Introduction
Breast cancer (BRCA) is the most commonly diagnosed
malignancy among women worldwide (1). Approximately 2.1 million patients
with BRCA were newly diagnosed, and >0.62 million of those
patients succumbed to the disease in 2018 (2). Based on the absence or presence of
human epidermal growth factor (HER)2, estrogen receptors (ERs) or
progesterone receptors (PRs), BRCA has been divided into three
subtypes, namely the ER+/HER2−,
HER+ and ER−/PR−/HER2−
subtypes (3). Although the
majority of patients are diagnosed at the non-metastatic and
curable stage, the remaining women with advanced-stage BRCA are
confronted with a dire predicament, as there are no effective
therapies available. Therefore, identifying novel molecular
biomarkers or essential contributors for BRCA may aid in the
development of effective therapies against this malignancy.
The histidine triad (HIT) protein superfamily is
classified into at least three subgroups, containing histidine
triad nucleotide-binding protein (HINT), fragile histidine triad
diadenosine triphosphatase and galactose-1-phosphate
uridylyltransferase (4).
Increasing evidence has indicated that the HINT subfamily function
as tumor suppressors in various types of cancer. The depletion of
HINT1 in mice has been shown to promote the development of
N-nitrosomethylbenzylamine-induced squamous tumors in the
forestomach and 7,12-dimethylbenz[a]anthracene-induced BRCA
(5,6). Long-term observations have revealed
that HINT1-knockout mice exhibit a higher incidence of spontaneous
tumors compared with wild-type mice (6). The decreased expression of HINT1 has
been found in gastric cancer and hepatocellular carcinoma (HCC)
(7,8). Similar to HINT1, HINT2 has also been
identified as a tumor suppressor in various types of cancer. HINT2
expression has been found to be downregulated in HCC and colorectal
cancer (9,10). The decreased expression of HINT2
has been shown to promote the proliferation, migration and
tumorigenesis of cancer cells (9,11).
Nevertheless, the significance of HINT3 in cancer, including in
BRCA, remains unclear.
In the present study, the clinical relevance of
HINT3 and its potential role in BRCA were investigated. In
vitro studies using cell lines and in vivo experiments
using a nude mouse model were performed to investigate the
functions of HINT3 in BRCA growth. A potential role of HINT3 in
regulating AKT/mammalian target of rapamycin (mTOR) signaling was
also explored using reverse transcription-quantitative PCR
(RT-qPCR), western blot analysis and dual-luciferase activity
assays. Taken together, the results of the present study
demonstrate that HINT3 functions as a tumor suppressor, and
inhibits the activation of the AKT/mTOR signaling cascade in
BRCA.
Materials and methods
The cancer genome atlas (TCGA)
database analysis
The clinical significance of HINT3 in BRCA was
analyzed using the UALCAN website (http://ualcan.path.uab.edu), which provided in-depth
analyses of TCGA (http://cancergenome.nih.gov) for 31 different cancer
subtypes (12). First, 1,097
cancerous and 114 normal tissues were used to analyze HINT3
expression in BRCA and normal tissues. Secondly, 114 normal, 183
stage I, 615 stage II, 247 stage III and 20 stage IV BRCA tissues
were used to analyze the correlation between HINT3 expression and
the BRCA stage. Thirdly, 114 normal, 516 N0 (no regional lymph node
metastasis), 362 N1 (metastases in 1–3 axillary lymph nodes), 120
N2 (metastases in 4–9 axillary lymph nodes) and 77 N3 (metastases
in ≥10 axillary lymph nodes) BRCA tissues were used to analyze the
association between HINT3 expression and nodal metastasis status.
Fourthly, 114 normal tissues, 566 luminal, 37 HER2-positive and 116
triple-negative cancer tissues from TCGA database were used to
analyze the expression of HINT3 and BRCA subclasses. Finally,
Spearman's correlation analysis was used to investigate the
correlation between HINT3 and PTEN in patients with BRCA from TCGA
database.
Human BRCA samples
Human BRCA samples were collected from 14 patients
prior to their receiving any treatment intervention at the General
Hospital of Ningxia Medical University (Yinchuan, China) between
January, 2019 to September, 2020. The patient characteristics are
presented in Table I. Written
informed consent was obtained from all the patients. The present
study was approved by the Ethics Committee of General Hospital of
Ningxia Medical University (no. 2022-68). The ethics approval was
obtained in January, 2019.
 | Table I.Association of HINT3 expression with
the clinicopathological characteristics of 14 patients with breast
cancer. |
Table I.
Association of HINT3 expression with
the clinicopathological characteristics of 14 patients with breast
cancer.
|
| HINT3
expression |
|
|---|
|
|
|
|
|---|
| Characteristic | High | Low | P-value |
|---|
| Age, years |
|
|
|
|
<60 | 5 | 5 | NS |
|
≥60 | 2 | 2 |
|
| Stage I/II/III |
|
|
|
| I | 3 | 3 | NS |
|
II/III | 4 | 4 |
|
| Ki-67 status |
|
|
|
|
Positive | 6 | 2 | NS |
|
Negative | 1 | 5 |
|
| Estrogen receptor
status |
|
|
|
|
Positive | 6 | 2 | NS |
|
Negative | 1 | 5 |
|
| Progesterone
receptor status |
|
|
|
|
Positive | 6 | 2 | NS |
|
Negative | 1 | 5 |
|
|
Triple-negative |
|
|
|
|
Yes | 1 | 5 | NS |
| No | 6 | 2 |
|
Cells and cell culture
The human. The MCF-7 and MDA-MB-231 cells were
cultured in HyClone® medium (Cytiva), the MDA-MB-436 and
MDA-MB-468 cells were cultured in L15 medium (Gibco; Thermo Fisher
Scientific, Inc.), and the MCF10A cells were cultured in DMEM
(Gibco; Thermo Fisher Scientific, Inc.), which was supplemented
with 10% Gibco® fetal bovine serum (Thermo Fisher
Scientific, Inc.) and 1% penicillin and streptomycin solution
(HyClone; Cytiva). The cell culture was maintained at 37°C in an
atmosphere of 5% CO2.
Lentivirus-mediated HINT3 knockdown
and overexpression
A lentivirus vector (GeneChem, Inc.) was used to
knock down HINT3 in the MCF-7 and MDA-MB-231 cells due to the
expression of HINT3. This system comprised three vectors:
pGCSIL-GFP (inserted with the targeted shRNA), pHelper1.0 (gag/pol)
and Helper2.0 (VSVG). The sequences of the shRNAs were as follows:
Negative control, 5′-TTCTCCGAACGTGTCACGT-3′; shHINT3-1,
5′-GCGAGAATGAGGACCTAATTT-3′; and shHINT3-2,
5′-GAGTCAATTCCTATTGGTTTA-3′. After cloning the shRNA into the
pGCSIL-GFP vectors, 15 µg of pGCSIL-GFP-shHINT3-1/2 vectors were
co-transfected with 5 µg of pHelper1.0 and 5 µg of Helper2.0 into
293FT cells (cat. no. R7007, Thermo Fisher Scientific, Inc.) using
Invitrogen™ Lipofectamine™ 2000 reagent
(Thermo Fisher Scientific, Inc.). To overexpress HINT3, the coding
sequence of HINT3 was synthesized and inserted into the pCDH-EGFP
lentivirus vectors (GeneChem, Inc.). The lentivirus was packaged by
co-transfecting 10 µg of pCDH-empty or 10 µg of pCDH-HINT3, 10 µg
of PSPAX2 and 10 µg of PDM2G in 293FT cells using Lipofectamine
2000 for 72 h. Viral supernatants were harvested at 48 and 72 h
following transfection, and then filtered and centrifuged at 72,000
× g/min for 2 h at 4°C for subsequent infection. The cells were
infected with virus at a multiplicity of infection of 3 for 24 h.
The knockdown and overexpression efficiency was subsequently
assessed using western blot analysis.
Western blot analysis
Total proteins were extracted from the shCtrl,
shHINT3-1 and shHINT3-2, Ctrl and HINT3-overexpressing MCF-7 and
MDA-MB-231 cells by lysing them in lysis buffer (Beyotime Institute
of Biotechnology). BCA protein assay, using a BCA kit provided by
the Beyotime Institute of Biotechnology, was performed to detect
the amounts of protein. The proteins were loaded on to 10% SDS-PAGE
gels for separation, and subsequently transferred onto PVDF
membranes (ISEQ00010, MilliporeSigma). Subsequently, the membranes
were blocked with 5% non-fat milk for 60 min at room temperature
and incubated with the following primary antibodies at 4°C
overnight: Anti-HINT3 (1:800; cat. no. ab121960, Abcam),
anti-phosphatase and tensin homolog (PTEN; 1:1,000; cat. no.
CST9188), phosphorylated (p)-AKT (1:1,000; cat. no. CST13038), AKT
(1:1,000; cat. no. CST4691), p-S6 (1:1,000; cat. no. CST2215) and
S6 (1:1,000; cat. no. CST2217) (all from Cell Signaling Technology,
Inc.), mTOR (1:1,000, cat. no. ab32028, Abcam) and p-mTOR (1:1,000,
cat. no. ab109268, Abcam). The loading control antibodies used
were: GAPDH (1:1,000, sc-32233; Santa Cruz Biotechnology, Inc.) and
β-actin (1:3,000; cat. no. 66009-1-Ig, ProteinTech Group, Inc.).
The secondary antibodies, HRP conjugates (1:6,000; cat. nos.
SA00001-1 and SA00001-2) were from ProteinTech Group, Inc. The
membranes were then washed with TBST containing 1% Tween-20 three
times and incubated with the secondary antibodies for 2 h at room
temperature. After washing with PBS for three times, the membranes
were subjected enhanced chemiluminescence using an ECL kit (Promega
Corporation). ImageJ software (V1.8.0, National Institutes of
Health) was performed to analyze the intensity of target
proteins.
RT-qPCR analysis
The shCtrl, shHINT3-1 and shHINT3-2, Ctrl and
HINT3-overexpressing MCF-7 and MDA-MB-231 cells were lysed using
Invitrogen® TRIzol™ reagent (Thermo Fisher
Scientific, Inc.). Total RNA was extracted using an Ultrapure RNA
kit from CoWin BioAciences (cat. no. CW0581), following the
manufacturer's protocol. After reverse transcribing the RNA into
cDNA using an RT-PCR kit (K1002S, Promega Corporation), the qPCR
reaction was performed on a Bio-Rad Laboratories CFX96 machine with
SYBR™-Green PCR Master Mix (Thermo Fisher Scientific,
Inc.). The sequences of the qPCR primers were as follows: HINT3
forward, 5′-CTGGTTGAGAACATGGTAACT-3′ and reverse,
5′-TGATCAGCTGTGATAAACCAAT-3′; PTEN forward,
5′-TTTGAAGACCATAACCCACCAC-3′, and reverse,
5′-ATTACACCAGTTCGTCCCTTTC-3′; and GAPDH forward,
5′-TGACTTCAACAGCGACACCCA-3′, and reverse,
5′-CACCCTGTTGCTGTAGCCAAA-3′. GAPDH served as the internal control.
The thermocycling conditions were as follows: An initial
denaturation at 94°C for 10 min, followed by 45 of cycles of
denaturation at 94°C for 30 sec, annealing at 60°C for 30 sec, and
extension at 72°C for 30 sec. The 2−ΔΔCq method was
performed for quantification (13).
Cell Counting Kit-8 (CCK-8) assay
Equal numbers (3,000 cells per well) of shCtrl,
shHINT3-1, shHINT3-2, Ctrl and HINT3-overexpressing MCF-7 and
MDA-MB-231 cells were seeded into 96-well plates, which contained
200 µl HyClone culture medium (HyClone; Cytiva) with 10% FBS (cat.
no. 26010074, Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin and streptomycin solution (cat. no. 10378016, Gibco;
Thermo Fisher Scientific, Inc.). After 1, 2, 3 and 4 days, 20 µl
CCK-8 reagent (Beyotime Institute of Biotechnology) were added to
each well, and the cells were incubated at 37°C for 3 h. The
optical density (OD) was detected at 450 nm using a
SPECTROstar® Nano microplate reader (BMG LABTECH).
Colony formation assay
Equal numbers of shCtrl, shHINT3-1, shHINT3-2 (500
cells per well for HINT3 knockdown), Ctrl and HINT3-overexpressing
(2,000 cells per well for HINT3 overexpression) MCF-7 and
MDA-MB-231 cells were seeded into six-well plates. Following
culture for 10 days, the colonies were fixed with methanol for 10
min at room temperature and stained using Giemsa solution (48900;
MilliporeSigma) for 20 min at room temperature. Images of the
colonies were captured using a camera (D810A; Nikon Corporation)
and the numbers of the colonies were counted using Photoshop CS5
(Adobe Systems, Inc.).
Transwell assay
Filter migration chambers (8.0 µm; Corning, Inc.)
were used to detect the migration of the shCtrl, shHINT3-1 and
shHINT3-2-transfected MCF-7 and MDA-MB-231 cells. In brief, 60,000
cells were seeded onto the upper surface of the chambers. The lower
chamber was filled with 200 µl HyClone culture medium (HyClone;
Cytiva) containing 10% FBS (cat. no. 26010074, Gibco; Thermo Fisher
Scientific, Inc.) and 1% penicillin and streptomycin solution (cat.
no. 10378016, Gibco; Thermo Fisher Scientific, Inc.). Following
incubation of the chambers at 37°C for 24 h, the cells on the upper
surface were removed using cotton tips. The cells on the lower
surface were fixed with methanol for 10 min at room temperature and
stained using crystal violet (cat. no. 94448, MilliporeSigma) for
20 min at room temperature. The stained cells were photographed
using a light microscope (Ei-BI-1000X, Nikon Corporation). The
magnification was ×20.
Cell apoptosis detection
Cell apoptosis was assessed using an
Invitrogen® PI/Annexin V-APC assay kit (cat. no. V35133,
Thermo Fisher Scientific, Inc.). First, the cells were collected
from six-well plates using EDTA-free trypsin (Corning, Inc.).
Subsequently, cell apoptosis was performed according to the
manufacturer's protocol. Briefly, the cells were incubated with
Annexin-V and PI at room temperature for 20 min in the dark. The
samples were analyzed using a FACScan (BD Biosciences) flow
cytometer.
Dual luciferase activity reporter
assay
The promoter sequence of PTEN was inserted into the
pGL3.basic reporter vector (cat. no. E1751, Promega Corporation),
whereas the coding sequence of HINT3 was inserted into pCDNA3.1
vectors. The MCF-7 cells were transfected with 0.1 µg pGL3.basic,
TK and shRNAs or pCDNA3.1 vectors using Lipofectamine®
2000(Thermo Fisher Scientific, Inc.). Following transfection for 48
h, the firefly and Renilla luciferase activities were
measured using the dual luciferase reporter kit (cat. No. E1910,
Promega Corporation), following the manufacturer's protocol.
5-Ethynyl-2′-deoxyuridine (EdU)
incorporation assay
EdU incorporation assay was performed to determine
DNA synthesis. Briefly, the cells were incubated with 50 µM EdU
(Guangzhou RiboBio Co., Ltd.) for 24 h. The cells were then
collected and centrifuged at 300 × g for 5 min at room temperature.
Red fluorescent reactive dye (Thermo Fisher Scientific, Inc.) was
added to the cells for 30 min at room temperature in the dark,
followed by washing with PBS with 1% BSA and fixing with 4%
paraformaldehyde (Beyotime Institute of Biotechnology) for 15 min
at room temperature. The fixation solution was then removed, and
the cells were resuspended in saponin-based permeabilization buffer
(MilliporeSigma) and incubated for 15 min at room temperature.
Subsequently, the cells were analyzed under a fluorescence
microscope (Olympus Corporation).
Immunohistochemistry (IHC)
A total of 24 paired breast cancer tissues and
adjacent normal tissues were collected without any intervention.
The paraffin-embedded tissues (4-µm-thick) derived from the
patients were deparaffinized, dehydrated and treated with 3%
hydrogen peroxide for 10 min to neutralize endogenous peroxidase
activity. Antigen retrieval was performed by immersing the slides
in 10 mM of boiling citrate buffer (cat. no. ab64214, Abcam) (pH 6)
for 22 min using a microwave oven and then cooling with cold
Tris-buffered saline (TBS, pH 7.4) for 20 min. The sections were
pre-incubated in 0.1 M Tris-HCl buffer (cat. no. 26-575, Moltox)
(pH 7.5) containing 3% BSA at room temperature, and then in TBS
containing 0.02% biotin for 15 min to reduce nonspecific staining.
The following antibodies were used to assess HINT3 and Ki-67
expression: Anti-HINT3 (1:100; cat. no. ab121960, Abcam) and
anti-KI-67 (1:100, IR626, clone MIB-1; Dako; Agilent Technologies,
Inc.) at 4°C for 30 min. After washing with PBS three times, the
immunoreactions were visualized using the rabbit/mouse EnVision-HRP
and DAB + kit (Dako; Agilent Technologies, Inc.). IHC was performed
to analyze the expression of HINT3. HINT3 expression was examined
using signal intensity as follows: 0 (negative), 1 (low), 2
(moderate) and 3 (high) using a Nikon microscope (ECLIPSE E200,
Nikon Corporation).
In vivo xenograft assay
Female Balb/c nude mice (weight, 14–16 g, 6–8 weeks)
were purchased from the Animal Center of General Hospital of
Ningxia Medical University, and were fed a standard diet and water
in a specific pathogen-free room. The temperature was maintained at
25°C and the room was lit with a 12-h light/12-h dark cycle. A
total of 10 mice were randomly divided into two groups (n=5 in each
group). A total of 1×107 MDA-MB-231 cells and a total of
107 MCF-7 cells were subcutaneously implanted into the
right armpit of 5-week-old female Balb/c nude mice, as previously
described (14–18). The volume of the xenograft tumors
was calculated using the following formula: v=ab2/2,
where ‘a’ represents the long diameter and ‘b’ is the short
diameter. Animal health and behavior were monitored at least twice
a week. None of the mice died during the experiment. All the
animals were euthanized when the tumor volume reached approximately
1,500 mm3. The mice implanted with MDA-MB-231 cells were
euthanized using CO2 (a flow rate of 50% chamber
volume/min) after 45 days, whereas those implanted with the MCF-7
cells were euthanized after 48 days. The death of the mice was
confirmed when the mice exhibited loss of breathing, and no
response to stimuli. The animal experiments were approved by the
Ethics Committee of General Hospital of Ningxia Medical University
(no. 2022-68, which was the same ethics approval document as that
for the human experiments), and were performed according to the
animal guidelines of the General Hospital of Ningxia Medical
University. The ethics approval for the study was obtained in
January, 2019.
Statistical analysis
Statistical analyses were performed using GraphPad
prism 8 software (GraphPad Software, Inc.). The data are presented
as the mean ± standard error of mean (SEM). An unpaired Student's
t-test was used for analyzing differences between two groups, and
one-way ANOVA followed by Tukey's post hoc test for multiple group
comparisons. The χ2 test or Fisher's exact test were
used to analyze categorical variables. P<0.05 was considered to
indicate a statistically significant difference.
Results
Expression of HINT3 is reduced in BRCA
tissues, and is inversely associated with tumor stage
TCGA database contains a large amount of information
on the transcript abundance of various genes in a variety of cancer
types. In the present study, to evaluate the clinical relevance of
HINT3 in BRCA, the mRNA expression levels of HINT3 were analyzed
using TCGA database. The analysis revealed that HINT3 expression
was decreased in BRCA tissues compared with normal tissues
(Fig. 1A). Subsequently, normal
and BRCA tissues were collected, and subjected to RT-qPCR in order
to assess the HINT3 mRNA level. The results obtained demonstrated
that the mRNA level of HINT3 was evidently decreased in BRCA
tissues (Fig. 1B and C). Since
BRCA is a malignancy with a rapid rate of disease progression and a
high potential for metastasis, HINT3 expression in BRCA tissues was
subsequently analyzed according to different cancer stages and the
metastasis status. Based on the samples analyzed, HINT3 expression
was significantly decreased in breast cancer tissues, compared with
adjacent normal tissues (Fig. 1D
and Table II). In addition, it
was found that the low expression of HINT3 was associated with
Ki-67 positive staining, although this was not associated with the
cancer stage of the patients (Table
I). Based on TCGA database, the median expression levels of
HINT3 in normal tissues, and in cancer tissues of stages I–IV were
31.635, 26.23, 25.56, 26.527 and 22.656, respectively. Compared
with the normal tissues, HINT3 expression was found at lower levels
in the cancer tissues. The expression of HINT3 was lowest in cancer
tissues of stage IV, although compared with the other stages, this
difference was not found to be significant (Fig. 1E). In addition, the median
expression level of HINT3 in normal and cancer tissues of N0, N1,
N2 and N3 (see the Materials and methods section for a fuller
explanation of these terms) was 31.635, 25.716, 25.738, 27.973 and
25.045, respectively. Compared with the normal tissues, HINT3
expression was lower in cancer tissues with a varied metastatic
status. Nevertheless, the P-values were >0.05 among cancer
tissues with N2 metastatic status and normal tissues (Fig. 1F); thus, these results were not
statistically significant. Finally, the median expression level of
HINT3 in normal tissues, and in luminal, HER2-positive and
triple-negative cancer tissues was 31.635, 26.597, 20.385 and
26.338, respectively. Although HINT3 was decreased in cancer
tissues compared with normal tissues, no significant differences
were observed among the subgroups (Fig. 1G). In addition, the expression of
HINT3 was examined in several BRCA cell lines, and it was found
that its expression was decreased in MCF7, MDA-MB-231 and
MDA-MB-436 cell lines, and increased in MCF10A and MDA-MB468 cell
lines (Fig. 1H). Therefore, HINT3
expression was clearly shown to be reduced in BRCA tissues,
although the change in levels was not significantly associated with
either the stage of cancer or the metastatic status of the
patients. Taken together, these findings indicate that HINT3 may
participate in the development of BRCA.
 | Figure 1.HINT3 expression is decreased in BRCA
tissues. (A) The transcript of HINT3 was analyzed in BRCA tissues
(n=1,097; median, 25.962) and normal tissues (n=114; median,
31.635) from TCGA database (***P<0.001). (B) The mRNA expression
of HINT3 was examined using RT-qPCR in BRCA tissues and paired
normal tissues. Statistical significance was analyzed using the
Mann-Whitney test (*P<0.05). (C) HINT3 mRNA expression was
determined using RT-qPCR in BRCA tissues and normal tissues.
Statistical significance was analyzed using the Mann-Whitney test
(*P<0.05). (D) The expression of HINT3 was analyzed in BRCA
tissues using immunohistochemistry. (E) The expression of HINT3 was
analyzed in BRCA tissues of different individual stages from TCGA
database. The median expression levels of HINT3 in normal tissues,
and stage I, stage II, stage III and stage IV cancer tissues were
found to be 31.635, 26.23, 25.56, 26.527 and 22.656, respectively;
*P<0.05. (F) The expression of HINT3 was analyzed in BRCA
tissues with a different metastatic status from TCGA database. The
median expression of HINT3 in normal tissues, N0, N1, N2 and N3
cancer tissues was found to be 31.635, 25.716, 25.738 and 27.9733,
respectively (**P<0.01, ***P<0.001). (G) The expression of
HINT3 was analyzed in BRCA tissues based on BRCA subclasses from
TCGA database. The median expression of HINT3 in normal tissues,
luminal, HER2-positive and triple-negative cancer tissues was
31.635, 26.597, 20.385 and 26.338, respectively (*P<0.05). (H)
The expression of HINT3 in different breast cancer cell lines was
examined using western blot analysis. BRCA, breast cancer; TCGA,
The Cancer Genome Atlas; HINT3, histidine triad nucleotide-binding
3; RT-qPCR, reverse transcription-quantitative PCR. |
 | Table II.The expression of HINT3 in normal and
breast cancer tissues determined using immunohistochemistry. |
Table II.
The expression of HINT3 in normal and
breast cancer tissues determined using immunohistochemistry.
| HINT3 expression
status | Tumor tissue | Adjacent normal
tissue | χ2 test
value | P-value |
|---|
| High
expression | 3 | 10 | 7.04 | 0.008 |
| Low expression | 11 | 4 |
|
|
| Total | 14 | 14 |
|
|
HINT3 inhibits the proliferation and
colony formation of BRCA cells
To explore the role of HINT3 in BRCA cells, the
lentivirus-mediated overexpression of HINT3 in BRCA cells was first
performed. Since HINT3 expression was found to be decreased in BRCA
cells, including MCF-7 and MDA-MB-231 cells, these cell lines were
selected for further investigations. The results of western blot
analysis revealed that HINT3 was overexpressed in both BRCA cell
lines transfected with overexpression vector (i.e., MCF-7 and
MDA-MB-231 cells) (Fig. 2A).
Subsequently, the effects of HINT3 on BRCA cell proliferation were
examined using CCK-8 and colony formation assays. HINT3
overexpression was found to inhibit the proliferation of both the
MCF-7 and MDA-MB-231 cells (Fig.
2B). The overexpression of HINT3 also led to the suppression of
the colony formation ability of both cell lines (Fig. 2C). Subsequently, the knockdown
efficiency of HINT3 was assessed using western blot analysis of
these cells. The results revealed that the protein levels of HINT3
were significantly reduced in the shHINT3-1- and
shHINT3-2-transfected MCF-7 and MDA-MB-231 cells (Fig. 2D). The decreased levels of HINT3
clearly promoted the proliferation of both MCF-7 and MDA-MB-231
cells (Fig. 2E). Consistently,
the knockdown of HINT3 enhanced the proliferation and
colony-forming ability of the MCF-7 and MDA-MB-231 cells (Fig. 2F and G). Taken together, these
results suggest that HINT3 functions as a tumor suppressor protein
in BRCA.
 | Figure 2.HINT3 suppresses the proliferation of
BRCA cells. (A) Western blot analysis of HINT3 in Ctrl and
HINT3-overexpressing MCF-7 and MDA-MB-231 cells. GAPDH served as
the internal control (***P<0.001). (B) Cell proliferation was
determined using CCK-8 assay (*P<0.05 and **P<0.01, vs.
control). (C) Ctrl and HINT3-overexpressing MCF-7 and MDA-MB-231
cells were subjected to colony formation assay (colony images are
presented on the left panel, and the quantification of the results
is shown on the right panel (*P<0.05, vs. control). (D) Western
blot analysis of HINT3 in shCtrl, shHINT3-1 and shHINT3-2 MCF-7 and
MDA-MB-231 cells. GAPDH served as the internal control. (E) Cell
proliferation was determined using CCK-8 assay (*P<0.05 and
**P<0.01, vs. control). (F and G) shCtrl, shHINT3-1 and
shHINT3-2 MCF-7 and MDA-MB-231cells were subjected to colony
formation assay; (F) colony images, and (G) the quantification of
the same results (*P<0.05 and **P<0.01, vs. control). BRCA,
breast cancer; HINT3, histidine triad nucleotide-binding 3; Ctrl,
control. |
HINT3 reduces EdU incorporation and
promotes the apoptosis of BRCA cells
Proliferative cells require increased DNA synthesis,
and EdU incorporation is an assay that enables the analysis of DNA
synthesis to be made (19). Thus,
in the present study, DNA synthesis was then examined using EdU
staining in BRCA cells. Compared with the EdU staining density in
the shCtrl MCF-7 and MDA-MB-231 cells, the positive signal of EdU
was markedly increased in the shHINT3-1 and shHINT3-2 cells
(Fig. 3A). By contrast, the
incorporation of EdU was suppressed by the overexpression of HINT3
in the MCF-7 and MDA-MB-231 cells (Fig. 3B). Subsequently, the present study
examined whether HINT3 regulates the apoptosis of BRCA cells. These
experiments revealed that HINT3 knockdown significantly reduced the
apoptosis of the MCF-7 and MDA-MB-231 cells (Fig. 3C). On the other hand, HINT3
overexpression significantly promoted the apoptosis of both cell
lines (Fig. 3D). Taken together,
these experiments confirmed that HINT3 suppressed DNA synthesis,
and promoted the apoptosis of BRCA cells.
 | Figure 3.HINT3 inhibits the EdU incorporation
of BRCA cells. (A) EdU staining was performed in shCtrl, shHINT3-1
and shHINT3-2 MCF-7 and MDA-MB-231 cells. Staining images are
presented on the left panel, whereas the quantification of the same
results is presented on the right panel (*P<0.05 and
**P<0.01, vs. control). (B) EdU staining was performed in Ctrl
and HINT3-overexpressing MCF-7 and MDA-MB-231 cells. Again,
staining of the images is presented on the left panel, and the
quantification of the same results is presented on the right panel
(**P<0.01, vs. control). (C) Cell apoptosis was examined using
PI/Annexin V-APC staining in shCtrl, shHINT3-1 and shHINT3-2 MCF-7
and MDA-MB-231 cells. Cell apoptosis images are presented on the
left panel, and the quantification of the results is presented on
the right panel (**P<0.01, vs. control). (D) Cell apoptosis was
examined using by PI/Annexin V-APC staining in the Ctrl and
HINT3-overexpressing MCF-7 and MDA-MB-231 cells The cell apoptosis
images are presented on the left, and the quantification of the
results is presented on the right panel (**P<0.01, vs. control).
BRCA, breast cancer; HINT3, histidine triad nucleotide-binding 3;
Ctrl, control; EdU, 5-ethynyl-2′-deoxyuridine. |
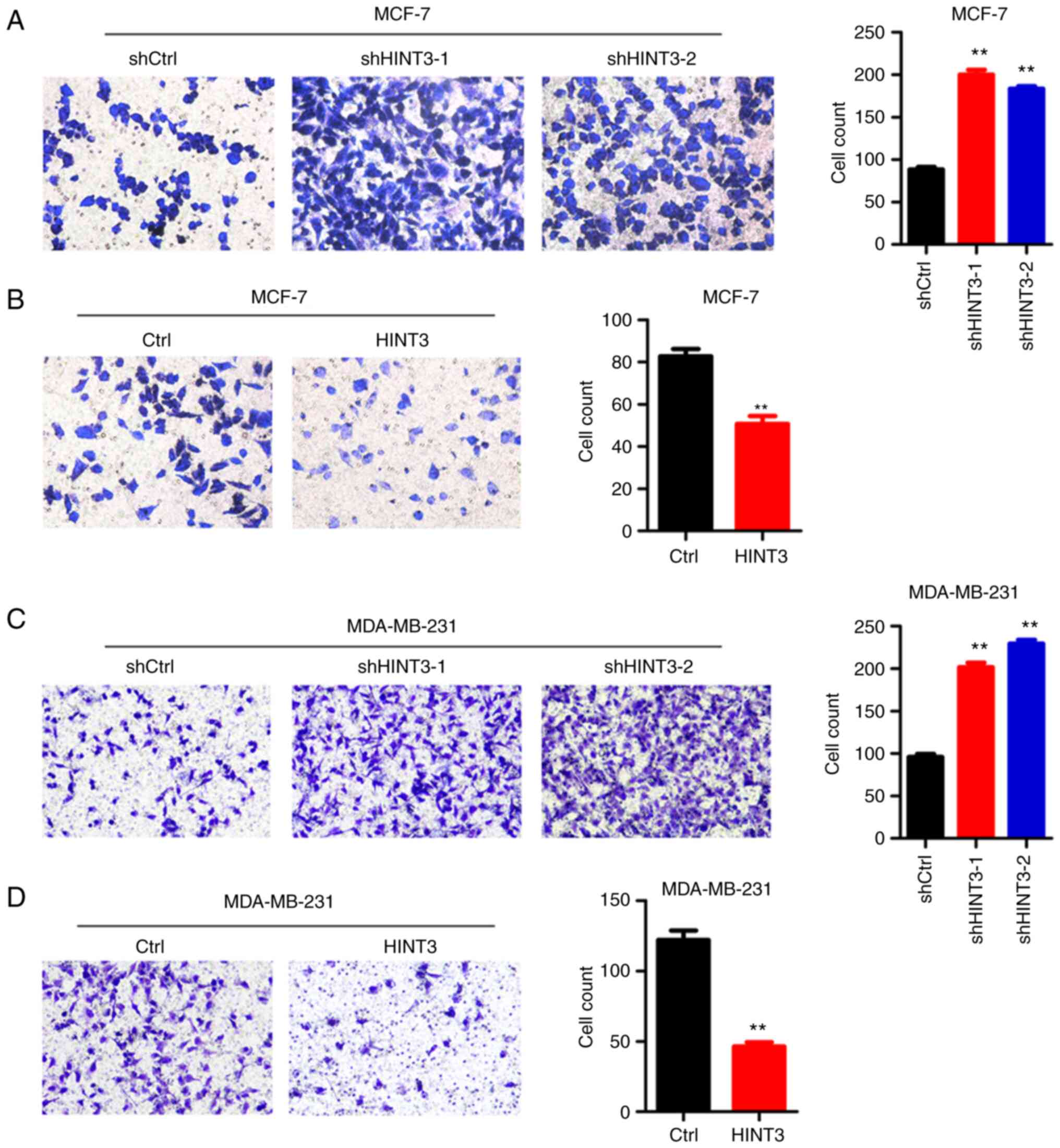
HINT3 suppresses the migration of
MCF-7 and MDA-MB-231 cells
BRCA is a malignancy with a high potential for
metastasis. Subsequently, the role of HINT3 in BRCA metastasis was
investigated using Transwell assay. The results obtained revealed
that HINT3 knockdown increased the migration of MCF-7 cells,
whereas the ectopic expression of HINT3 suppressed the migratory
capacity of the cells (Fig. 4A and
B). Similar results were obtained in HINT3-silenced and
-overexpressing MDA-MB-231 cells (Fig. 4C and D).
HINT3 suppresses PTEN/AKT/mTOR
signaling in BRCA cells
Subsequently, the molecular changes in MCF-7 and
MDA-MB-231 cells that were associated with the knockdown or
overexpression of HINT3 were explored. It was found that HINT3
knockdown and overexpression led to the down- and upregulation of
PTEN at the mRNA level, respectively (Fig. 5A). Spearman's correlation analysis
in patients with BRCA from TCGA database revealed that HINT3
inversely correlated with PTEN in BRCA cancer samples (Fig. 5B). HINT3 is a nuclear protein that
may participate in regulating the transcriptional activity of
downstream genes (20) (PMID:
17870088). Subsequently, in the present study, luciferase activity
reporter assay was performed to examine whether HINT3 regulates the
transcriptional activity of PTEN. The results obtained revealed
that HINT3 promoted the transcriptional activity of PTEN in the
MCF-7 and MDA-MB-231 cells (Fig.
5C). The results of western blot analysis also revealed that
PTEN protein expression was decreased following the silencing of
HINT3, and was increased following the ectopic expression of HINT3
(Figs. 5D and S1). Consistently, it was found that the
phosphorylation, but not the protein expression, of AKT, mTOR and
S6 were enhanced and suppressed by HINT3 knockdown and
overexpression, respectively in the MCF-7 and MDA-MB-231 cells
(Figs. 5D and S1). Taken together, these experimental
findings suggested that PTEN/AKT signaling may be involved in the
tumor-suppressive role of HINT3 in BRCA.
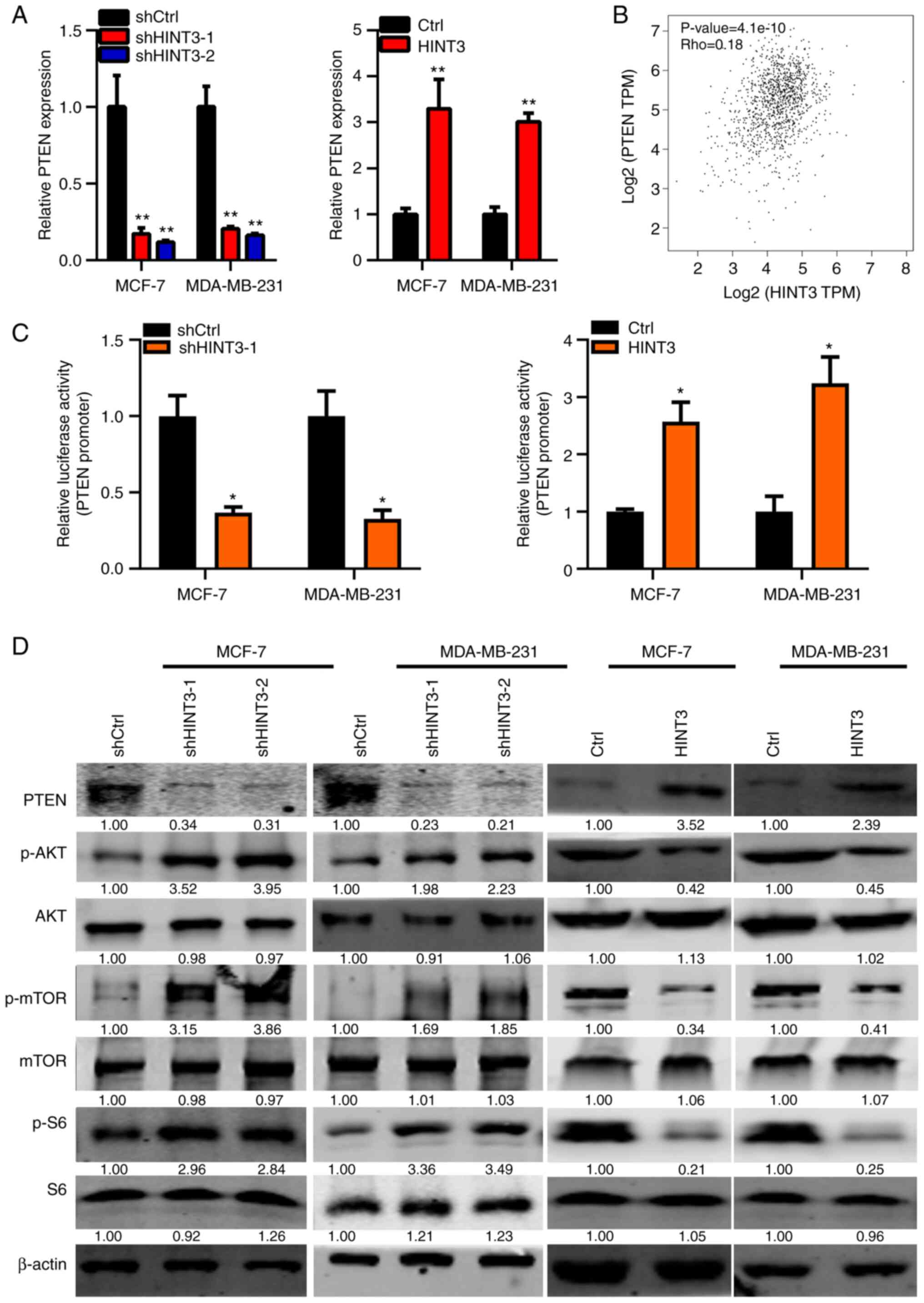 | Figure 5.HINT3 inhibits AKT signaling in BRCA
cells. (A) RT-qPCR analysis of PTEN in shCtrl, shHINT3-1 and
shHINT3-2, and in Ctrl and HINT3-overexpressing MCF-7 and
MDA-MB-231 cells (**P<0.01, vs. control). (B) The correlation
analysis between PTEN and HINT3 expression in TCGA database is
shown. (C) Dual luciferase activity was examined in MCF-7 and
MDA-MB-231 cells transfected with HINT3-overexpressing vectors or
shHINT3-1, pGL3.Basic and TK vectors (*P<0.05, vs. control). (D)
shCtrl, shHINT3-1 and shHINT3-2, Ctrl and HINT3-overexpressing
MCF-7 and MDA-MB-231 cells were subjected to western blot analysis
to assess the protein levels of PTEN, p-AKT, AKT, p-mTOR, mTOR,
p-S6, S6 and β-actin. β-actin served as the internal control. The
quantification of proteins is presented in Fig. S1. PTEN, phosphatase and tensin
homolog; BRCA, breast cancer; HINT3, histidine triad
nucleotide-binding 3; mTOR, mammalian target of rapamycin; Ctrl,
control; p-, phosphorylated. |
HINT3 inhibits AKT activity and tumor
xenograft growth
To explore the functions of HINT3 in vivo,
MDA-MB-231 and MCF-7 cells infected with Ctrl or
HINT3-overexpressing lentivirus were subcutaneously implanted into
the right armpits of 5-week-old female nude mice. The mice were
sacrificed either on day 45 (for the MDA-MB-21 cells) or day 48
(for the MCF-7 cells), as detailed in the Materials and methods
section. In the present study, the maximum tumor volume obtained
was measured. These results revealed that HINT3 overexpression
inhibited the tumorigenesis and progression of MDA-MB-231 cells
(Fig. 6A and B). Consistent
results were observed in the MCF-7 cells with the overexpression of
HINT3 (Fig. 6C and D). It was
also found that HINT3 overexpression suppressed AKT and S6
phosphorylation in the tumors obtained following the implantation
of the MDA-MB-231 and MCF-7 cells (Figs. 6E and F, and S2). Taken together, these results
suggest that HINT3 can block AKT activity and the tumor growth of
BRCA cells in nude mice.
Discussion
Unlike HINT1 and HINT2, the role of HINT3 has yet to
be fully elucidated in cancer development. In the present study, it
was demonstrated that HINT3 functioned as a tumor suppressor in
BRCA. In vitro, HINT3 knockdown promoted the proliferation,
colony growth, EdU incorporation and migration of BRCA cells. By
contrast, opposite results were obtained in HINT3-overexpressing
BRCA cells. In vivo, the tumorigenesis of MDA-MB-231 and
MCF-7 cells was suppressed via the ectopic expression of HINT3.
In total, ~10% of patients with BRCA exhibit
inherited genetic variants, and among these, mutations of BRCA1 and
BRCA2, which are involved in DNA repair, play crucial roles in BRCA
development (21–24). Furthermore, the most commonly
mutated genes or amplified genes, such as TP53, PIK3CA, MYC, PTEN,
CCND1, ERBB2, FGFR1 and GATA3, play critical roles in the
progression of BRCA (25). These
genetic alterations provide possible avenues for therapeutic
intervention in patients with BRCA. One representative drug is
poly(ADP-ribose) polymerase (PARP) inhibitor, which is used to
treat patients with BRCA who have BRCA mutations (26). However, the effectiveness of this
treatment remains unsatisfactory, and therefore the use of PARP
inhibitor is limited. It is thus necessary to identify novel drug
targets to cure this malignancy. The HINT family contains 3 family
members, including HINT1, HINT2 and HINT3. A previous study
demonstrated that HINT1 displayed tumor-suppressive properties and
regulated tumorigenesis signaling pathways, and may thus function
as a diagnostic target in tumors (27). Another member, HINT2 has been
reported to be downregulated in hepatocellular carcinoma tissues
from patients with a poor prognosis (28). To date, at least to the best of
our knowledge, no studies have investigated the role of HINT3 in
tumor formation or progression. In the present study, it was found
that HINT3 functioned as a tumor suppressor in BRCA. HINT3
expression was found to be decreased in BRCA tissues. Functional
assays revealed that the ectopic expression of HINT3 suppressed the
proliferative and migratory capacity of the MCF-7 and MDA-MB-231
cells. DNA synthesis, which is the hallmark of cell cycle
progression, was also suppressed by HINT3. Moreover, cell apoptosis
was promoted by HINT3 overexpression, and was suppressed by HINT3
knockdown. In vivo, HINT3-overexpressing MDA-MB-231 and
MCF-7 cells developed xenograft tumors at a slower rate compared
with Ctrl MDA-MB-231 and MCF-7 cells. Taken together, these
findings indicated that HINT3 exhibited tumor-suppressive functions
in BRCA cells; thus, it may be suitable as a diagnostic target for
breast cancer.
PTEN is a well-known tumor suppressor (29). The inactivation of PTEN leads to
the enhanced phosphorylation and activity of AKT. The activation of
AKT contributes to cancer development via the regulation of
distinct downstream targets, including glycogen synthase kinase 3β,
FOXO, p21 and caspase 9 (30).
The AKT/mTOR pathway is involved in the regulation of cell growth,
differentiation, motility and survival, processes which are usually
enhanced in various types of cancer, such as colorectal cancer,
ovarian and cervical cancer; the inhibitor of mTOR signaling,
suppresses colorectal cancer development, and thus, it has been
considered as a potential target for cancer therapy (31,32).
The absence of PTEN, or mutations of PTEN are
commonly observed in BRCA (33–35). However, the upstream regulator of
PTEN is not yet well known in BRCA. In the present study, it was
found that HINT3 negatively regulated the mRNA and protein
expression of PTEN. HINT3 knockdown and overexpression up- and
downregulated the phosphorylation level of AKT, respectively.
Furthermore, the results of dual luciferase activity reporter assay
demonstrated that HINT3 promoted the transcriptional activity of
PTEN gene in BRCA cells. Of note, HINT3 overexpression also
suppressed the activity of AKT in BRCA tumors, suggesting that the
HINT3-induced inactivation of AKT is involved in the tumorigenesis
of BRCA. Collectively, these results suggest that HINT3 suppresses
the development of BRCA, at least partly by inactivating the AKT
signaling pathway. Accordingly, owing to the key roles of PTEN and
the AKT/mTOR pathway, HINT3 may play crucial roles in other
diseases, such as human immunodeficiency virus and coronavirus
disease 2019, as well as in autoimmune diseases, such as multiple
sclerosis and systemic lupus erythematosus (36–39).
However, the present study had several limitations
which should be mentioned. The molecular mechanisms through which
HINT3 downregulation may operate in BRCA remain unknown. Whether
HINT3 downregulation is associated with promoter hypermethylation,
or whether it is regulated by other factors at the transcriptional
level in BRCA requires further investigation. Moreover, the
association between HINT3 and the stage/metastasis and
status/molecular subtype of patients with BRCA was found not to be
significant, as shown from TCGA database. Two reasons may account
for these observations: i) The patients originated from different
countries; or ii) the numbers of patients with BRCA differed when
comparing the different groups. In addition, the small sample size
limited the analysis of HINT3 in the clinicopathological
characteristics of these patients. In the future, a larger number
of patients with BRCA in China need to be examined in order to
analyze the association between HINT3 expression and the
stage/metastasis and status/molecular subtype of the patients.
Additionally, HINT3-knockout mice need to be established to explore
the functional roles of HINT3 in vivo. Furthermore, clinical
analyses will be required in the future. This will be crucial in
terms of evaluating the diagnostic value of HINT3 in BRCA.
In the future, the authors aim to explore the role
of HINT3 knockdown in the development of BRCA cell tumorigenesis,
in which drugs, including AKT inhibitors, will be applied for
rescue experiments. Moreover, the molecular mechanisms, as well as
the clinical application of HINT3 also need to be investigated in
the future.
In conclusion, the present study demonstrated that
HINT3 expression was decreased in BRCA tissues. HINT3 knockdown
contributed to the development of BRCA via the activation of the
PTEN/AKT signaling pathway. The present study supports the role of
HINT3 as a novel tumor suppressor in BRCA cells.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was financially supported by the National
Natural Science Foundation of China (grant no. 81760482) and the
Natural Science Foundation of Ningxia (grant nos. NZ17138,
2019AAC03214, 2018AAC03162, 2019AAC03232 and 2018AAC03165). The
funding provided contributed to the procurement of the experiment
materials used in the present study.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YL, BL and JL designed the study. YL, BL, HL, DC, YG
and YW conducted the experiments and analyzed the results. wrote
and revised the manuscript. All authors have read and approved the
final manuscript. JL and YL confirm the authenticity of all the raw
data.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of General Hospital of Ningxia Medical University (no.
2022-68). Written informed consents were collected from all
patients. The animal experiments were approved by the Ethics
Committee of General Hospital of Ningxia Medical University (no.
2022-68), and were performed according to the animal guidelines of
General Hospital of Ningxia Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Harbeck N, Penault-Llorca F, Cortes J,
Gnant M, Houssami N, Poortmans P, Ruddy K, Tsang J and Cardoso F:
Breast cancer. Nat Rev Dis Primers. 5:662019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Waks AG and Winer EP: Breast cancer
treatment: A review. JAMA. 321:288–300. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Brenner C: Hint, Fhit, and GalT: Function,
structure, evolution, and mechanism of three branches of the
histidine triad superfamily of nucleotide hydrolases and
transferases. Biochemistry. 41:9003–9014. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Su T, Suzui M, Wang L, Lin CS, Xing WQ and
Weinstein IB: Deletion of histidine triad nucleotide-binding
protein 1/PKC-interacting protein in mice enhances cell growth and
carcinogenesis. Proc Natl Acad Sci USA. 100:7824–7829. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li H, Zhang Y, Su T, Santella RM and
Weinstein IB: Hint1 is a haplo-insufficient tumor suppressor in
mice. Oncogene. 25:713–721. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Huang H, Wei X, Su X, Qiao F, Xu Z, Gu D,
Fan H and Chen J: Clinical significance of expression of Hint1 and
potential epigenetic mechanism in gastric cancer. Int J Oncol.
38:1557–1564. 2011.PubMed/NCBI
|
|
8
|
Zhang YJ, Li H, Wu HC, Shen J, Wang L, Yu
MW, Lee PH, Bernard Weinstein I and Santella RM: Silencing of
Hint1, a novel tumor suppressor gene, by promoter hypermethylation
in hepatocellular carcinoma. Cancer Lett. 275:277–284. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li W, Cai S, Wang L, Yang C, Zhou B and
Wang H: HINT2 downregulation promotes colorectal carcinoma
migration and metastasis. Oncotarget. 8:13521–13531. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhou DK, Qian XH, Cheng J, Chen LH and
Wang WL: Clinical significance of down-regulated HINT2 in
hepatocellular carcinoma. Medicine (Baltimore). 98:e178152019.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Martin J, Magnino F, Schmidt K, Piguet AC,
Lee JS, Semela D, St-Pierre MV, Ziemiecki A, Cassio D, Brenner C,
et al: Hint2, a mitochondrial apoptotic sensitizer down-regulated
in hepatocellular carcinoma. Gastroenterology. 130:2179–2188. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Du L, Li X, Zhen L, Chen W, Mu L, Zhang Y
and Song A: Everolimus inhibits breast cancer cell growth through
PI3K/AKT/mTOR signaling pathway. Mol Med Rep. 17:7163–7169.
2018.PubMed/NCBI
|
|
15
|
Li F, Shi Y, Yang X, Luo Z, Zhang G, Yu K,
Li F, Chen L, Zhao Y, Xie Y, et al: Anhydroicaritin inhibits EMT in
breast cancer by enhancing GPX1 expression: A research based on
sequencing technologies and bioinformatics analysis. Front Cell Dev
Biol. 9:7644812022. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang Y, Wu N, Zhang J, Wang H and Men X:
MiR-153-5p enhances the sensitivity of triple-negative breast
cancer cells to paclitaxel by inducing G2M phase arrest. Onco
Targets Ther. 13:4089–4097. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gao C, Xiao G, Piersigilli A, Gou J,
Ogunwobi O and Bargonetti J: Context-dependent roles of MDMX (MDM4)
and MDM2 in breast cancer proliferation and circulating tumor
cells. Breast Cancer Res. 21:52019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bachmeier B, Fichtner I, Killian PH,
Kronski E, Pfeffer U and Efferth T: Development of resistance
towards artesunate in MDA-MB-231 human breast cancer cells. PLoS
One. 6:e205502011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Salic A and Mitchison TJ: A chemical
method for fast and sensitive detection of DNA synthesis in vivo.
Proc Natl Acad Sci USA. 105:2415–2420. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chou TF, Cheng J, Tikh IB and Wagner CR:
Evidence that human histidine triad nucleotide binding protein 3
(Hint3) is a distinct branch of the histidine triad (HIT)
superfamily. J Mol Biol. 373:978–989. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Huen MSY, Sy SMH and Chen J: BRCA1 and its
toolbox for the maintenance of genome integrity. Nat Rev Mol Cell
Biol. 11:138–148. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kuchenbaecker KB, Hopper JL, Barnes DR,
Phillips KA, Mooij TM, Roos-Blom MJ, Jervis S, van Leeuwen FE,
Milne RL, Andrieu N, et al: Risks of breast, ovarian, and
contralateral breast cancer for BRCA1 and BRCA2 mutation carriers.
JAMA. 317:2402–2416. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Balmaña J, Diez O, Rubio IT and Cardoso F;
ESMO Guidelines Working Group, : BRCA in breast cancer: ESMO
clinical practice guidelines. Ann Oncol. 22 (Suppl 6):vi31–vi34.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Paluch-Shimon S, Cardoso F, Sessa C,
Balmana J, Cardoso MJ, Gilbert F and Senkus E; ESMO Guidelines
Committee, : Prevention and screening in BRCA mutation carriers and
other breast/ovarian hereditary cancer syndromes: ESMO clinical
practice guidelines for cancer prevention and screening. Ann Oncol.
27 (Suppl 5):v103–v110. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nik-Zainal S, Davies H, Staaf J,
Ramakrishna M, Glodzik D, Zou X, Martincorena I, Alexandrov LB,
Martin S, Wedge DC, et al: Landscape of somatic mutations in 560
breast cancer whole-genome sequences. Nature. 534:47–54. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Litton JK, Rugo HS, Ettl J, Hurvitz SA,
Gonçalves A, Lee KH, Fehrenbacher L, Yerushalmi R, Mina LA, Martin
M, et al: Talazoparib in patients with advanced breast cancer and a
germline BRCA mutation. N Engl J Med. 379:753–763. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ozga M: HINT1-a novel tumor suppressor
protein of the HIT superfamily. Postepy Biochem. 56:55–60. 2010.(In
Polish). PubMed/NCBI
|
|
28
|
Martin J, St-Pierre MV and Dufour JF: Hit
proteins, mitochondria and cancer. Biochim Biophys Acta.
1807:626–632. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Worby CA and Dixon JE: Pten. Annu Rev
Biochem. 83:641–669. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Manning BD and Cantley LC: AKT/PKB
signaling: Navigating downstream. Cell. 129:1261–1274. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fan H, Wu Y, Yu S, Li X, Wang A, Wang S,
Chen W and Lu Y: Critical role of mTOR in regulating aerobic
glycolysis in carcinogenesis (review). Int J Oncol. 58:9–19. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang J, Liang D, Zhang XP, He CF, Cao L,
Zhang SQ, Xiao X, Li SJ and Cao YX: Novel PI3K/Akt/mTOR signaling
inhibitor, W922, prevents colorectal cancer growth via the
regulation of autophagy. Int J Oncol. 58:70–82. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Costa C, Wang Y, Ly A, Hosono Y, Murchie
E, Walmsley CS, Huynh T, Healy C, Peterson R, Yanase S, et al: PTEN
loss mediates clinical cross-resistance to CDK4/6 and PI3Kα
inhibitors in breast cancer. Cancer Discov. 10:72–85. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pandolfi PP: Breast cancer-loss of PTEN
predicts resistance to treatment. N Engl J Med. 351:2337–2338.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li J, Yen C, Liaw D, Podsypanina K, Bose
S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, et al:
PTEN, a putative protein tyrosine phosphatase gene mutated in human
brain, breast, and prostate cancer. Science. 275:1943–1947. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Donia M, McCubrey JA, Bendtzen K and
Nicoletti F: Potential use of rapamycin in HIV infection. Br J Clin
Pharmacol. 70:784–793. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Basile MS, Cavalli E, McCubrey J,
Hernández-Bello J, Muñoz-Valle JF, Fagone P and Nicoletti F: The
PI3K/Akt/mTOR pathway: A potential pharmacological target in
COVID-19. Drug Discov Today. 27:848–856. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Mammana S, Bramanti P, Mazzon E, Cavalli
E, Basile MS, Fagone P, Petralia MC, McCubrey JA, Nicoletti F and
Mangano K: Preclinical evaluation of the PI3K/Akt/mTOR pathway in
animal models of multiple sclerosis. Oncotarget. 9:8263–8277. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ji L, Xie W and Zhang Z: Efficacy and
safety of sirolimus in patients with systemic lupus erythematosus:
A systematic review and meta-analysis. Semin Arthritis Rheum.
50:1073–1080. 2020. View Article : Google Scholar : PubMed/NCBI
|