, Zhu RY and Jin
J: ATF2 translation is induced under chemotherapeutic drug-mediated
cellular stress via an IRES-dependent mechanism in human hepatic
cancer Bel7402 cells. Oncol Lett. 12:4795–4802. 2016. View Article : Google Scholar
|
|
28
|
Lin Y, Wang Y, Li H, Chen Y, Qiao W, Xie
Z, Tan J and Yang Z: Simultaneous and systematic analysis of
cellular and viral gene expression during Enterovirus 71-induced
host shutoff. Protein Cell. 10:72–77. 2019. View Article : Google Scholar :
|
|
29
|
Coldwell MJ, Mitchell SA, Stoneley M,
MacFarlane M and Willis AE: Initiation of Apaf-1 translation by
internal ribosome entry. Oncogene. 19:899–905. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ohno Y, Kihara A, Sano T and Igarashi Y:
Intracellular localization and tissue-specific distribution of
human and yeast DHHC cysteine-rich domain-containing proteins.
Biochim Biophys Acta. 1761:474–483. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Caceres CJ, Contreras N, Angulo J,
Vera-Otarola J, Pino-Ajenjo C, Llorian M, Ameur M, Lisboa F, Pino
K, Lowy F, et al: Polypyrimidine tract-binding protein binds to the
5′ untranslated region of the mouse mammary tumor virus mRNA and
stimulates cap-independent translation initiation. FEBS J.
283:1880–1901. 2016. View Article : Google Scholar
|
|
32
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
33
|
Gao G, Dhar S and Bedford MT: PRMT5
regulates IRES-dependent translation via methylation of hnRNP A1.
Nucleic Acids Res. 45:4359–4369. 2017.PubMed/NCBI
|
|
34
|
Wein N, Vulin A, Falzarano MS, Szigyarto
CA, Maiti B, Findlay A, Heller KN, Uhlén M, Bakthavachalu B,
Messina S, et al: Translation from a DMD exon 5 IRES results in a
functional dystrophin isoform that attenuates dystrophinopathy in
humans and mice. Nat Med. 20:992–1000. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Thompson SR: So you want to know if your
message has an IRES? Wiley Interdiscip Rev RNA. 3:697–705. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kunze MM, Benz F, Brauss TF, Lampe S,
Weigand JE, Braun J, Richter FM, Wittig I, Brüne B and Schmid T:
sST2 translation is regulated by FGF2 via an hnRNP A1-mediated
IRES-dependent mechanism. Biochim Biophys Acta. 1859:848–859. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lin JY, Li ML, Huang PN, Chien KY, Horng
JT and Shih SR: Heterogeneous nuclear ribonuclear protein K
interacts with the enterovirus 71 5′ untranslated region and
participates in virus replication. J Gen Virol. 89:2540–2549. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang X, Hua L, Yan D, Zhao F, Liu J, Zhou
H, Liu J, Wu M, Zhang C, Chen Y, et al: Overexpression of PCBP2
contributes to poor prognosis and enhanced cell growth in human
hepatocellular carcinoma. Oncol Rep. 36:3456–3464. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kumar A, Ray U and Das S: Human La protein
interaction with GCAC near the initiator AUG enhances hepatitis C
Virus RNA replication by promoting linkage between 5′ and 3′
untranslated regions. J Virol. 87:6713–6726. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Dave P, George B, Sharma DK and Das S:
Polypyrimidine tract-binding protein (PTB) and PTB-associated
splicing factor in CVB3 infection: An ITAF for an ITAF. Nucleic
Acids Res. 45:9068–9084. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Plevka P, Perera R, Cardosa J, Kuhn RJ and
Rossmann MG: Crystal structure of human enterovirus 71. Science.
336:12742012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Le Quesne JP, Stoneley M, Fraser GA and
Willis AE: Derivation of a structural model for the c-myc IRES. J
Mol Biol. 310:111–126. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Fros JJ and Pijlman GP: Alphavirus
Infection: Host cell shut-off and inhibition of antiviral
responses. Viruses. 8:1662016. View Article : Google Scholar :
|
|
44
|
Hoffmann M, Wu YJ, Gerber M,
Berger-Rentsch M, Heimrich B, Schwemmle M and Zimmer G:
Fusion-active glycoprotein G mediates the cytotoxicity of vesicular
stomatitis virus M mutants lacking host shut-off activity. J Gen
Virol. 91:2782–2793. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Vreede FT and Fodor E: The role of the
influenza virus RNA polymerase in host shut-off. Virulence.
1:436–439. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Dremel SE and DeLuca NA: Herpes simplex
viral nucleoprotein creates a competitive transcriptional
environment facilitating robust viral transcription and host shut
off. Elife. 8:e511092019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Rutkowski AJ, Erhard F, L'Hernault A,
Bonfert T, Schilhabel M, Crump C, Rosenstiel P, Efstathiou S,
Zimmer R, Friedel CC and Dölken L: Widespread disruption of host
transcription termination in HSV-1 infection. Nat Commun.
6:71262015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Bai J, Chen X, Liu Q, Zhou X and Long JE:
Characteristics of enterovirus 71-induced cell death and genome
scanning to identify viral genes involved in virus-induced cell
apoptosis. Virus Res. 265:104–114. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ho BC, Yu SL, Chen JJ, Chang SY, Yan BS,
Hong QS, Singh S, Kao CL, Chen HY, Su KY, et al:
Enterovirus-induced miR-141 contributes to shutoff of host protein
translation by targeting the translation initiation factor eIF4E.
Cell Host Microbe. 9:58–69. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Shi W, Hou X, Li X, Peng H, Shi M, Jiang
Q, Liu X, Ji Y, Yao Y, He C and Lei X: Differential gene
expressions of the MAPK signaling pathway in enterovirus
71-infected rhabdomyosarcoma cells. Braz J Infect Dis. 17:410–417.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wu TY, Hsieh CC, Hong JJ, Chen CY and Tsai
YS: IRSS: A web-based tool for automatic layout and analysis of
IRES secondary structure prediction and searching system in silico.
BMC Bioinformatics. 10:1602009. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhao J, Wu J, Xu T, Yang Q, He J and Song
X: IRESfinder: Identifying RNA internal ribosome entry site in
eukaryotic cell using framed k-mer features. J Genet Genomics.
45:403–406. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wang J and Gribskov M: IRESpy: An XGBoost
model for prediction of internal ribosome entry sites. BMC
Bioinformatics. 20:4092019. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Kolekar P, Pataskar A, Kulkarni-Kale U,
Pal J and Kulkarni A: IRESPred: Web server for prediction of
cellular and viral internal ribosome entry site (IRES). Sci Rep.
6:274362016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Plank TD and Kieft JS: The structures of
nonprotein-coding RNAs that drive internal ribosome entry site
function. Wiley Interdiscip Rev RNA. 3:195–212. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Li ML, Lin JY, Chen BS, Weng KF, Shih SR,
Calderon JD, Tolbert BS and Brewer G: EV71 3C protease induces
apoptosis by cleavage of hnRNP A1 to promote apaf-1 translation.
PLoS One. 14:e02210482019. View Article : Google Scholar : PubMed/NCBI
|