Bladder cancer (BLCA) is one of the top 10 most
common cancers worldwide, with an annual incidence rate of
>500,000 individuals and an annual mortality rate of >200,000
individuals (1,2) In the process of tumor development,
the metabolism of tumor cells also changes, which is now referred
to as metabolic reprogramming. Similarly, metabolic reprogramming
serves a crucial role in the development and progression of BLCA.
To some extent, BLCA is also a metabolic disease. Glucose is mainly
metabolized by glycolysis in tumor cells (3,4),
whether or not they are well oxygenated and this has prompted
continued exploration and research into the role of metabolism in
tumorigenesis (5). When cells are
hypoxic, BLCA cells can adapt their own glucose metabolism in the
oxidative phosphorylation pathway into glycolysis, a phenomenon
called the Warburg effect (6).
This metabolic switch meets the needs of tumor cells to absorb
nutrients and provides sufficient energy supply for tumor
progression. The growth and membrane synthesis of BLCA cells also
require lipid function and the growth of cells under hypoxia is
also inseparable from the modification of lipid metabolism, which
allows bladder tumor cells to continue to survive cellular hypoxia
and chemotherapeutic drugs (7).
Amino acid metabolism also serves an important role in the
proliferation of tumor cells and the maintenance of oxidative
homeostasis. In conclusion, metabolic reprogramming of BLCA cells
will become an important metabolic marker of bladder carcinogenesis
through changes in metabolic patterns.
Noncoding RNA (ncRNAs) have not been evaluated
because they do not encode proteins. They were previously
considered to be noise generated during transcription and to have
no biological function (8). In
the human genome, genes encoding proteins are only ~2% of the
total, and the remaining fraction is transcribed to ncRNA,
Therefore, a number of researchers hypothesize that ncRNAs serve an
important role in biological processes. As more research has begun
to focus on ncRNAs in BLCA, a number of studies have showed their
role and function (9–13). For example, miR-183-5p has low
expression in BLCA and enhances cisplatin-induced apoptosis in BLCA
cells by regulating PNPT 1 (14).
miR-665 is downregulated by upstream methylation in BLCA and
suppresses epithelial-stromal transformation in BLCA via the
SMAD3/zinc finger protein SNAIL axis (15). miR-125a-5p inhibits BLCA
progression by targeting fucosyltransferase 4 (16). miR-125b-5p inhibits BLCA
progression by targeting hexokinase (HK)2 and inhibiting the
PI3K/AKT pathway (17). Aberrant
expression of long ncRNAs (lncRNAs) is also critical in the
metabolic process of BLCA. For example, lncRNA RP11-89 promotes
tumorigenesis and iron death resistance via sponging miR-129-5p
through pro 2-activated iron export in BLCA (18). Exosomal lncRNA LNMAT2 promotes
lymphatic metastasis of BLCA (19). As a target and coactivator of E2F
transcription factor 1, lncRNA-SLC16A1-AS1 induces metabolic
reprogramming during BLCA progression (20). lncRNA BLACAT2 promotes
BLCA-related lymphangiogenesis and lymphatic metastasis (21). In addition, a large number of
dysregulated circular RNAs (circRNAs) have been found in BLCA. A
recent study showed that circFAM13B is dysregulated in BLCA and
inhibits glycolysis via the IGF2 mRNA binding protein 1/pyruvate
kinase M2 (PKM2) pathway, increasing immunotherapy sensitivity in
BLCA (22). Another study found
that circRNA_0071196 promotes BLCA proliferation and migration
through the miRNA-19b-3p/CIT axis (15). Exosome-derived circTRPS1 promotes
the malignant phenotype and CD8+ T cell exhaustion in the BLCA
microenvironment (23).
circ0008399 interacts with WT1 associated protein to promote the
assembly and activity of the m6A methyltransferase complex and
promotes cisplatin resistance in BLCA (24).
BLCA cells often vary because of genetic changes and
changes in the surrounding microenvironment (20). During BLCA development, ncRNAs
regulate metabolic pathways in cancer cells. With the study of
metabolomics, researchers have gained a new understanding of
metabolic reprogramming and developed new alternative treatments
for BLCA (25,26). Therefore, how ncRNA functions in
BLCA metabolism and its mechanism of action need to be studied in
depth. In the present review, ncRNA classification, abnormal ncRNA
expression in BLCA and ncRNAs involved in metabolic reprogramming
in BLCA are summarized. In addition, the ncRNAs involved in
glucose, lipid and amino acid metabolism in BLCA are discussed.
RNAs were first identified some time ago and are
classified into coding RNAs and ncRNAs based on whether they encode
proteins (27,28). Coding RNA mainly refers to mRNA,
while ncRNA encompasses more types, such as microRNA (miRNA),
lncRNA, small nuclear RNA (snRNA), small nucleolar RNA (snoRNA),
piwi-interacting RNA (piRNA), circRNA, small interfering RNA
(siRNA) and signal recognition particle RNA (srpRNA) (29–33). According to their length, ncRNAs
are divided into lncRNAs and small ncRNA (sncRNAs). lncRNAs are
ncRNAs with nucleotide lengths >200 bp and sncRNAs are ncRNAs
with nucleotide lengths <200 bp (34,35), including miRNAs, circRNAs, siRNAs,
snRNAs, srpRNAs, snoRNAs and telomerase RNAs (11,36,37).
With increased research into ncRNAs, their various
roles in cellular activities have been identified, such as
substance metabolism, protein transport and translation and RNA
splicing, modification and editing (38–40). In particular, the role of ncRNAs
in tumor metabolism has been widely recognized (41–43). Studies have implicated ncRNAs in
regulating metabolic reprogramming in BLCA, including miRNAs, such
as miRNA-195-5p, miR-16, miRNA-21 and miR-210 (44–47). lncRNAs, such as lncRNA-SLC16A1-AS,
lncRNA-P21, lncRNA-UCA1, lncDBET and lncRNA-TPRG1-AS1, have also
been implicated in the metabolic reprogramming of BLCA (48–51).
With the further study of miRNAs and lncRNAs,
another type of ncRNA has gradually gained attention: circRNAs,
which have gradually become a research hot spot. circRNA is
generated by mRNA or linear ncRNA ‘reverse splicing’. It has a
closed-loop structure, is stably expressed, is not susceptible to
degradation, and is not influenced by RNA exonucleases (52). circRNAs generally exert their
effects using sponge miRNAs to regulate protein function and
translation (53–55). Numerous circRNAs have been
discovered, but exactly how they perform their biological functions
remains to be elucidated. However, a number of recent studies on
circRNAs have focused on tumors and circRNA_0088036, circRIP2,
circNCOR1, circRNA-ST6GALNAC6, circEHBP1, circRNA-ST6GALNAC6,
hsa_circ_0014130 and circRNA_0071196 have been found to serve
oncogenic or suppressor gene roles in the development of BLCA
(15,56–61). Studies have also found that
circ_0020394, circFAM13B and circANKHD1 are strongly linked to
metabolic reprogramming in BLCA (22,62,63).
At present, studies on snoRNAs and piRNAs are
relatively rare in research related to metabolic reprogramming in
BLCA. snoRNA is an sncRNA of 60–300 nt in length that forms an
snoRNP complex by binding to nucleolar ribonucleoproteins (64). snoRNAs are mainly involved in rRNA
processing, transcription regulation, RNA splicing and oxidative
stress (65). In previous
studies, it has been found that Rpl13a, SNORD12B and SNORA73 are
involved in BLCA metabolism (66–68). piRNAs are sncRNAs 24–31 nt in
length and have been found in up to 15,000 species (69). piRNA complexes are formed by
binding to piwi proteins, which mainly regulate translocon
expression in mammalian germ cells (70,71). piRABC regulates bladder function
(72). However, there have been
no relevant studies on the piRNAs involved in BLCA metabolism. In
conclusion, various ncRNAs serve important roles in the metabolic
reprogramming of BLCA.
Studies have found that multiple ncRNAs are closely
associated with glucose metabolism in BLCA (73–79). The most common mode of
participation is the promotion of the degradation of the mRNA or
the inhibition of protein translation. Glucose metabolism mainly
refers to glycolysis, the tricarboxylic acid (TCA) cycle and the
pentose phosphate pathway. Bladder tumor cells rely on aerobic
glycolysis-dependent metabolism (the Warburg effect) to promote
their proliferation. Enhanced glycolytic metabolism generally
promotes the increased production of pyruvate, alanine and lactate
through the increased expression of its related genes, such as
glucose transporter 1, lactate dehydrogenase (LDHA), hexokinase 1
and pyruvate kinase M (PKM) (78,81). In addition, the expression of
genes related to the pentose phosphate pathway and fatty acid
synthesis increases during BLCA metabolism, promoting tumor cell
proliferation (73). Numerous
studies have demonstrated that changes in tumor oncogenes and tumor
suppressor genes influence metabolic reprogramming by regulating
key metabolic enzymes (74–87). The role of ncRNAs in glucose
metabolism is discussed in detail below.
Studies have indicated that the ncRNAs involved in
glucose metabolism in BLCA are mainly miRNAs, lncRNAs and circRNAs,
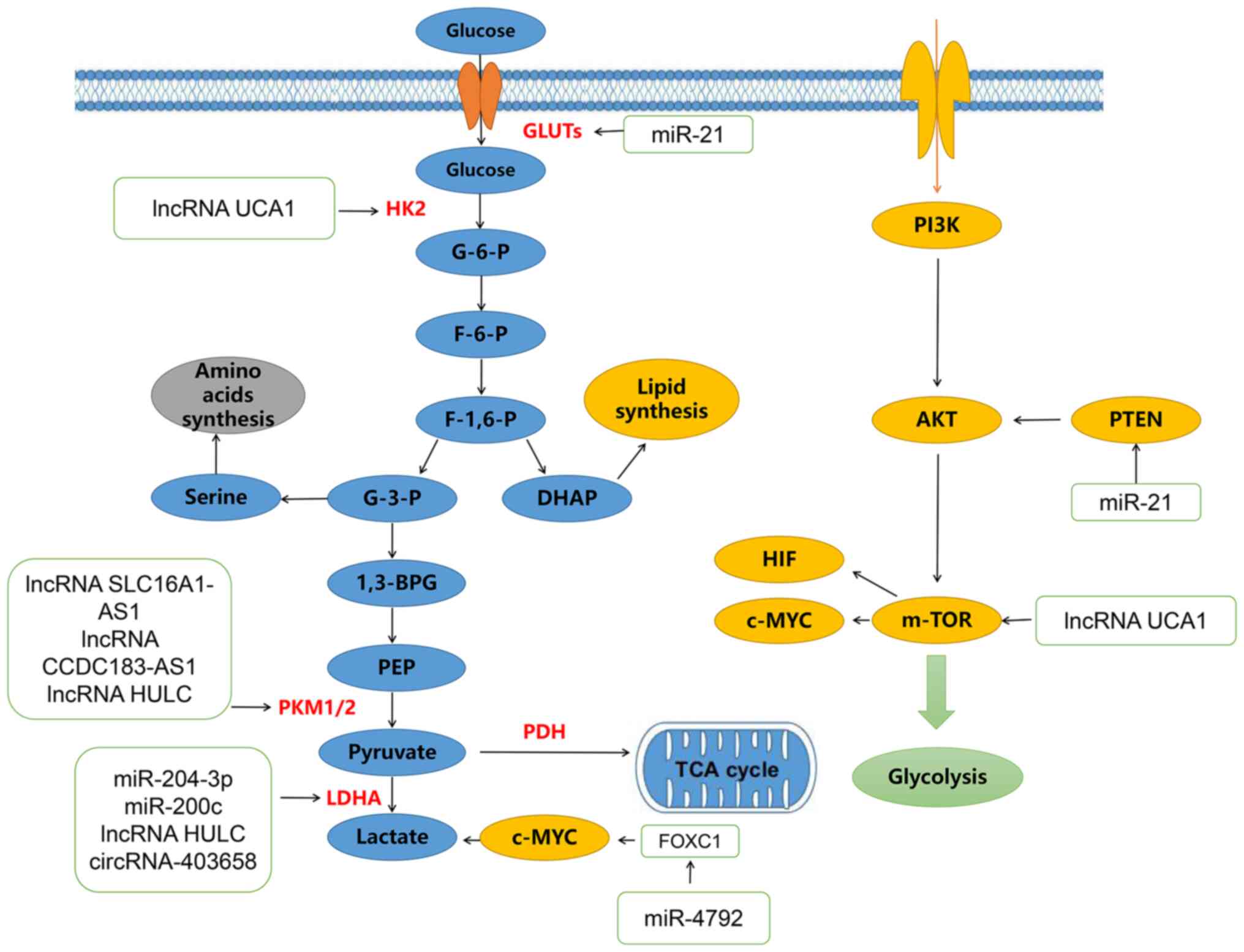
with few studies on piRNAs and snoRNAs (75–84). miR-204-3p binds to the 3′-UTR of
LDHA in BLCA to reduce its mRNA and protein expression. Glucose
depletion and lactate formation are depleted in BLCA cells after
overexpression of miR-204-3p (75). Enhanced growth inhibition and
apoptosis of BLCA cells induced by miR-204-3p are restored by LDHA
overexpression (Table I).
miR-204-3p can inhibit BLCA cell growth by regulating LDHA-mediated
glycolysis (75). Low expression
of miR-21 in BLCA cells leads to reduced tumor aerobic glycolysis
and reduced glucose uptake and lactate production, which increases
the expression of phosphatase and tensin homologue, inhibits
phosphorylated AKT and inactivates mTOR to regulate aerobic
glycolysis in BLCA cells (76).
Low expression of miR-4792 in BLCA tissues and its overexpression
in BLCA cells inhibits the expression levels of Forkhead box C1 and
c-Myc, inhibits cell proliferation and decreases aerobic glycolysis
and lactate content (77).
miR-200c can inhibit glycolysis, cell growth and invasion of BLCA
cells by targeting LDHA in BLCA cells (78).
lncRNA SLC16A1-AS1 promotes ATP production by
increasing aerobic glycolysis and mitochondrial respiration and
through fatty acid β-oxidation. In addition, complex formation of
lncRNA-SLC16A1 by E2F1-induced AS1 with its transcription factors
promotes cancer metabolic reprogramming and increases oxidative
phosphorylation and glycolysis to promote BLCA invasion (20). The lncRNA CCDC183-AS1 serves an
oncogenic role in BLCA by increasing aerobic glycolysis (79). The lncRNA HULC can bind to key
enzymes of glycolysis, LDHA and PKM2, to enhance the binding of
fibroblast growth factor receptor type 1, thus, increasing the
phosphorylation levels of these two enzymes and promoting
glycolysis (80). Ho et al
(81) identified a group of
glycolysis-related lncRNAs using The Cancer Genome Atlas database
and found that these molecular-related BLCA patients had a poor
prognosis and high immune infiltration, while epithelial
mesenchymal transition was activated to promote tumor progression.
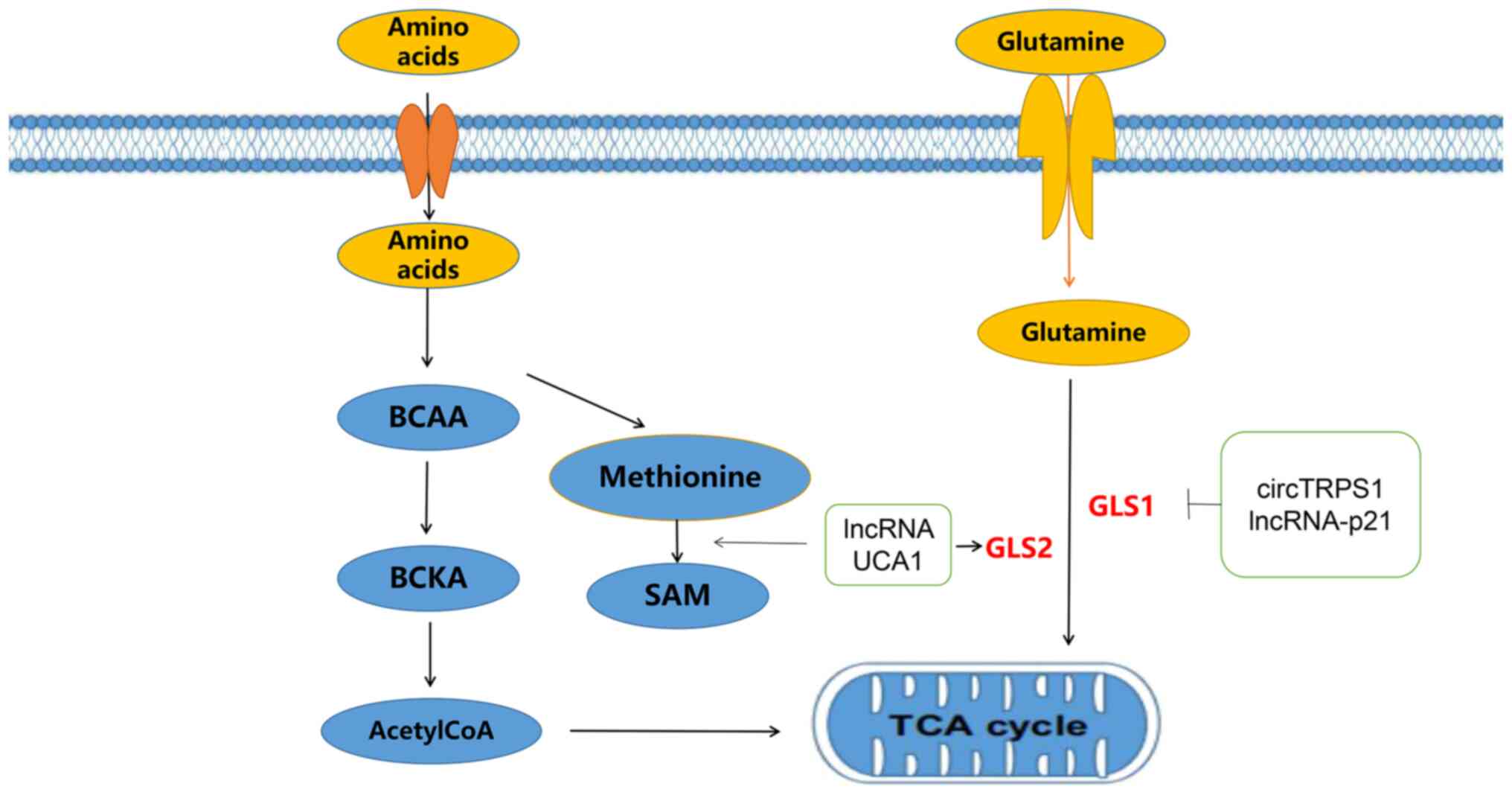
The lncRNA UCA1 can bind to heterogeneous nuclear ribonucleoprotein
(hnRNP), promote its binding to the glutamate transaminotransferase
(GPT)2 promoter, enhance the expression of glutamine-derived carbon
in the TCA cycle and affect glycolysis, the TCA cycle, glutamine
metabolism and proliferation in BLCA (49). Another study on lncRNA UCA1 found
that it could regulate hexokinase2 (HK2) to promote glycolysis in
BLCA cells by activating STAT3, inhibiting miRNA143 and activating
mTOR, revealing a new relationship between lncRNAs and glucose
metabolism in BLCA (82).
Regulated by puerarin, circ_0020394 inhibits
glycolysis and promotes apoptosis in BLCA cells (62). circFAM13B is present in low levels
in BLCA and is associated with glycolysis and CD8+ T cell
activation. circFAM13B can promote the function of CD8+ T cells,
weaken glycolysis in BLCA cells and reverse the acidic tumor
microenvironment (TME) (22).
Inhibition of the acidic TME inhibits immune evasion and enhances
immunotherapy sensitivity. Has-circRNA-403658 is highly expressed
in BLCA cells under hypoxia and silencing its expression inhibits
BLCA cell proliferation by inhibiting LDHA-mediated aerobic
glycolysis (83). In conclusion,
ncRNAs are associated with the metabolic reprogramming of BLCA
cells in multiple ways (Fig.
1).
Unlike glucose metabolism, there have been few
reports on lipid metabolism in BLCA; however, with the gradual
understanding of fatty acid metabolism, the importance of
reprogramming lipid metabolism in cancer cells is being
increasingly appreciated (84).
Lipids are primarily comprised of triglycerides, phospholipids,
sphingolipids and cholesterol (85). Lipids serve important roles in
cellular metabolism, including energy storage, fatty acid
synthesis, biofilm formation and signal transmission (86). In tumor cells, migration is mainly
promoted through lipid synthesis, storage and metabolism. For
example, cholesterol lipids are synthesized from acetyl-CoA and
NADPH through the TCA cycle and the pentose phosphate pathway
(87).
How ncRNAs regulate lipid metabolism was detailed in
a recent study. In BLCA, the m6A-related enzyme
methyltransferase-like 14 promotes lncDBET expression through
methylation modification and activates the peroxisome
proliferators-activated receptor (PPAR) signaling pathway,
promoting lipid metabolism in BLCA cells by directly interacting
with fatty acid binding protein 5, thus, promoting the malignant
progression of BCa in vitro and in vivo (50). lncRNA SLC16A1-AS1 promotes ATP
production through the β-oxidation of fatty acids in BLCA,
accompanied by changes in the expression of the SLC16A1-AS1:E2F1
response gene PPARA, which is critical for fatty acid β-oxidation
(Table II). E2F1-induced
lncRNA-SLC16A1-AS1 forms a complex with its transcription factors,
promoting metabolic reprogramming of BLCA and increasing BLCA
invasion (20). In summary, the
ncRNA and lipid metabolism in BLCA are closely related (Fig. 2).
In cells, amino acid metabolism mainly includes
anabolism and catabolism. Anabolism is primarily the supply of
peptides, proteins and other nitrogenous substances to the human
body. Catabolism involves the breakdown of alpha-keto acids,
glutamate by transamination, deamination, or carbon dioxide. The
alpha-keto acids generated by amino acid metabolism can also be
transformed into sugars or lipids, or resynthesized into a number
of nonessential amino acids. They can also release energy from the
TCA cycle to form carbon dioxide and water (88,89). With the development of current
understanding of amino acid metabolism, the role of ncRNAs in amino
acid metabolism in BLCA has gained greater attention (90). Glutamine is a type of free
nonessential amino acid. It participates in the anabolism of BLCA
cells by converting glutaminase (GLS) to glutamate and is also an
energy source for tumor cell proliferation (91).
As metabolism has become an important research
topic, the participation of ncRNAs in tumor metabolic reprogramming
has received extensive attention in the fields of biology and
oncology. Numerous studies have shown that ncRNAs are closely
related with BLCA metabolism (49,50,77,83). Metabolic reprogramming has become
an important aspect of malignancy. ncRNAs affect the progression of
BLCA, such as proliferation, invasion and metastasis, by
participating in the metabolism of BLCA cells. Thus, these key
ncRNAs involved in BLCA metabolism could serve as potential
biomarkers for BLCA.
However, there are still some limitations in the
current metabolic reprogramming of bladder cancer. For example,
what are the causes of metabolic abnormalities? How can we treat
bladder cancer by affecting the metabolic reprogramming of cancer
cells? What is the mechanism of action in the metabolic
reprogramming of bladder cancer? Furthermore, we lack the ncRNA
knockout in animal models to study metabolic reprogramming in
bladder cancer and have not investigated this model in metabolic
reprogramming in bladder cancer. These are the problems to be
solved by future research. An ncRNA knockout mouse model will help
identify the key molecules that regulate the metabolic
reprogramming of bladder cancer and provide a rationale for
clinical applications.
ncRNAs regulate the metabolic reprogramming of BLCA
in various ways; however, a large number of ncRNA functions and
mechanisms of action still require further study, as metabolic
reprogramming of BLCA not only provides an energy source for tumor
progression, but also creates suitable conditions for its
development. Metabolic reprogramming acts mainly through
metabolism-related enzymes, which are modulated by ncRNAs, offering
new options for the treatment of BLCA (45,82,83). Current research on the metabolic
reprogramming of lncRNAs in BLCA is relatively extensive and offers
good theoretical support for preclinical studies; however, there
remain a number of areas of uncertainty, such as the cause of
metabolic abnormalities. Extensive studies are needed to identify
how treatment can be more directly targeted to BLCA and how
ncRNA-based therapies can be applied clinically. This requires
further study of the mechanism of ncRNAs in the metabolic
reprogramming of BLCA as well as theoretical support for clinical
treatments in the thriving field of precision medicine.
Not applicable.
The present study was supported by Cuiying Science and
Technology Innovation plan project of Lanzhou University Second
Hospital (grant no. CY2021-MS-B16) and the Medical Innovation and
Development Project of Lanzhou University (grant no.
lzuyxcx-2022-106).
Not applicable.
ZB conceived and supervised the present study and
wrote, reviewed and edited the original draft of the manuscript. YM
wrote, reviewed and edited the manuscript. HY, HL and QP performed
data curation. SF supervised the present study, project
administration and funding. All authors read and approved the final
manuscript. Data sharing is not applicable to this article.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Jubber I, Ong S, Bukavina L, Black PC,
Compérat E, Kamat AM, Kiemeney L, Lawrentschuk N, Lerner SP, Meeks
JJ, et al: Epidemiology of bladder cancer in 2023: A systematic
review of risk factors. Eur Urol. May 15–2023.(Epub ahead of
print). View Article : Google Scholar : PubMed/NCBI
|
|
2
|
World Cancer Research Fund (WCRF)
International, . Bladder cancer statistics. WCRF International,
London, 2020. https://www.wcrf.org/cancer-trends/bladder-cancer-statistics
|
|
3
|
Warburg O: On the origin of cancer cells.
Science. 123:309–314. 1956. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Warburg O, Wind F and Negelein E: The
metabolism of tumors in the body. J Gen Physiol. 8:519–530. 1927.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Upadhyay M, Samal J, Kandpal M, Singh OV
and Vivekanandan P: The Warburg effect: Insights from the past
decade. Pharmacol Ther. 137:318–330. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pascale RM, Calvisi DF, Simile MM, Feo CF
and Feo F: The Warburg effect 97 years after its discovery. Cancers
(Basel). 12:28192020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Massari F, Ciccarese C, Santoni M,
Iacovelli R, Mazzucchelli R, Piva F, Scarpelli M, Berardi R,
Tortora G, Lopez-Beltran A, et al: Metabolic phenotype of bladder
cancer. Cancer Treat Rev. 45:46–57. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Adnane S, Marino A and Leucci E: LncRNAs
in human cancers: Signal from noise. Trends Cell Biol. 32:565–573.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang J, Zhu S, Meng N, He Y, Lu R and Yan
GR: ncRNA-encoded peptides or proteins and cancer. Mol Ther.
27:1718–1725. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Anastasiadou E, Jacob LS and Slack FJ:
Non-coding RNA networks in cancer. Nat Rev Cancer. 18:5–18. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li Y, Li G, Guo X, Yao H, Wang G and Li C:
Non-coding RNA in bladder cancer. Cancer Lett. 485:38–44. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chandra Gupta S and Nandan Tripathi Y:
Potential of long non-coding RNAs in cancer patients: From
biomarkers to therapeutic targets. Int J Cancer. 140:1955–1967.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Martens-Uzunova ES, Böttcher R, Croce CM,
Jenster G, Visakorpi T and Calin GA: Long noncoding RNA in
prostate, bladder, and kidney cancer. Eur Urol. 65:1140–1151. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hu QG, Yang Z, Chen JW, Kazobinka G, Tian
L and Li WC: MiR-183-5p-PNPT1 axis enhances cisplatin-induced
apoptosis in bladder cancer cells. Curr Med Sci Aug. 42:785–796.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu Z, Yang Y, Yang Z, Xia S, Lin D, Xiao
B and Xiu Y: Novel circRNA_0071196/miRNA-19b-3p/CIT axis is
associated with proliferation and migration of bladder cancer. Int
J Oncol. 57:767–779. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang Y, Zhang D, Lv J, Wang S and Zhang
Q: MiR-125a-5p suppresses bladder cancer progression through
targeting FUT4. Biomed Pharmacother. 108:1039–1047. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu S, Chen Q and Wang Y: MiR-125b-5p
suppresses the bladder cancer progression via targeting HK2 and
suppressing PI3K/AKT pathway. Hum Cell. 33:185–194. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Luo W, Wang J, Xu W, Ma C, Wan F, Huang Y,
Yao M, Zhang H, Qu Y, Ye D and Zhu Y: LncRNA RP11-89 facilitates
tumorigenesis and ferroptosis resistance through PROM2-activated
iron export by sponging miR-129-5p in bladder cancer. Cell Death
Dis. 12:10432021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen C, Luo Y, He W, Zhao Y, Kong Y, Liu
H, Zhong G, Li Y, Li J, Huang J, et al: Exosomal long noncoding RNA
LNMAT2 promotes lymphatic metastasis in bladder cancer. J Clin
Invest. 130:404–421. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Logotheti S, Marquardt S, Gupta SK,
Richter C, Edelhäuser BAH, Engelmann D, Brenmoehl J, Söhnchen C,
Murr N, Alpers M, et al: LncRNA-SLC16A1-AS1 induces metabolic
reprogramming during Bladder Cancer progression as target and
co-activator of E2F1. Theranostics. 10:9620–9643. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
He W, Zhong G, Jiang N, Wang B, Fan X,
Chen C, Chen X, Huang J and Lin T: Long noncoding RNA BLACAT2
promotes bladder cancer-associated lymphangiogenesis and lymphatic
metastasis. J Clin Invest. 128:861–875. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lv J, Li K, Yu H, Han J, Zhuang J, Yu R,
Cheng Y, Song Q, Bai K, Cao Q, et al: HNRNPL induced circFAM13B
increased bladder cancer immunotherapy sensitivity via inhibiting
glycolysis through IGF2BP1/PKM2 pathway. J Exp Clin Cancer Res.
42:412023. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang C, Wu S, Mou Z, Zhou Q, Dai X, Ou Y,
Chen X, Chen Y, Xu C, Hu Y, et al: Exosome-derived circTRPS1
promotes malignant phenotype and CD8+ T cell exhaustion in bladder
cancer microenvironments. Mol Ther. 30:1054–1070. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wei W, Sun J, Zhang H, Xiao X, Huang C,
Wang L, Zhong H, Jiang Y, Zhang X and Jiang G: Circ0008399
Interaction with WTAP promotes assembly and activity of the m6A
Methyltransferase complex and promotes cisplatin resistance in
bladder cancer. Cancer Res. 81:6142–6156. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
McConkey DJ and Choi W: Molecular subtypes
of bladder cancer. Curr Oncol Rep. 20:772018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ko YH, Verhoeven HA, Lee MJ, Corbin DJ,
Vogl TJ and Pedersen PL: A translational study ‘case report’ on the
small molecule ‘energy blocker’ 3-bromopyruvate (3BP) as a potent
anticancer agent: from bench side to bedside. J Bioenerg Biomembr.
44:163–170. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mattick JS: Challenging the dogma: The
hidden layer of non-proteincoding RNAs in complex organisms.
Bioessays. 25:930–939. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mattick JS: The hidden genetic program of
complex organisms. Sci Am. 291:60–67. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hombach S and Kretz M: Non-coding RNAs:
Classification, biology and functioning. Adv Exp Med Biol.
937:3–17. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Heinrichs A: MicroRNAs get a boost. Nat
Rev Mol Cell Biol. 10:302–303. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Baumann K: Gene expression: RNAi as a
global transcriptional activator. Nat Rev Mol Cell Biol.
15:2982014.PubMed/NCBI
|
|
32
|
Sato K and Siomi MC: Piwi-interacting
RNAs: Biological functions and biogenesis. Essays Biochem.
54:39–52. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ross RJ, Weiner MM and Lin HF: PIWI
proteins and PIWI-interacting RNAs in the soma. Nature.
505:353–359. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kim VN, Han J and Siomi MC: Biogenesis of
small RNAs in animals. Nat Rev Mol Cell Biol. 10:126–139. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bonasio R and Shiekhattar R: Regulation of
transcription by long noncoding RNAs. Annu Rev Genet. 48:433–455.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Brown JW, Marshall DF and Echeverria M:
Intronic noncoding RNAs and splicing. Trends Plant Sci. 13:335–342.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Shankaraiah RC, Veronese A, Sabbioni S and
Negrini M: Non-coding RNAs in the reprogramming of glucose
metabolism in cancer. Cancer Lett. 419:167–174. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Fu XD: Non-coding RNA: A new frontier in
regulatory biology. Natl Sci Rev. 1:190–204. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kugel JF and Goodrich JA: Non-coding RNAs:
Key regulators of mammalian transcription. Trends Biochem Sci.
37:144–151. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Mondal T and Kanduri C: Maintenance of
epigenetic information: A noncoding RNA perspective. Chromosome
Res. 21:615–625. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chan B, Manley J, Lee J and Singh SR: The
emerging roles of microRNAs in cancer metabolism. Cancer Lett.
356:301–308. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Pulito C, Donzelli S, Muti P, Puzzo L,
Strano S and Blandino G: MicroRNAs and cancer metabolism
reprogramming: The paradigm of metformin. Ann Transl Med.
2:582014.PubMed/NCBI
|
|
43
|
Zhao XY and Lin JD: Long non-coding RNAs:
A new regulatory code in metabolic control. Trends Biochem Sci.
40:586–596. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Fei X, Qi M, Wu B, Song Y, Wang Y and Li
T: MicroRNA-195-5p suppresses glucose uptake and proliferation of
human bladder cancer T24 cells by regulating GLUT3 expression. FEBS
Lett. 586:392–397. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Li HJ, Li X, Pang H, Pan JJ, Xie XJ and
Chen W: Long non-coding RNA UCA1 promotes glutamine metabolism by
targeting miR-16 in human bladder cancer. Jpn J Clin Oncol.
45:1055–1063. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Sekar D: miRNA 21: A novel biomarker in
the treatment of bladder cancer. Biomark Med. 14:1065–1067. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Irlam-Jones JJ, Eustace A, Denley H,
Choudhury A, Harris AL, Hoskin PJ and West CM: Expression of
miR-210 in relation to other measures of hypoxia and prediction of
benefit from hypoxia modification in patients with bladder cancer.
Br J Cancer. 115:571–578. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhou Q, Zhan H, Lin F, Liu Y, Yang K, Gao
Q, Ding M, Liu Y, Huang W and Cai Z: LincRNA-p21 suppresses
glutamine catabolism and bladder cancer cell growth through
inhibiting glutaminase expression. Biosci Rep. 39:BSR201823722019.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhao H, Wu W, Li X and Chen W: Long
noncoding RNA UCA1 promotes glutamine-driven anaplerosis of bladder
cancer by interacting with hnRNP I/L to upregulate GPT2 expression.
Transl Oncol. 17:1013402022. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Liu P, Fan B, Othmane B, Hu J, Li H, Cui
Y, Ou Z, Chen J and Zu X: m6A-induced lncDBET promotes the
malignant progression of bladder cancer through FABP5-mediated
lipid metabolism. Theranostics. 12:6291–6307. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
He J, Dong C, Zhang H, Jiang Y, Liu T and
Man X: The oncogenic role of TFAP2A in bladder urothelial carcinoma
via a novel long noncoding RNA TPRG1-AS1/DNMT3A/CRTAC1 axis. Cell
Signal. 102:1105272023. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Salzman J: Circular RNA expression: Its
potential regulation and function. Trends Genet. 32:309–316. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Jeck WR and Sharpless NE: Detecting and
characterizing circular RNAs. Nat Biotechnol. 32:453–461. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Yang J, Qi M, Fei X, Wang X and Wang K:
Hsa_circRNA_0088036 acts as a ceRNA to promote bladder cancer
progression by sponging miR-140-3p. Cell Death Dis. 13:3222022.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Su Y, Feng W, Shi J, Chen L, Huang J and
Lin T: circRIP2 accelerates bladder cancer progression via
miR-1305/Tgf-β2/smad3 pathway. Mol Cancer. 19:232020. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
An M, Zheng H, Huang J, Lin Y, Luo Y, Kong
Y, Pang M, Zhang D, Yang J, Chen J, et al: Aberrant nuclear export
of circNCOR1 underlies SMAD7-Mediated lymph node metastasis of
bladder cancer. Cancer Res. 82:2239–2253. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Wang L, Wu S, He H, Ai K, Xu R, Zhang L
and Zhu X: CircRNA-ST6GALNAC6 increases the sensitivity of bladder
cancer cells to erastin-induced ferroptosis by regulating the
HSPB1/P38 axis. Lab Invest. 102:1323–1334. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Zhu J, Luo Y, Zhao Y, Kong Y, Zheng H, Li
Y, Gao B, Ai L, Huang H, Huang J, et al: circEHBP1 promotes
lymphangiogenesis and lymphatic metastasis of bladder cancer via
miR-130a-3p/TGFβR1/VEGF-D signaling. Mol Ther. 29:1838–1852. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Li G, Guo BY, Wang HD, Lin GT, Lan TJ,
Ying H and Xu J: CircRNA hsa_circ_0014130 function as a miR-132-3p
sponge for playing oncogenic roles in bladder cancer via
upregulating KCNJ12 expression. Cell Biol Toxicol. 38:1079–1096.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Du L, Zhang L and Sun F: Puerarin inhibits
the progression of bladder cancer by regulating
circ_0020394/miR-328-3p/NRBP1 axis. Cancer Biother Radiopharm.
37:435–450. 2022.PubMed/NCBI
|
|
63
|
Wei WS, Wang N, Deng MH, Dong P, Liu JY,
Xiang Z, Li XD, Li ZY, Liu ZH, Peng YL, et al: LRPPRC regulates
redox homeostasis via the circANKHD1/FOXM1 axis to enhance bladder
urothelial carcinoma tumorigenesis. Redox Biol. 48:1022012021.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Williams GT and Farzaneh F: Are snoRNAs
and snoRNA host genes new players in cancer? Nat Rev Cancer.
12:84–88. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Lee J, Harris AN, Holley CL, Mahadevan J,
Pyles KD, Lavagnino Z, Scherrer DE, Fujiwara H, Sidhu R, Zhang J,
et al: Rpl13a small nucleolar RNAs regulate systemic glucose
metabolism. J Clin Invest. 126:4616–4625. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Dong W, Liu X, Yang C, Wang D, Xue Y, Ruan
X, Zhang M, Song J, Cai H, Zheng J and Liu Y: Glioma glycolipid
metabolism: MSI2-SNORD12B-FIP1L1-ZBTB4 feedback loop as a potential
treatment target. Clin Transl Med. 11:e4112021. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Sletten AC, Davidson JW, Yagabasan B,
Moores S, Schwaiger-Haber M, Fujiwara H, Gale S, Jiang X, Sidhu R,
Gelman SJ, et al: Loss of SNORA73 reprograms cellular metabolism
and protects against steatohepatitis. Nat Commun. 12:52142021.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Farazi TA, Juranek SA and Tuschl T: The
growing catalog of small RNAs and their association with distinct
Argonaute/Piwi family members. Development. 135:1201–1214. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Luteijn MJ and Ketting RF:
PIWI-interacting RNAs: From generation to transgenerational
epigenetics. Nat Rev Genet. 14:523–534. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Siomi MC, Sato K, Pezic D and Aravin AA:
PIWIinteracting small RNAs: The vanguard of genome defence. Nat Rev
Mol Cell Biol. 12:246–258. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Chu H, Hui G, Yuan L, Shi D, Wang Y, Du M,
Zhong D, Ma L, Tong N, Qin C, et al: Identification of novel piRNAs
in bladder cancer. Cancer Lett. 356:561–567. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Hu S, Balakrishnan A, Bok RA, Anderton B,
Larson PE, Nelson SJ, Kurhanewicz J, Vigneron DB and Goga A:
13C-pyruvate imaging reveals alterations in glycolysis that precede
c-Myc-induced tumor formation and regression. Cell Metab.
14:131–142. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Gao P, Sun L, He X, Cao Y and Zhang H:
MicroRNAs and the Warburg effect: New players in an old arena. Curr
Gene Ther. 12:285–291. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Guo J, Zhao P, Liu Z, Li Z, Yuan Y, Zhang
X, Yu Z, Fang J and Xiao K: MiR-204-3p inhibited the proliferation
of bladder cancer cells via modulating lactate
dehydrogenase-mediated glycolysis. Front Oncol. 9:12422019.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Yang X, Cheng Y, Li P, Tao J, Deng X,
Zhang X, Gu M, Lu Q and Yin C: A lentiviral sponge for miRNA-21
diminishes aerobic glycolysis in bladder cancer T24 cells via the
PTEN/PI3K/AKT/mTOR axis. Tumour Biol. 36:383–391. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Wu JH, Sun KN, Chen ZH, He YJ and Sheng L:
Exosome-mediated miR-4792 transfer promotes bladder cancer cell
proliferation via enhanced FOXC1/c-Myc signaling and Warburg
effect. J Oncol. 2022:56803532022.PubMed/NCBI
|
|
78
|
Yuan D, Zheng S, Wang L, Li J, Yang J,
Wang B, Chen X and Zhang X: MiR-200c inhibits bladder cancer
progression by targeting lactate dehydrogenase A. Oncotarget.
8:67663–67669. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Cai W, Wei M and Su Z: MITF-Mediated
lncRNA CCDC183-As1 promotes the tumorigenic properties and aerobic
glycolysis of bladder cancer via upregulating TCF7L2. J Oncol.
2022:67859562022. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Wang C, Li Y, Yan S, Wang H, Shao X, Xiao
M, Yang B, Qin G, Kong R, Chen R and Zhang N: Interactome analysis
reveals that lncRNA HULC promotes aerobic glycolysis through LDHA
and PKM2. Nat Commun. 11:31622020. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Ho KH, Huang TW, Shih CM, Lee YT, Liu AJ,
Chen PH and Chen KC: Glycolysis-associated lncRNAs identify a
subgroup of cancer patients with poor prognoses and a
high-infiltration immune microenvironment. BMC Med. 19:592021.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Li Z, Li X, Wu S, Xue M and Chen W: Long
non-coding RNA UCA1 promotes glycolysis by upregulating hexokinase
2 through the mTOR-STAT3/microRNA143 pathway. Cancer Sci.
105:951–955. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Wei Y, Zhang Y, Meng Q, Cui L and Xu C:
Hypoxia-induced circular RNA has_circRNA_403658 promotes bladder
cancer cell growth through activation of LDHA. Am J Transl Res.
11:6838–6849. 2019.PubMed/NCBI
|
|
84
|
Fernandez-Hernando C, Suarez Y, Rayner KJ
and Moore KJ: MicroRNAs in lipid metabolism. Curr Opin Lipidol.
22:86–92. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Santos CR and Schulze A: Lipid metabolism
in cancer. FEBS J. 279:2610–2623. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Currie E, Schulze A, Zechner R, Walther TC
and Farese RV Jr: Cellular fatty acid metabolism and cancer. Cell
Metab. 18:153–161. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Bian X, Liu R, Meng Y, Xing D, Xu D and Lu
Z: Lipid metabolism and cancer. J Exp Med. 218:e202016062021.
View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Vettore L, Westbrook RL and Tennant DA:
New aspects of amino acid metabolism in cancer. Br J Cancer.
122:150–156. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Li Z and Zhang H: Reprogramming of
glucose, fatty acid and amino acid metabolism for cancer
progression. Cell Mol Life Sci. 73:377–392. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Hu Q, Li Y, Li D, Yuan Y, Wang K, Yao L,
Cheng Z and Han T: Amino acid metabolism regulated by lncRNAs: The
propellant behind cancer metabolic reprogramming. Cell Commun
Signal. 21:872023. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Xu Y, Xia Z, Sun X, Wei B, Fu Y, Shi D and
Zhu Y: Identification of a glutamine metabolism reprogramming
signature for predicting prognosis, immunotherapy efficacy, and
drug candidates in bladder cancer. Front Immunol. 14:11113192023.
View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Chen CW: Comment on ‘Long noncoding RNA
UCA1 promotes glutamine-driven anaplerosis of bladder cancer by
interacting with hnRNP I/L to upregulate GPT2 expression’ by Chen
et al.’”. Transl Oncol. 18:1013722022. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Roh J, Im M, Chae Y, Kang J and Kim W: The
involvement of long non-coding RNAs in glutamine-metabolic
reprogramming and therapeutic resistance in cancer. Int J Mol Sci.
23:148082022. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Ortiz-Pedraza Y, Muñoz-Bello JO,
Olmedo-Nieva L, Contreras-Paredes A, Martínez-Ramírez I, Langley E
and Lizano M: Non-coding RNAs as key regulators of glutaminolysis
in cancer. Int J Mol Sci. 21:28722020. View Article : Google Scholar : PubMed/NCBI
|