Breast cancer, the most frequently diagnosed cancer
in women globally, accounted for up to 2 million new cases in
recent years, making up 25% of all cancer cases in women (1). The complexity of breast cancer is
highlighted by its high degree of heterogeneity, with current
classifications based on three molecular markers: The estrogen
receptor (ER), the progesterone receptor (PR) and the human
epidermal growth factor receptor 2 (HER2) (2). Therapeutic approaches and outcomes
differ significantly among the various subtypes of breast cancer.
The ER-negative (ER-) group, which includes the HER2-positive
(HER2+) and triple-negative breast cancer (TNBC)
subtypes, is less responsive to neoadjuvant chemotherapy compared
with the ER-positive (ER+) group (3,4).
In the past decades, numerous studies utilizing
genome sequencing of breast cancers have revealed increasing
numbers, up to thousands, of somatic mutations during the onset and
progression of cancer (5).
However, only a minor proportion of these mutations inherently
provide a growth advantage to cancer cells, and are referred to as
'driver mutations' (6). Some of
the driver mutations frequently observed in a particular DNA
segment are known as 'hotspot mutations' and serve as targets for
diagnosis and drug development (7). Finding a 'neoantigen', a
tumor-specific antigen present exclusively in tumors, is needed to
elicit an antitumor immune response. So far, most studies of
neoantigen-targeted immunotherapies have focused on private
neoantigens that are patient-specific. Such studies are
time-consuming and costly. Neoantigens found in multiple patients
with the same type of cancer, namely shared or public neoantigens,
could be utilized in a broader spectrum of patients with cancer and
aid corresponding cost savings. In the present review, an overview
of neoantigens was provided and the public ones in clinical use
were highlighted as immunotherapeutic targets in breast cancer
treatment.
Globally, breast cancer is the most common cancer in
women and the leading cause of cancer death among women (1). In 2020, the estimated number of new
breast cancer cases was 2,261,419, accounting for 12% of all new
cancer cases, and the estimated number of deaths was 684,996
individuals, ~7% of all cancer deaths worldwide (1,8).
There are several histological subtypes of invasive breast cancer,
of which infiltrating ductal carcinoma is the most common,
accounting for ~80% of invasive breast cancers, infiltrating
lobular carcinoma accounts for 8%, ductal lobular carcinomas make
up another 7%, and other less common histologic subtypes comprising
medullary, metaplastic, mucinous and papillary carcinomas (9).
Breast cancer is an extremely heterogeneous disease
caused by a diversity of genetic alterations that result in various
biological and clinical behaviors within cancers themselves and in
differences in disease progression and therapeutic response between
patients (10,11). Based on the expression profiles of
ER, PR, HER2 and Ki-67, four major molecular subtypes have been
classified: Luminal A, luminal B, HER-2-enriched and basal-like.
Each subtype has a different prognosis and potential therapeutic
target (12,13). The luminal subtype is the most
common subtype and is characterized by the expression of ER and
other genes associated with resemblance to luminal epithelial
cells. Cancer type luminal A is characterized by high
ER+, PR+, HER2-, and low Ki-67
expression and comprises ~40-50% of all breast cancers, while
luminal B, which accounts for 20%, is classified by high
ER+, low PR, HER2-/+ and high Ki-67 (14). Luminal A cancers present as low
grade with a favorable prognosis, whereas luminal B cancers tend to
be high-grade with poor prognosis (15,16). The HER2-enriched subtype makes up
~15% of all invasive breast cancer and is characterized by
upregulation of HER2 expression and no expression of ER-related
genes (ER-/PR-/HER2-) (12,17). HER2-enriched tumors tend to
manifest a high-grade and more aggressive form with decreased
disease-free survival and overall survival (OS) rates compared with
subtypes that do not overexpress HER2. However, this subtype
responds well to monoclonal antibody-targeted HER2 therapy, which
may improve survival outcomes (18). The fourth subtype, the basal-like
subtype or TNBC, accounts for 15% of all breast cancer subtypes and
has the least favorable prognosis; relapses may occur within five
years after diagnosis (19). TNBC
is associated with positive expression of cytokeratin 5, 14, and 17
genes in mammary basal cells and high expression of
proliferation-related genes, but no expression of ER, PR and HER2
(20).
Management of patients with breast cancer is diverse
and varies according to each patient's specific condition. The
primary aim of treating early-stage breast cancer is to prevent its
progression to invasive breast cancer. This involves a range of
management strategies, such as surgery, radiation therapy, and, for
suitable patients, adjuvant endocrine therapy to minimize the risk
of cancer recurrence (21).
Several studies of early-stage breast cancer have demonstrated that
incorporating whole breast radiation after surgery reduces the
recurrence of breast cancer (22,23). However, this approach does not
significantly impact longer-term metastasis-free survival (24). Several drugs targeting the
overexpressed or mutated genes have been used in the treatment of
breast cancers (25). For
example, the US Food and Drug Administration (FDA) has approved the
use of tamoxifen to reduce the risk of ER+ breast cancer
recurrence (26), and buparlisib
has been shown to be safe and efficacious in HER2-,
PIK3CA-mutated, advanced, or metastatic breast cancer (27). However, some targeting drugs did
not significantly improve progression-free survival (PFS), and
their dosing is limited by toxicity, which potentially limits their
efficacy (28). Immunotherapy is
a newer form of treatment that helps the immune system fight
cancer. It is now frequently used in certain subtypes of advanced
breast cancer (29).
Atezolizumab, a human monoclonal antibody against programmed cell
death-ligand 1 (PD-L1), has shown efficacy in enhancing PFS when
used in combination with nab-paclitaxel for first-line chemotherapy
in patients with metastatic TNBC or in cases of recurrent TNBC that
are inoperable (30). The
development and early successes in active immunotherapy techniques
have led to therapeutic cancer vaccines. The first cancer vaccine,
approved by the FDA in 2010, was for treating metastatic prostate
cancer (31). However, the
availability of specific immunotherapies or cancer vaccines for the
treatment of breast cancer remains limited.
Immunotherapy to escalate the host's immune response
has become a reputable pillar of cancer treatment, aiming to
improve the therapeutic outcome of patients with certain types of
hematological and solid tumors (32). Stimulation is achieved by using
tumor antigens directly presentable by cancer cells to the host
adaptive immune system (33).
Strategies for targeting tumor antigens include cancer vaccines,
which stimulate the immune system to produce an immune response
against specific tumor antigens, and adoptive T-cell therapy, which
involves modifying T-cells from a patient's blood to recognize
specific tumor antigens and then infusing them back into the
patient's body to attack the cancer cells (34,35).
Tumor antigens can be processed and presented with
the major histocompatibility complex (MHC) on the cell membrane and
recognized by receptors on the cell surface of T-cells to elicit an
antitumor immune response (36).
The properties of tumor antigens can differ depending on the
specific antigen and the type of cancer. Common properties of tumor
antigens include: Overexpression-tumor antigens are usually
expressed at higher levels in cancer cells than in normal cells,
which can make them more detectable by the immune system;
mutation-some tumor antigens are mutated versions of normal
proteins, which can render cancer cells more distinct from the
normal cells and more vulnerable to immune attack;
specificity-tumor antigens are specific to certain types of cancer,
or even to certain subtypes of cancer which means that therapies
targeting these tumor antigens can be less toxic than more
broad-spectrum cancer treatments (37); and heterogeneity-when different
cancer cells have unique properties and antigens it becomes more
difficult to develop effective immunotherapies that target a broad
range of tumor cells (38).
There are two main types of tumor antigens:
Tumor-associated antigens (TAAs) and tumor-specific antigens
(TSAs). TAAs are self proteins that are highly expressed in cancer
cells but typically found at minimal levels in normal cells
(39). TAAs naturally arise
through genetic amplification or post-translational modification,
leading to the aberrant expression of proteins that are then no
longer shielded from the MHC-processing machinery and consequently
acquire immunogenic properties (40). TAA categories are based on the
pattern of expression in normal cells, including overexpressed
antigens, oncofetal antigens and cancer-testis antigens. The
overexpressed antigens are proteins present in higher levels in
cancer cells compared with normal tissues due to amplification of
their DNA or mRNA and ultimately protein levels. Examples of
overexpressed antigens include HER2/neu in breast cancer or ovarian
cancer (41) and mesothelin in
brain metastasis tumors (42).
Oncofetal antigens are normal proteins that are expressed at high
levels during fetal development and increase in adult cancer, but
not in normal adult tissues. Examples of these antigens are
MART-1/Melan-A in melanoma (43)
and carcinoembryonic antigen in colorectal cancer (44). Cancer-testis antigens or tumor
germline antigens are proteins that are normally highly expressed
only in germ cells of ovaries, testis and placenta but are also
expressed in cancer cells. Examples are New York esophageal
squamous cell carcinoma 1 (45)
and melanoma antigen gene protein (46).
Even though TAAs are great potential targets for
cancer immunotherapy, there are several difficulties in their use.
The heterogeneity of cancer cells can lead to variability in the
mutation and expression of TAAs across different tumor types,
stages and patients. This heterogeneity can make it difficult to
develop a single immunotherapy effective for all patients with a
particular type of cancer (47).
TAAs are typically removed from the immune repertoire by central
and peripheral tolerance mechanisms, so they are not recognized as
foreign by the immune system. This results in an immune tolerance
that prevents the immune system from attacking the cancer cells
(48). Thus, a cancer vaccine
that uses these antigens may not be potent enough. In addition,
some normal tissues may share epitopes with or express low levels
of TAAs, which can lead to autoimmune reactions and on-target
off-tumor toxicities when targeted by immunotherapy. To mitigate
this effect, and to further improve the efficacy and safety of
TAA-targeted immunotherapies, several approaches have been taken,
including selection of only the highly expressed targeting TAA,
adjustment of the affinity and avidity of the chimeric receptor in
chimeric antigen receptor T-cells to respond only to cells
expressing a high level of TAA (49), and the use of immune checkpoint
inhibitors (ICIs) in combination with the TAA vaccine (50). Further development of these
strategies may strengthen the promise shown by TAAs as targets for
cancer immunotherapy.
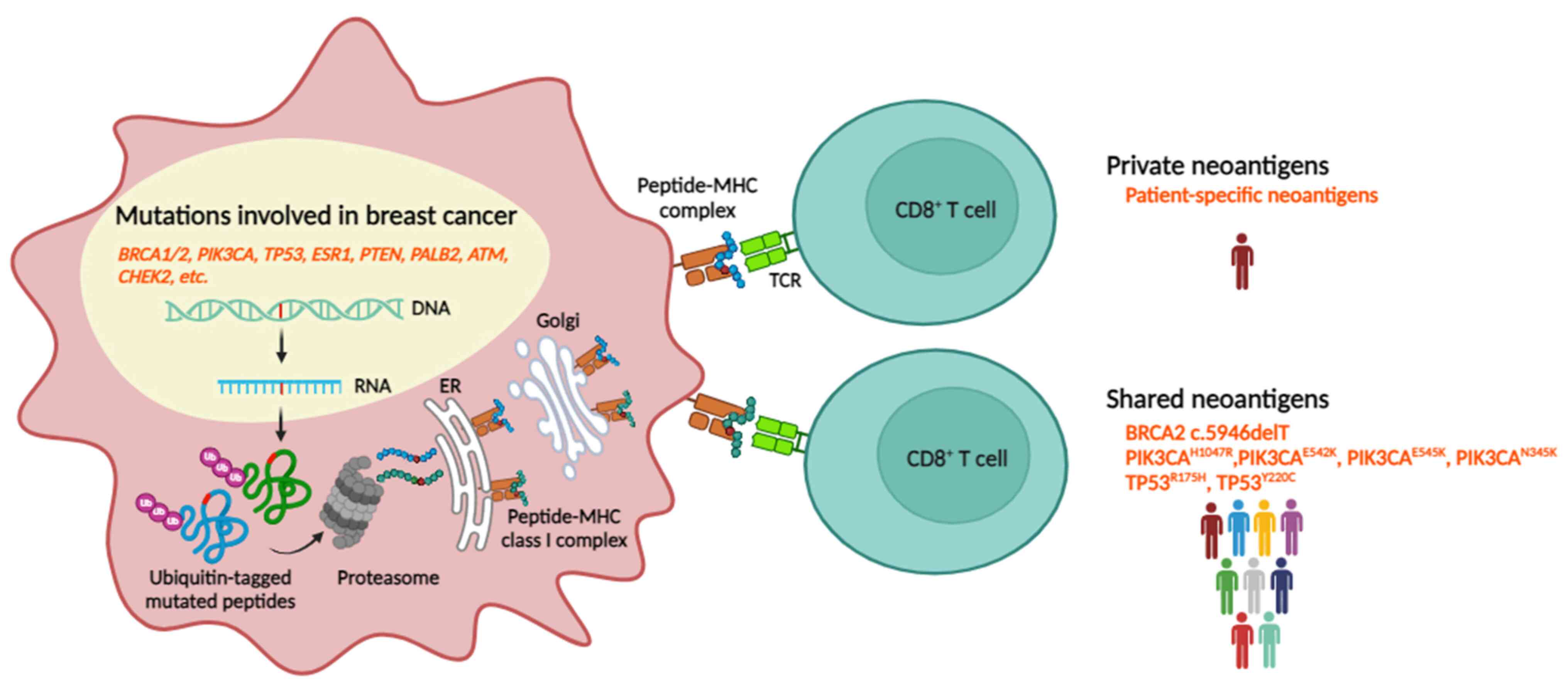
TSAs are detected as foreign non-self proteins that
are presented exclusively on cancer cell surfaces and are
particularly potent in eliciting an immune response (51). TSAs primarily originate from
mutations in passenger genes, with only a small fraction
originating from mutations in driver genes, and are heterogeneous
between, and within, patients. The identifying common passenger
gene mutations may generate immunogenic TSAs acting as the shared
neoantigens among patients. Moreover, the variability of TSAs
within individual patients may impact the efficacy of personalized
treatments tailored to target specific antigens while minimizing
adverse effects associated with non-specific targeting.
Since TSAs are solely expressed in cancer cells,
they are ideal targets for cancer immunotherapy. Numerous TSAs are
oncovirus antigens, viral proteins derived from oncogenic cancer
viruses that have integrated into the genome in cancer cells and
induce cell transformation and tumorigenesis (52). These include the E6 and E7
proteins from human papillomavirus that elicit strong
antigen-specific T-cell responses in cervical cancer and head and
neck cancer (53). Another
important group of TSAs comprises neoantigens: A specific type of
tumor antigen resulting from genetic and epigenetic aberrations
that arise during cancer initiation and progression (54). Multiple types of mutations can
give rise to neoantigens when they occur in protein-coding regions
and appear as non-synonymous polymorphisms. Well-defined sources of
neoantigens are somatic missense mutations that can generate single
nucleotide variants (SNVs) and insertions or deletions (INDELs).
These may create new open reading frames by frameshifts, as well as
other genomic alterations, including gene fusions (55). Frameshift mutations may be more
immunogenic than missense mutations due to the lack of similarity
to normal protein sequences and an increased probability of
generating neoantigens (56).
Interestingly, neoantigens derived from gene fusions are now
highlighted for immunotherapeutic treatments, especially in cancers
that have low tumor mutational burden (TMB) and less immune
infiltration (57).
Although inherited genetic mutations are associated
with a high risk for breast cancer development, <15% of women
with breast cancer come from families with a history of breast
cancer diagnosis (58). The
majority of breast cancer development in women who do not have a
family history of breast cancer may originate from somatic
mutations. The important genetic mutations in breast cancer are as
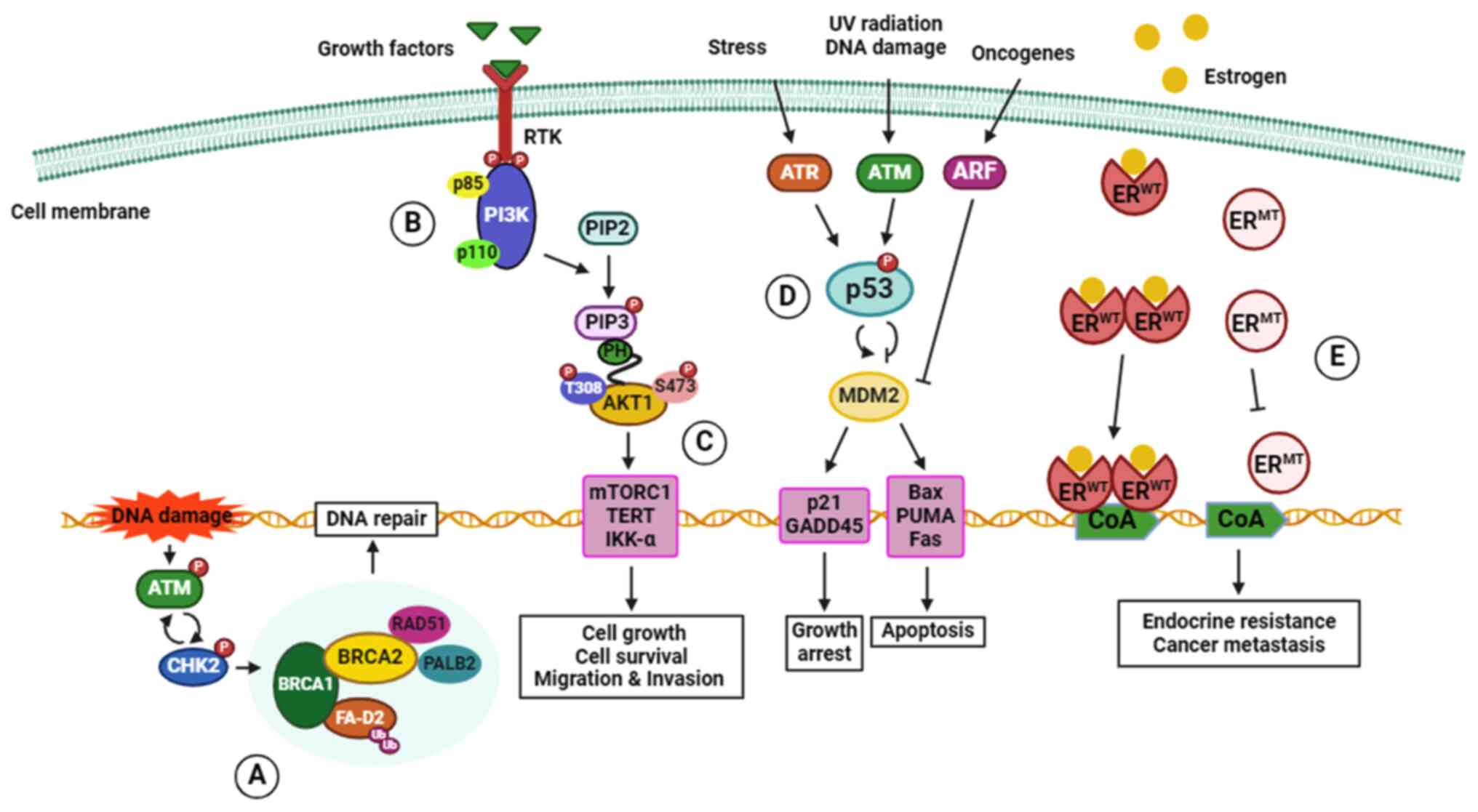
follows, and the mechanisms of action are summarized in Fig. 1.
In total, ~20% of women with a family history of
breast cancer have mutations in breast cancer susceptibility genes
1 or 2 (BRCA1 or BRCA2) (59). BRCA1 and BRCA2 proteins are tumor
suppressors that are crucial for homologous recombinant
(HR)-mediated repair of DNA double-strand breaks (DSBs). They
protect the integrity of the genome in proliferating cells to
prevent tumorigenesis (60).
BRCA1 involvement with DSBs requires recruitment of DNA repair
protein RAD51 homolog 1 (RAD51) to the sites of DNA damage through
interactions with partner and localizer of BRCA2 (PALB2), a
stabilizer of the replication fork, and BRCA2 (61). The BRCA mutations impair
HR-mediated repair of DSBs, raising the risk of developing
inherited and sporadic breast cancer (62,63). Although BRCA mutations can be
inherited and are found in a certain breast cancer family
histories, Metcalfe et al (64) have reported no difference in the
incidence of BRCA gene mutation carriers with or without a family
history of breast cancer (64).
Other gene mutations are involved with breast cancer
development; for example, the phosphatase and tensin homolog
(PTEN) gene serves as a tumor suppressor that regulates
various biological functions such as cell cycle progression, cell
proliferation and apoptosis regulation through negative regulation
of PI3K and AKT pathways (79).
The alterations or mutations of PTEN promote proliferation,
migration and invasion, leading to breast cancer development and
metastasis (80). Partner and
localizer of BRCA2 (PALB2) plays a role together with BRCA2 in DSB
repair (81). Mutations that
result in a loss of function in PALB2 are associated with the
occurrence of hereditary breast cancer (82). ATM, ataxia-telangiectasia
mutated, is a tumor suppressor gene that plays a role in repairing
damaged DNA by triggering repair mechanisms in response to the DNA
damage. Data collected by Gao et al (83) indicated that ATM missense
mutations increase the risk of breast cancer; notably, the V2424G
(c. 7271 T>G) missense variant exhibits the highest association
with breast cancer in all subtypes (83). In addition, the CHEK2 gene
encodes checkpoint kinase 2 (CHK2) protein, a serine/threonine
kinase involved in DSBs repair mechanisms. Failure of kinase
functions associated with missense mutation or deletion is found in
various types of cancer and frequently in breast cancer (84). The mediator of DNA damage
checkpoint 1 gene (MDC1) encodes the MDC1 scaffold protein which
plays a crucial role in regulating precise DNA repair in DNA damage
response (85). Loss of the MDC1
protein may serve as an indicator of recurrent metastases in
patients with breast cancer (86), and mutations in the MDC1 gene have
been associated with poor prognoses among patients with breast
cancer (87). Hence, mutations of
MDC1 may also give rise to plausible neoantigens. Numerous
additional mutated genes associated with an increased risk of
breast cancer development have been identified and reported
(88,89).
The frequency of somatic mutations varies
substantially within and between cancer classes, ranging from
~0.001 mutations per megabase (Mut/Mb) to over 400 Mut/Mb (90). TMB indicates the total number of
non-synonymous mutations (NSMs) per coding area of a cancer genome
and varies widely between individual patients and cancer types;
breast cancer exhibits a noticeably lower TMB than other cancers,
such as melanoma and lung cancer (54,91). No significant difference in OS or
PFS between the high TMB (>5 Mut/Mb) and low TMB (≤5 Mut/Mb)
groups have been reported, but high TMB may be associated with more
prolonged survival of patients undergoing ICI-based treatment
(92). Among breast cancer
subtypes, the highest median number of NSMs was found in patients
with TNBC (63, range 2-765 NSMs), followed by HER2+ (39,
range 1-1206), and the lowest median in
ER+/PR+/HER2- patients (32, range
1-2,860) (93). The association
between high TMB and NSMs in TNBC indicates that TNBC is the most
suitable subtype of breast cancer for high neoantigen loading.
The mutations that provide a clonal selective
advantage to cancer cells, are strongly associated with
oncogenesis, and play a causative role in the development and
progression of cancer are known as 'driver genes' (94). Some driver mutations are inherited
in the germline, but the majority of them arise in somatic cells
during the lifetime of a patient with cancer, along with numerous
'passenger mutations' that are not linked to cancer development
(94). Several driver gene
mutations in breast cancer have been reported (95,96). The most frequent or hotspot
mutations of the driver genes in breast cancer are summarized in
Table I.
A clinical study of a private neoantigen-based
vaccine against TNBC has been reported. Zhang et al
(134) identified neoantigens in
patients with TNBC with distinct HLA phenotypes. PALB2
(SPVTEIRTDL) and
ROBO3 (RVAGSMSSL) were predicted with
HLA-B*07:02, and PTPRS (RQLEVPWPYI) and ZDHHC16
(ALGALTVWL) were
predicted with HLA-A*02:01) (134). After stimulating each patient's
peripheral blood mononuclear cells with neoantigens, increased
neoantigen-specific immune responses were obtained, and there was
no cross-reaction with the corresponding wild-type peptides. These
neoantigens entered a Phase I clinical trial (NCT02348320) as a
neoantigen polyepitope DNA vaccine applied to patients by
electroporation. The presence of T-cells that specifically target
neoantigens was induced in both the pre-and post-vaccination
phases, and at 36-month follow-up, the median of responding
patients was 87.5% [95% confidence interval (CI): 72.7-100%]
compared with 49% (95% CI: 36.4-65.9%) of institutional historical
control patients (135).
In a clinical trial, NCT04105582, a total of 25
neoantigen peptides from patients with TNBC were administered in
autologous dendritic cells (DCs) over 16 weeks. The aforementioned
study proved the safety and immunogenicity of these vaccines. In
another clinical trial, NCT04879888, DC vaccines pulsed with
synthetic neoantigen peptides are being tested in nine patients
with TNBC. The trial's primary goal is to ensure safety, with a
secondary focus on evaluating the vaccine's immunogenicity using
interferon-γ (IFNγ) ELISPOT. However, these trials have not
examined the stability of the neoantigen peptides. Disis et
al (136) reported that ~60%
of patients with breast cancer who receive a DNA vaccine had
persistent DNA at week 16 following vaccination. Currently, no
trial results have been made available. In another clinical study,
Morisaki et al (137)
identified neoantigens in primary cancer cells from the pleural
fluid of a patient with TNBC. In total, 2/10 of predicted
neoantigens had a high binding affinity for HLA-A*02:06:
GEMIN2 (AQCPDVLV) and PARP10
(SISCHVFCL). Both
caused a significant increase in the response of IFNγ-producing
T-cells to autologous cancer cells observed after inducing
cytotoxic T lymphocytes with peptide-pulsed autologous DCs
(137). However, the
aforementioned study also reported that AKT1, ARAP3, NOTCH3,
PIK3CA and SLC35E2 contained identical non-synonymous
SNVs without testing immunogenicity. Additionally, there are other
ongoing clinical trials to evaluate personalized neoantigen-based
immunotherapy in breast cancer (Table II).
Cancer-driver mutations, which are typically found
in oncogenes or specific tumor suppressor genes, often occur in
genomic hotspots. These hotspots lead to changes in protein
function and are frequently observed across different patients
(138). The mutated peptides
produced by these genetic alterations, when presented by HLA
alleles, can generate neoantigens that are common among individuals
with the same genetic changes and HLA types (139). This phenomenon marks these
neoantigens as shared among cancer patients, giving rise to
'public' neoantigens (140).
Setting aside R157H, other p53 mutants could serve as neoantigens,
such as TP53Y220C found in breast cancer. The
TP53Y220C neoepitopes, VVPCEPPEV, RNTFRHSVVVPCE and NTFRHSVVVPCEPPE, were recognized by
HLA-A*02:01-, HLA-DRB1*04:01 and
HLA-DRB3*02:02-expressing tumor-infiltrating lymphocytes
(TILs), respectively (141).
Consequently, the results of current research support efforts to
create off-the-shelf anticancer therapies based on the public or
shared neoantigens, which could potentially benefit a broad group
of patients with breast cancer.
In breast cancer, most shared target antigens have
been reported as TAAs; for example, a Phase I/II clinical trial of
autologous DC transfected with the cDNA of mucin (MUC1) in patients
with advanced breast cancer (142). The trial demonstrated the
feasibility and safety of this vaccine approach and demonstrated
immunologic responses in patients who had previously received
treatment and had advanced-stage disease (142). In situ testing of a
HER2-pulsed DC vaccine in 27 patients with HER2/neu-overexpressing
ductal carcinoma was well-tolerated and demonstrated reduction or
elimination of HER2/neu expression (143).
Previously, a small number of public neoantigens
were explored in breast cancer (Table III). Pettitt et al
(144) analyzed the prediction
of an out-of-frame sequence resulting from common founder
BRCA2 reversion (c.5946delT: RENLSRYQMLHYKTQ) that can be
presented by numerous HLA class I alleles such as
HLA-A*0101, HLA-A*3303,
HLA-B*3501, HLA-B*4001, HLA-C*0602
and HLA-C*1203 (144). The results showed a revertant
neoepitope sequence that can be presented with a high probability
by the MHC class I across the general population and therefore
provides an option for targeting public neoantigens (144).
Previous research has indicated that TP53 hotspot
mutations can be utilized in a shared neoantigen-based cancer
vaccine to treat breast cancers (148,149). Lo et al (148) described the identification and
characterization of HLA-A*0201-restricted TCRs that
react with TP53R175H and recognize various cancer cell
lines, including breast cancer endogenously expressing
TP53R175H. When a patient with the HLA-A*0201
allele and chemorefractory breast cancer was treated by adoptive
transfer using TP53R175H-TCR-engineered autologous
peripheral blood lymphocytes, a significant reduction in
subcutaneous tumor deposits occurred, and all skin lesions wholly
resolved by day 60 after cell therapy (149). This finding may indicate a
suitable shared neoantigen target for the treatment of patients
whose cancers convey HLA-A*0201 and contain a
TP53R175H mutation (148). Kim et al (149) and Zacharakis et al
(150) reported that patients
with breast cancer who received adoptive transfer of ex
vivo-expanded autologous TILs targeting TP53Y220C
exhibited a partial response. There was complete resolution of two
right subpectoral lymph nodes, subcutaneous breast nodules, and
substantial regression of the primary breast tumor with response
durations of 6 months (150).
The summary of hotspot mutations recognized as public neoantigens
sharing among patients with breast cancer is shown in Fig. 2.
Despite multiple studies that have identified
shared neoantigens in breast cancer and demonstrated their ability
to stimulate anti-tumor immune responses in vitro, clinical
investigations that have focused on exploiting the immunogenicity
of the associated shared neoantigens in breast cancer are limited.
Ongoing clinical trials testing the immunogenicity of shared
neoantigen vaccines are in non-small cell lung, colorectal and
pancreatic cancer (Clinical trial NCT03953235). The aforementioned
study has been measuring PFS and OS for ~4 years (2019-2023). There
remains a crucial necessity for further clinical investigation into
the implications and outcomes of these shared neoantigens within
breast cancer. The broader application across a diverse spectrum of
cancer patients suggests that they may be promising targets for
off-the-shelf immunotherapy.
Cancer mutations are patient-specific, giving
neoantigens a highly personalized character. Yet, some neoepitopes
are consistently found across multiple patients with cancer, making
them shared neoantigens. These common neoantigens hold promise for
treating a substantial number of patients who share several
specific HLA types, offering viable targets for off-the-shelf
cancer immunotherapies. Current clinical research into breast
cancer predominantly investigates personalized neoantigens, whereas
the study of shared neoantigens in breast cancer is limited due to
the low proportion of neoantigen burden, limited studies of hotspot
mutations in breast cancer, and the challenges in locating patients
who have both a specific HLA allele and a particular common
neoantigen. The true potential of broad HLA-specific public
neoantigen prediction remains to be fully explored through clinical
trials involving diverse patient groups with various HLA types.
This presents a significant challenge for the development of
broadly applicable ready-to-use immunotherapies for cancers in
general and for patients with breast cancer in particular.
Not applicable.
NS designed and wrote the manuscript. CT, PT and PY
provided the research direction. NS and CT edited the manuscript.
All authors read and approved the final manuscript. Data
authentication is not applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
The authors would like to thank Dr Jan Davies,
Faculty of Graduate Studies, Mahidol University, for English
editing.
The present study was supported by the Research and Innovation
Grant, the National Research Council of Thailand, Ministry of
Higher Education, Science, Research and Innovation, Thailand (grant
no. R016341038) and the Siriraj Research Grant, Faculty of Medicine
Siriraj Hospital, Mahidol University (grant no. R016334002).
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cancer Genome Atlas Network: Comprehensive
molecular portraits of human breast tumours. Nature. 490:61–70.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rouzier R, Perou CM, Symmans WF, Ibrahim
N, Cristofanilli M, Anderson K, Hess KR, Stec J, Ayers M, Wagner P,
et al: Breast cancer molecular subtypes respond differently to
preoperative chemotherapy. Clin Cancer Res. 11:5678–5685. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bhargava R, Beriwal S, Dabbs DJ, Ozbek U,
Soran A, Johnson RR, Brufsky AM, Lembersky BC and Ahrendt GM:
Immunohistochemical surrogate markers of breast cancer molecular
classes predicts response to neoadjuvant chemotherapy: A single
institutional experience with 359 cases. Cancer. 116:1431–1439.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nik-Zainal S, Davies H, Staaf J,
Ramakrishna M, Glodzik D, Zou X, Martincorena I, Alexandrov LB,
Martin S, Wedge DC, et al: Landscape of somatic mutations in 560
breast cancer whole-genome sequences. Nature. 534:47–54. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pon JR and Marra MA: Driver and passenger
mutations in cancer. Annu Rev Pathol. 10:25–50. 2015. View Article : Google Scholar
|
|
7
|
Chen T, Wang Z, Zhou W, Chong Z,
Meric-Bernstam F, Mills GB and Chen K: Hotspot mutations
delineating diverse mutational signatures and biological utilities
across cancer types. BMC Genomics. 17(Suppl 2): S3942016.
View Article : Google Scholar
|
|
8
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2022. CA Cancer J Clin. 72:7–33. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li CI, Uribe DJ and Daling JR: Clinical
characteristics of different histologic types of breast cancer. Br
J Cancer. 93:1046–1052. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Caswell-Jin JL, Lorenz C and Curtis C:
Molecular heterogeneity and evolution in breast cancer. Annu Rev
Cancer Biol. 5:79–94. 2021. View Article : Google Scholar
|
|
11
|
Wagner J, Rapsomaniki MA, Chevrier S,
Anzeneder T, Langwieder C, Dykgers A, Rees M, Ramaswamy A, Muenst
S, Soysal SD, et al: A single-cell atlas of the tumor and immune
ecosystem of human breast cancer. Cell. 177:1330–1345.e18. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tsang JYS and Tse GM: Molecular
classification of breast cancer. Adv Anat Pathol. 27:27–35. 2020.
View Article : Google Scholar
|
|
13
|
Goldhirsch A, Wood WC, Coates AS, Gelber
RD, Thürlimann B and Senn HJ; Panel members: Strategies for
subtypes-dealing with the diversity of breast cancer: Highlights of
the St. Gallen international expert consensus on the primary
therapy of early breast cancer 2011. Ann Oncol. 22:1736–1747. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li J, Chen Z, Su K and Zeng J:
Clinicopathological classification and traditional prognostic
indicators of breast cancer. Int J Clin Exp Pathol. 8:8500–8505.
2015.PubMed/NCBI
|
|
15
|
Prat A, Cheang MCU, Martín M, Parker JS,
Carrasco E, Caballero R, Tyldesley S, Gelmon K, Bernard PS, Nielsen
TO and Perou CM: Prognostic significance of progesterone
receptor-positive tumor cells within immunohistochemically defined
luminal A breast cancer. J Clin Oncol. 31:203–209. 2013. View Article : Google Scholar
|
|
16
|
Maisonneuve P, Disalvatore D, Rotmensz N,
Curigliano G, Colleoni M, Dellapasqua S, Pruneri G, Mastropasqua
MG, Luini A, Bassi F, et al: Proposed new clinicopathological
surrogate definitions of luminal A and luminal B (HER2-negative)
intrinsic breast cancer subtypes. Breast Cancer Res. 16:R652014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tang P and Tse GM: Immunohistochemical
surrogates for molecular classification of breast carcinoma: A 2015
update. Arch Pathol Lab Med. 140:806–814. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dawood S, Broglio K, Buzdar AU, Hortobagyi
GN and Giordano SH: Prognosis of women with metastatic breast
cancer by HER2 status and trastuzumab treatment: An
institutional-based review. J Clin Oncol. 28:92–98. 2010.
View Article : Google Scholar :
|
|
19
|
Badve S, Dabbs DJ, Schnitt SJ, Baehner FL,
Decker T, Eusebi V, Fox SB, Ichihara S, Jacquemier J, Lakhani SR,
et al: Basal-like and triple-negative breast cancers: A critical
review with an emphasis on the implications for pathologists and
oncologists. Mod Pathol. 24:157–167. 2011. View Article : Google Scholar
|
|
20
|
Prat A, Pineda E, Adamo B, Galván P,
Fernández A, Gaba L, Díez M, Viladot M, Arance A and Muñoz M:
Clinical implications of the intrinsic molecular subtypes of breast
cancer. Breast. 24(Suppl 2): S26–S35. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gradishar WJ, Moran MS, Abraham J, Aft R,
Agnese D, Allison KH, Anderson B, Burstein HJ, Chew H, Dang C, et
al: Breast cancer, version 3.2022, NCCN clinical practice
guidelines in oncology. J Natl Compr Canc Netw. 20:691–722. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
EORTC Breast Cancer Cooperative Group;
EORTC Radiotherapy Group; Bijker N, Meijnen P, Peterse JL, Bogaerts
J, Van Hoorebeeck I, Julien JP, Gennaro M, Rouanet P, et al:
Breast-conserving treatment with or without radiotherapy in ductal
carcinoma-in-situ: Ten-year results of European organisation for
research and treatment of cancer randomized phase III trial 10853-A
study by the EORTC breast cancer cooperative group and EORTC
radiotherapy group. J Clin Oncol. 24:3381–3387. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
McCormick B, Winter K, Hudis C, Kuerer HM,
Rakovitch E, Smith BL, Sneige N, Moughan J, Shah A, Germain I, et
al: RTOG 9804: A prospective randomized trial for good-risk ductal
carcinoma in situ comparing radiotherapy with observation. J Clin
Oncol. 33:709–715. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Holmberg L, Garmo H, Granstrand B,
Ringberg A, Arnesson LG, Sandelin K, Karlsson P, Anderson H and
Emdin S: Absolute risk reductions for local recurrence after
postoperative radiotherapy after sector resection for ductal
carcinoma in situ of the breast. J Clin Oncol. 26:1247–1252. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bhushan A, Gonsalves A and Menon JU:
Current state of breast cancer diagnosis, treatment, and
theranostics. Pharmaceutics. 13:7232021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cuzick J, Sestak I, Bonanni B, Costantino
JP, Cummings S, DeCensi A, Dowsett M, Forbes JF, Ford L, LaCroix
AZ, et al: Selective oestrogen receptor modulators in prevention of
breast cancer: An updated meta-analysis of individual participant
data. Lancet. 381:1827–1834. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Rodon J, Braña I, Siu LL, De Jonge MJ,
Homji N, Mills D, Di Tomaso E, Sarr C, Trandafir L, Massacesi C, et
al: Phase I dose-escalation and -expansion study of buparlisib
(BKM120), an oral pan-class I PI3K inhibitor, in patients with
advanced solid tumors. Invest New Drugs. 32:670–681. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Krop IE, Mayer IA, Ganju V, Dickler M,
Johnston S, Morales S, Yardley DA, Melichar B, Forero-Torres A, Lee
SC, et al: Pictilisib for oestrogen receptor-positive, aromatase
inhibitor-resistant, advanced or metastatic breast cancer (FERGI):
A randomised, double-blind, placebo-controlled, phase 2 trial.
Lancet Oncol. 17:811–821. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Schneble E, Jinga DC and Peoples G: Breast
cancer immunotherapy. Maedica (Bucur). 10:185–191. 2015.PubMed/NCBI
|
|
30
|
Kang C and Syed YY: Atezolizumab (in
combination with nab-paclitaxel): A review in advanced
triple-negative breast cancer. Drugs. 80:601–607. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kantoff PW, Schuetz TJ, Blumenstein BA,
Glode LM, Bilhartz DL, Wyand M, Manson K, Panicali DL, Laus R,
Schlom J, et al: Overall survival analysis of a phase II randomized
controlled trial of a Poxviral-based PSA-targeted immunotherapy in
metastatic castration-resistant prostate cancer. J Clin Oncol.
28:1099–1105. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hu ZI, Ho AY and McArthur HL: Combined
radiation therapy and immune checkpoint blockade therapy for breast
cancer. Int J Radiat Oncol Biol Phys. 99:153–164. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Boon T, Cerottini JC, Van den Eynde B, van
der Bruggen P and Van Pel A: Tumor antigens recognized by T
lymphocytes. Annu Rev Immunol. 12:337–365. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Butterfield LH: Cancer vaccines. BMJ.
350:h9882015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Perica K, Varela JC, Oelke M and Schneck
J: Adoptive T cell immunotherapy for cancer. Rambam Maimonides Med
J. 6:e00042015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ilyas S and Yang JC: Landscape of tumor
antigens in T cell immunotherapy. J Immunol. 195:5117–5122. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Srinivasan R and Wolchok JD: Tumor
antigens for cancer immunotherapy: Therapeutic potential of
xenogeneic DNA vaccines. J Transl Med. 2:122004. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Benvenuto M, Focaccetti C, Izzi V,
Masuelli L, Modesti A and Bei R: Tumor antigens heterogeneity and
immune response-targeting neoantigens in breast cancer. Semin
Cancer Biol. 72:65–75. 2021. View Article : Google Scholar
|
|
39
|
Alatrash G, Crain AK and Molldrem JJ:
Chapter 7-tumor-associated antigens. Immune Biology of Allogeneic
Hematopoietic Stem Cell Transplantation. Socié G, Zeiser R and
Blazar BR: 2nd edition. Academic Press; pp. 107–125. 2019,
View Article : Google Scholar
|
|
40
|
Tawara I, Kageyama S, Miyahara Y, Fujiwara
H, Nishida T, Akatsuka Y, Ikeda H, Tanimoto K, Terakura S, Murata
M, et al: Safety and persistence of WT1-specific T-cell receptor
gene-transduced lymphocytes in patients with AML and MDS. Blood.
130:1985–1994. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Baxevanis CN, Gritzapis AD, Tsitsilonis
OE, Katsoulas HL and Papamichail M: HER-2/neu-derived peptide
epitopes are also recognized by cytotoxic CD3(+)CD56(+) (natural
killer T) lymphocytes. Int J Cancer. 98:864–872. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhenjiang L, Rao M, Luo X, Sandberg E,
Bartek J Jr, Schoutrop E, von Landenberg A, Meng Q, Valentini D,
Poiret T, et al: Mesothelin-specific immune responses predict
survival of patients with brain metastasis. EBioMedicine. 23:20–24.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Pittet MJ, Valmori D, Dunbar PR, Speiser
DE, Liénard D, Lejeune F, Fleischhauer K, Cerundolo V, Cerottini JC
and Romero P: High frequencies of naive Melan-A/MART-1-specific
CD8(+) T cells in a large proportion of human histocompatibility
leukocyte antigen (HLA)-A2 individuals. J Exp Med. 190:705–716.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lesterhuis WJ, De Vries IJ, Schreibelt G,
Schuurhuis DH, Aarntzen EH, De Boer A, Scharenborg NM, Van De Rakt
M, Hesselink EJ, Figdor CG, et al: Immunogenicity of dendritic
cells pulsed with CEA peptide or transfected with CEA mRNA for
vaccination of colorectal cancer patients. Anticancer Res.
30:5091–5097. 2010.PubMed/NCBI
|
|
45
|
Parvanova I, Rettig L, Knuth A and Pascolo
S: The form of NY-ESO-1 antigen has an impact on the clinical
efficacy of anti-tumor vaccination. Vaccine. 29:3832–3836. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Mohsenzadegan M, Razmi M, Vafaei S,
Abolhasani M, Madjd Z, Saeednejad Zanjani L and Sharifi L:
Co-expression of cancer-testis antigens of MAGE-A6 and MAGE-A11 is
associated with tumor aggressiveness in patients with bladder
cancer. Sci Rep. 12:5992022. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Lawrence MS, Stojanov P, Polak P, Kryukov
GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH and
Roberts SA, et al: Mutational heterogeneity in cancer and the
search for new cancer-associated genes. Nature. 499:214–218. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Makkouk A and Weiner GJ: Cancer
immunotherapy and breaking immune tolerance: New approaches to an
old challenge. Cancer Res. 75:5–10. 2015. View Article : Google Scholar
|
|
49
|
Wang Y, Buck A, Piel B, Zerefa L, Murugan
N, Coherd CD, Miklosi AG, Johal H, Bastos RN, Huang K, et al:
Affinity fine-tuning anti-CAIX CAR-T cells mitigate on-target
off-tumor side effects. Mol Cancer. 23:562024. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Sanderson K, Scotland R, Lee P, Liu D,
Groshen S, Snively J, Sian S, Nichol G, Davis T, Keler T, et al:
Autoimmunity in a phase I trial of a fully human anti-cytotoxic
T-lymphocyte antigen-4 monoclonal antibody with multiple melanoma
peptides and montanide ISA 51 for patients with resected stages III
and IV melanoma. J Clin Oncol. 23:741–750. 2005. View Article : Google Scholar
|
|
51
|
Wölfel T, Hauer M, Schneider J, Serrano M,
Wölfel C, Klehmann-Hieb E, De Plaen E, Hankeln T, Meyer zum
Büschenfelde KH and Beach D: A p16INK4a-insensitive CDK4 mutant
targeted by cytolytic T lymphocytes in a human melanoma. Science.
269:1281–1284. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Plummer M, de Martel C, Vignat J, Ferlay
J, Bray F and Franceschi S: Global burden of cancers attributable
to infections in 2012: A synthetic analysis. Lancet Glob Health.
4:e609–e616. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Trimble CL, Morrow MP, Kraynyak KA, Shen
X, Dallas M, Yan J, Edwards L, Parker RL, Denny L, Giffear M, et
al: Safety, efficacy, and immunogenicity of VGX-3100, a therapeutic
synthetic DNA vaccine targeting human papillomavirus 16 and 18 E6
and E7 proteins for cervical intraepithelial neoplasia 2/3: A
randomised, double-blind, placebo-controlled phase 2b trial.
Lancet. 386:2078–2088. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Castle JC, Uduman M, Pabla S, Stein RB and
Buell JS: mutation-derived neoantigens for cancer immunotherapy.
Front Immunol. 10:18562019. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Capietto AH, Hoshyar R and Delamarre L:
Sources of cancer neoantigens beyond single-nucleotide variants.
Int J Mol Sci. 23:101312022. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Roudko V, Bozkus CC, Orfanelli T, McClain
CB, Carr C, O'Donnell T, Chakraborty L, Samstein R, Huang KL, Blank
SV, et al: Shared immunogenic poly-epitope frameshift mutations in
microsatellite unstable tumors. Cell. 183:1634–1649.e17. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Yang W, Lee KW, Srivastava RM, Kuo F,
Krishna C, Chowell D, Makarov V, Hoen D, Dalin MG, Wexler L, et al:
Immunogenic neoantigens derived from gene fusions stimulate T cell
responses. Nat Med. 25:767–775. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Colditz GA, Kaphingst KA, Hankinson SE and
Rosner B: Family history and risk of breast cancer: Nurses' health
study. Breast Cancer Res Treat. 133:1097–1104. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Couch FJ, Nathanson KL and Offit K: Two
decades after BRCA: Setting paradigms in personalized cancer care
and prevention. Science. 343:1466–1470. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Narod SA and Foulkes WD: BRCA1 and BRCA2:
1994 And beyond. Nat Rev Cancer. 4:665–676. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Roy R, Chun J and Powell SN: BRCA1 and
BRCA2: Different roles in a common pathway of genome protection.
Nat Rev Cancer. 12:68–78. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Russo A, Calò V, Agnese V, Bruno L,
Corsale S, Augello C, Gargano G, Barbera F, Cascio S, Intrivici C,
et al: BRCA1 genetic testing in 106 breast and ovarian cancer
families from Southern Italy (Sicily): A mutation analyses. Breast
Cancer Res Treat. 105:267–276. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Armstrong N, Ryder S, Forbes C, Ross J and
Quek RG: A systematic review of the international prevalence of
BRCA mutation in breast cancer. Clin Epidemiol. 11:543–561. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Metcalfe KA, Lubinski J, Gronwald J,
Huzarski T, McCuaig J, Lynch HT, Karlan B, Foulkes WD, Singer CF,
Neuhausen SL, et al: The risk of breast cancer in BRCA1 and BRCA2
mutation carriers without a first-degree relative with breast
cancer. Clin Genet. 93:1063–1068. 2018. View Article : Google Scholar
|
|
65
|
Liu P, Cheng H, Roberts TM and Zhao JJ:
Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev
Drug Discov. 8:627–644. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Sun K, Luo J, Guo J, Yao X, Jing X and Guo
F: The PI3K/AKT/mTOR signaling pathway in osteoarthritis: A
narrative review. Osteoarthritis Cartilage. 28:400–409. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Fillbrunn M, Signorovitch J, André F, Wang
I, Lorenzo I, Ridolfi A, Park J, Dua A and Rugo HS: PIK3CA mutation
status, progression and survival in advanced HR + /HER2-breast
cancer: A meta-analysis of published clinical trials. BMC Cancer.
22:10022022. View Article : Google Scholar
|
|
68
|
Tonnessen-Murray CA, Lozano G and Jackson
JG: The regulation of cellular functions by the p53 protein:
Cellular senescence. Cold Spring Harb Perspect Med. 7:a0261122017.
View Article : Google Scholar
|
|
69
|
Hamroun D, Kato S, Ishioka C, Claustres M,
Béroud C and Soussi T: The UMD TP53 database and website: Update
and revisions. Hum Mutat. 27:14–20. 2006. View Article : Google Scholar
|
|
70
|
Ungerleider NA, Rao SG, Shahbandi A, Yee
D, Niu T, Frey WD and Jackson JG: Breast cancer survival predicted
by TP53 mutation status differs markedly depending on treatment.
Breast Cancer Res. 20:1152018. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Kato S, Han SY, Liu W, Otsuka K, Shibata
H, Kanamaru R and Ishioka C: Understanding the function-structure
and function-mutation relationships of p53 tumor suppressor protein
by high-resolution missense mutation analysis. Proc Natl Acad Sci
USA. 100:8424–8429. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Soussi T and Béroud C: Assessing TP53
status in human tumours to evaluate clinical outcome. Nat Rev
Cancer. 1:233–240. 2001. View Article : Google Scholar
|
|
73
|
Parker MG, Arbuckle N, Dauvois S,
Danielian P and White R: Structure and function of the estrogen
receptor. Ann N Y Acad Sci. 684:119–126. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Pejerrey SM, Dustin D, Kim JA, Gu G,
Rechoum Y and Fuqua SAW: The impact of ESR1 mutations on the
treatment of metastatic breast cancer. Horm Cancer. 9:215–228.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Jeselsohn R, Buchwalter G, De Angelis C,
Brown M and Schiff R: ESR1 mutations-a mechanism for acquired
endocrine resistance in breast cancer. Nat Rev Clin Oncol.
12:573–583. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Lei JT, Shao J, Zhang J, Iglesia M, Chan
DW, Cao J, Anurag M, Singh P, He X, Kosaka Y, et al: Functional
annotation of ESR1 gene fusions in estrogen receptor-positive
breast cancer. Cell Rep. 24:1434–1444.e7. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Siddika T, Balasuriya N, Frederick MI,
Rozik P, Heinemann IU and O'Donoghue P: Delivery of active AKT1 to
human cells. Cells. 11:38342022. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
George B, Gui B, Raguraman R, Paul AM,
Nakshatri H, Pillai MR and Kumar R: AKT1 transcriptomic landscape
in breast cancer cells. Cells. 11:22902022. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Keniry M and Parsons R: The role of PTEN
signaling perturbations in cancer and in targeted therapy.
Oncogene. 27:5477–5485. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Chen J, Sun J, Wang Q, Du Y, Cheng J, Yi
J, Xie B, Jin S, Chen G, Wang L, et al: Systemic deficiency of PTEN
accelerates breast cancer growth and metastasis. Front Oncol.
12:8254842022. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Sy SMH, Huen MSY, Zhu Y and Chen J: PALB2
regulates recombinational repair through chromatin association and
oligomerization. J Biol Chem. 284:18302–18310. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Antoniou AC, Casadei S, Heikkinen T,
Barrowdale D, Pylkäs K, Roberts J, Lee A, Subramanian D, De Leeneer
K, Fostira F, et al: Breast-cancer risk in families with mutations
in PALB2. N Engl J Med. 371:497–506. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Gao LB, Pan XM, Sun H, Wang X, Rao L, Li
LJ, Liang WB, Lv ML, Yang WZ and Zhang L: The association between
ATM D1853N polymorphism and breast cancer susceptibility: A
meta-analysis. J Exp Clin Cancer Res. 29:1172010. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Apostolou P and Papasotiriou I: Current
perspectives on CHEK2 mutations in breast cancer. Breast Cancer
(Dove Med Press). 9:331–335. 2017.PubMed/NCBI
|
|
85
|
Stewart GS, Wang B, Bignell CR, Taylor AMR
and Elledge SJ: MDC1 is a mediator of the mammalian DNA damage
checkpoint. Nature. 421:961–966. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Patel AN, Goyal S, Wu H, Schiff D, Moran
MS and Haffty BG: Mediator of DNA damage checkpoint protein 1
(MDC1) expression as a prognostic marker for nodal recurrence in
early-stage breast cancer patients treated with breast-conserving
surgery and radiation therapy. Breast Cancer Res Treat.
126:601–607. 2011. View Article : Google Scholar
|
|
87
|
Liu C, Chang H, Li XH, Qi YF, Wang JO,
Zhang Y and Yang XH: Network meta-analysis on the effects of DNA
damage response-related gene mutations on overall survival of
breast cancer based on TCGA database. J Cell Biochem.
118:4728–4734. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Wang YA, Jian JW, Hung CF, Peng HP, Yang
CF, Cheng HS and Yang AS: Germline breast cancer susceptibility
gene mutations and breast cancer outcomes. BMC Cancer. 18:3152018.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Breast Cancer Association Consortium;
Dorling L, Carvalho S, Allen J, González-Neira A, Luccarini C,
Wahlström C, Pooley KA, Parsons MT, Fortuno C, et al: Breast cancer
risk genes-association analysis in more than 113,000 women. N Engl
J Med. 384:428–439. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Alexandrov LB, Nik-Zainal S, Wedge DC,
Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A,
Børresen-Dale AL, et al: Signatures of mutational processes in
human cancer. Nature. 500:415–421. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Vogelstein B, Papadopoulos N, Velculescu
VE, Zhou S, Diaz LA Jr and Kinzler KW: Cancer genome landscapes.
Science. 339:1546–1558. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Ke L, Li S and Cui H: The prognostic role
of tumor mutation burden on survival of breast cancer: A systematic
review and meta-analysis. BMC Cancer. 22:11852022. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Narang P, Chen M, Sharma AA, Anderson KS
and Wilson MA: The neoepitope landscape of breast cancer:
Implications for immunotherapy. BMC Cancer. 19:2002019. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Stratton MR, Campbell PJ and Futreal PA:
The cancer genome. Nature. 458:719–724. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Zhang G, Wang Y, Chen B, Guo L, Cao L, Ren
C, Wen L, Li K, Jia M, Li C, et al: Characterization of frequently
mutated cancer genes in Chinese breast tumors: A comparison of
Chinese and TCGA cohorts. Ann Transl Med. 7:1792019. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Zhou S, Liu S, Zhao L and Sun HX: A
comprehensive survey of genomic mutations in breast cancer reveals
recurrent neoantigens as potential therapeutic targets. Front
Oncol. 12:7864382022. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Carraro DM, Koike Folgueira MAA, Garcia
Lisboa BC, Ribeiro Olivieri EH, Vitorino Krepischi AC, de Carvalho
AF, de Carvalho Mota LD, Puga RD, do Socorro Maciel M, Michelli RA,
et al: Comprehensive analysis of BRCA1, BRCA2 and TP53 germline
mutation and tumor characterization: A portrait of early-onset
breast cancer in Brazil. PLoS One. 8:e575812013. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Kurian AW: BRCA1 and BRCA2 mutations
across race and ethnicity: Distribution and clinical implications.
Curr Opin Obstet Gynecol. 22:72–78. 2010. View Article : Google Scholar
|
|
99
|
Winter C, Nilsson MP, Olsson E, George AM,
Chen Y, Kvist A, Törngren T, Vallon-Christersson J, Hegardt C,
Häkkinen J, et al: Targeted sequencing of BRCA1 and BRCA2 across a
large unselected breast cancer cohort suggests that one-third of
mutations are somatic. Ann Oncol. 27:1532–1538. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Staaf J, Glodzik D, Bosch A,
Vallon-Christersson J, Reuterswärd C, Häkkinen J, Degasperi A,
Amarante TD, Saal LH, Hegardt C, et al: Whole-genome sequencing of
triple-negative breast cancers in a population-based clinical
study. Nat Med. 25:1526–1533. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Larsen MJ, Kruse TA, Tan Q, Lænkholm AV,
Bak M, Lykkesfeldt AE, Sørensen KP, Hansen TV, Ejlertsen B, Gerdes
AM and Thomassen M: Classifications within molecular subtypes
enables identification of BRCA1/BRCA2 mutation carriers by RNA
tumor profiling. PLoS One. 8:e642682013. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
De Talhouet S, Peron J, Vuilleumier A,
Friedlaender A, Viassolo V, Ayme A, Bodmer A, Treilleux I, Lang N,
Tille JC, et al: Clinical outcome of breast cancer in carriers of
BRCA1 and BRCA2 mutations according to molecular subtypes. Sci Rep.
10:70732020. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Linger RJ and Kruk PA: BRCA1 16 years
later: risk-associated BRCA1 mutations and their functional
implications. FEBS J. 277:3086–3096. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Lavoro A, Scalisi A, Candido S, Zanghì GN,
Rizzo R, Gattuso G, Caruso G, Libra M and Falzone L: Identification
of the most common BRCA alterations through analysis of germline
mutation databases: Is droplet digital PCR an additional strategy
for the assessment of such alterations in breast and ovarian cancer
families. Int J Oncol. 60:582022. View Article : Google Scholar
|
|
105
|
Sabine VS, Crozier C, Brookes CL, Drake C,
Piper T, van de Velde CJ, Hasenburg A, Kieback DG, Markopoulos C,
Dirix L, et al: Mutational analysis of PI3K/AKT signaling pathway
in tamoxifen exemestane adjuvant multinational pathology study. J
Clin Oncol. 32:2951–2958. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Meyer DS, Brinkhaus H, Müller U, Müller M
and Cardiff RD; mBentires-Alj M: Luminal expression of PIK3CA
mutant H1047R in the mammary gland induces heterogeneous tumors.
Cancer Res. 71:4344–4351. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Forbes SA, Bindal N, Bamford S, Cole C,
Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A, et al:
COSMIC: Mining complete cancer genomes in the catalogue of somatic
mutations in cancer. Nucleic Acids Res. 39(Database Issue):
D945–D950. 2011. View Article : Google Scholar :
|
|
108
|
Mangone FR, Bobrovnitchaia IG, Salaorni S,
Manuli E and Nagai MA: PIK3CA exon 20 mutations are associated with
poor prognosis in breast cancer patients. Clinics (Sao Paulo).
67:1285–1290. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Arsenic R, Treue D, Lehmann A, Hummel M,
Dietel M, Denkert C and Budczies J: Comparison of targeted
next-generation sequencing and sanger sequencing for the detection
of PIK3CA mutations in breast cancer. BMC Clin Pathol. 15:202015.
View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Martínez-Sáez O, Chic N, Pascual T, Adamo
B, Vidal M, González-Farré B, Sanfeliu E, Schettini F, Conte B,
Brasó-Maristany F, et al: Frequency and spectrum of PIK3CA somatic
mutations in breast cancer. Breast Cancer Res. 22:452020.
View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Li X, Chen X, Wen L, Wang Y, Chen B, Xue
Y, Guo L and Liao N: Impact of TP53 mutations in breast cancer:
Clinicopathological features and prognosisImpact of TP53 mutations
in breast CA. Thorac Cancer. 11:1861–1868. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Behring M, Vazquez AI, Cui X, Irvin MR,
Ojesina AI, Agarwal S, Manne U and Shrestha S: Gain of function in
somatic TP53 mutations is associated with immune-rich breast tumors
and changes in tumor-associated macrophages. Mol Genet Genomic Med.
7:e10012019. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Perumal D, Imai N, Laganà A, Finnigan J,
Melnekoff D, Leshchenko VV, Solovyov A, Madduri D, Chari A, Cho HJ,
et al: Mutation-derived neoantigen-specific T-cell responses in
multiple myeloma. Clin Cancer Res. 26:450–464. 2020. View Article : Google Scholar
|
|
114
|
Walerych D, Napoli M, Collavin L and Del
Sal G: The rebel angel: Mutant p53 as the driving oncogene in
breast cancer. Carcinogenesis. 33:2007–2017. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Boyle DP, McArt DG, Irwin G,
Wilhelm-Benartzi CS, Lioe TF, Sebastian E, McQuaid S, Hamilton PW,
James JA, Mullan PB, et al: The prognostic significance of the
aberrant extremes of p53 immunophenotypes in breast cancer.
Histopathology. 65:340–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Weis KE, Ekena K, Thomas JA, Lazennec G
and Katzenellenbogen BS: Constitutively active human estrogen
receptors containing amino acid substitutions for tyrosine 537 in
the receptor protein. Mol Endocrinol. 10:1388–1398. 1996.PubMed/NCBI
|
|
117
|
Zhang QX, Borg A, Wolf DM, Oesterreich S
and Fuqua SA: An estrogen receptor mutant with strong
hormone-independent activity from a metastatic breast cancer.
Cancer Res. 57:1244–1249. 1997.PubMed/NCBI
|
|
118
|
De Mattos-Arruda L, Weigelt B, Cortes J,
Won HH, Ng CKY, Nuciforo P, Bidard FC, Aura C, Saura C, Peg V, et
al: Capturing intra-tumor genetic heterogeneity by de novo mutation
profiling of circulating cell-free tumor DNA: A proof-of-principle.
Ann Oncol. 25:1729–1735. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Toy W, Shen Y, Won H, Green B, Sakr RA,
Will M, Li Z, Gala K, Fanning S, King TA, et al: ESR1
ligand-binding domain mutations in hormone-resistant breast cancer.
Nat Genet. 45:1439–1445. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Carpten JD, Faber AL, Horn C, Donoho GP,
Briggs SL, Robbins CM, Hostetter G, Boguslawski S, Moses TY, Savage
S, et al: A transforming mutation in the pleckstrin homology domain
of AKT1 in cancer. Nature. 448:439–444. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Rudolph M, Anzeneder T, Schulz A, Beckmann
G, Byrne AT, Jeffers M, Pena C, Politz O, Köchert K, Vonk R and
Reischl J: AKT1 (E17K) mutation profiling in breast cancer:
Prevalence, concurrent oncogenic alterations, and blood-based
detection. BMC Cancer. 16:6222016. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Wu W, Chen Y, Huang L, Li W, Tao C and
Shen H: Effects of AKT1 E17K mutation hotspots on the biological
behavior of breast cancer cells. Int J Clin Exp Pathol. 13:332–346.
2020.PubMed/NCBI
|
|
123
|
Xie R, Yan Z, Jing J, Wang Y, Zhang J, Li
Y, Liu X, Yu X and Wu C: Functional defects of cancer-associated
MDC1 mutations in DNA damage repair. DNA Repair (Amst).
114:1033302022. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Ott PA, Hu Z, Keskin DB, Shukla SA, Sun J,
Bozym DJ, Zhang W, Luoma A, Giobbie-Hurder A, Peter L, et al: An
immunogenic personal neoantigen vaccine for patients with melanoma.
Nature. 547:217–221. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Tran E, Robbins PF, Lu YC, Prickett TD,
Gartner JJ, Jia L, Pasetto A, Zheng Z, Ray S, Groh EM, et al:
T-cell transfer therapy targeting mutant KRAS in cancer. N Engl J
Med. 375:2255–2262. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Barroso-Sousa R, Jain E, Cohen O, Kim D,
Buendia-Buendia J, Winer E, Lin N, Tolaney SM and Wagle N:
Prevalence and mutational determinants of high tumor mutation
burden in breast cancer. Ann Oncol. 31:387–394. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Zhu Y, Meng X, Ruan X, Lu X, Yan F and
Wang F: Characterization of neoantigen load subgroups in
gynecologic and breast cancers. Front Bioeng Biotechnol. 8:7022020.
View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Richters MM, Xia H, Campbell KM,
Gillanders WE, Griffith OL and Griffith M: Best practices for
bioinformatic characterization of neoantigens for clinical utility.
Genome Med. 11:562019. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Ren Y, Cherukuri Y, Wickland DP, Sarangi
V, Tian S, Carter JM, Mansfield AS, Block MS, Sherman ME, Knutson
KL, et al: HLA class-I and class-II restricted neoantigen loads
predict overall survival in breast cancer. Oncoimmunology.
9:17449472020. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Sahin U and Türeci Ö: Personalized
vaccines for cancer immunotherapy. Science. 359:1355–1360. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Parkhurst MR, Robbins PF, Tran E, Prickett
TD, Gartner JJ, Jia L, Ivey G, Li YF, El-Gamil M, Lalani A, et al:
Unique neoantigens arise from somatic mutations in patients with
gastrointestinal cancers. Cancer Discov. 9:1022–1035. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Sueangoen N, Grove H, Chuangchot N,
Prasopsiri J, Rungrotmongkol T, Sanachai K, Darai N, Thongchot S,
Suriyaphol P, Sa-Nguanraksa D, et al: Stimulating T cell responses
against patient-derived breast cancer cells with neoantigen
peptide-loaded peripheral blood mononuclear cells. Cancer Immunol
Immunother. 73:432024. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Jain KK: Personalized immuno-oncology. Med
Princ Pract. 30:1–16. 2021. View Article : Google Scholar :
|
|
134
|
Zhang X, Kim S, Hundal J, Herndon JM, Li
S, Petti AA, Soysal SD, Li L, McLellan MD, Hoog J, et al: Breast
cancer neoantigens can induce CD8+ T-cell responses and antitumor
immunity. Cancer Immunol Res. 5:516–523. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Xiuli Z, Goedegebuure SP, Myers NB,
Vickery T, McLellan MD, Gao F, Sturmoski MA, Chen MY, Kim SW, Chen
I, et al: Neoantigen DNA vaccines are safe, feasible, and capable
of inducing neoantigen-specific immune responses in patients with
triple negative breast cancer. medRxiv: 2021.2011.2019.21266466.
2021.
|
|
136
|
Disis MLN, Guthrie KA, Liu Y, Coveler AL,
Higgins DM, Childs JS, Dang Y and Salazar LG: Safety and outcomes
of a plasmid DNA vaccine encoding the ERBB2 intracellular domain in
patients with advanced-stage ERBB2-positive breast cancer: A phase
1 nonrandomized clinical trial. JAMA Oncol. 9:71–78. 2023.
View Article : Google Scholar
|
|
137
|
Morisaki T, Kubo M, Umebayashi M, Yew PY,
Yoshimura S, Park JH, Kiyotani K, Kai M, Yamada M, Oda Y, et al:
Neoantigens elicit T cell responses in breast cancer. Sci Rep.
11:135902021. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Chang MT, Asthana S, Gao SP, Lee BH,
Chapman JS, Kandoth C, Gao J, Socci ND, Solit DB, Olshen AB, et al:
Identifying recurrent mutations in cancer reveals widespread
lineage diversity and mutational specificity. Nat Biotechnol.
34:155–163. 2016. View Article : Google Scholar :
|
|
139
|
Neefjes J, Jongsma ML, Paul P and Bakke O:
Towards a systems understanding of MHC class I and MHC class II
antigen presentation. Nat Rev Immunol. 11:823–836. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Klebanoff CA and Wolchok JD: Shared cancer
neoantigens: Making private matters public. J Exp Med. 215:5–7.
2018. View Article : Google Scholar :
|
|
141
|
Malekzadeh P, Pasetto A, Robbins PF,
Parkhurst MR, Paria BC, Jia L, Gartner JJ, Hill V, Yu Z, Restifo
NP, et al: Neoantigen screening identifies broad TP53 mutant
immunogenicity in patients with epithelial cancers. J Clin Invest.
129:1109–1114. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Pecher G, Häring A, Kaiser L and Thiel E:
Mucin gene (MUC1) transfected dendritic cells as vaccine: results
of a phase I/II clinical trial. Cancer Immunol Immunother.
51:669–673. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Sharma A, Koldovsky U, Xu S, Mick R, Roses
R, Fitzpatrick E, Weinstein S, Nisenbaum H, Levine BL, Fox K, et
al: HER-2 pulsed dendritic cell vaccine can eliminate HER-2
expression and impact ductal carcinoma in situ. Cancer.
118:4354–4362. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Pettitt SJ, Frankum JR, Punta M, Lise S,
Alexander J, Chen Y, Yap TA, Haider S, Tutt ANJ and Lord CJ:
Clinical BRCA1/2 reversion analysis identifies hotspot mutations
and predicted neoantigens associated with therapy resistance.
Cancer Discov. 10:1475–1488. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Ruangapirom L, Sutivijit N, Teerapakpinyo
C, Mutirangura A and Doungkamchan C: Identification of shared
neoantigens in BRCA1-related breast cancer. Vaccines (Basel).
10:15972022. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Iiizumi S, Ohtake J, Murakami N, Kouro T,
Kawahara M, Isoda F, Hamana H, Kishi H, Nakamura N and Sasada T:
Identification of novel HLA class II-restricted neoantigens derived
from driver mutations. Cancers (Basel). 11:2662019. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Chandran SS, Ma J, Klatt MG, Dündar F,
Bandlamudi C, Razavi P, Wen HY, Weigelt B, Zumbo P, Fu SN, et al:
Immunogenicity and therapeutic targeting of a public neoantigen
derived from mutated PIK3CA. Nat Med. 28:946–957. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Lo W, Parkhurst M, Robbins PF, Tran E, Lu
YC, Jia L, Gartner JJ, Pasetto A, Deniger D, Malekzadeh P, et al:
Immunologic recognition of a shared p53 mutated neoantigen in a
patient with metastatic colorectal cancer. Cancer Immunol Res.
7:534–543. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Kim SP, Vale NR, Zacharakis N, Krishna S,
Yu Z, Gasmi B, Gartner JJ, Sindiri S, Malekzadeh P, Deniger DC, et
al: Adoptive cellular therapy with autologous tumor-infiltrating
lymphocytes and T-cell receptor-engineered T cells targeting common
p53 neoantigens in human solid tumors. Cancer Immunol Res.
10:932–946. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Zacharakis N, Huq LM, Seitter SJ, Kim SP,
Gartner JJ, Sindiri S, Hill VK, Li YF, Paria BC, Ray S, et al:
Breast cancers are immunogenic: Immunologic analyses and a phase II
pilot clinical trial using mutation-reactive autologous
lymphocytes. J Clin Oncol. 40:1741–1754. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
O'Connell MJ: Selection and the cell
cycle: Positive Darwinian selection in a well-known DNA damage
response pathway. J Mol Evol. 71:444–457. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Su YC, Lee WC, Wang CC, Yeh SA, Chen WH
and Chen PJ: Targeting PI3K/AKT/mTOR signaling pathway as a
radiosensitization in head and neck squamous cell carcinomas. Int J
Mol Sci. 23:157492022. View Article : Google Scholar : PubMed/NCBI
|