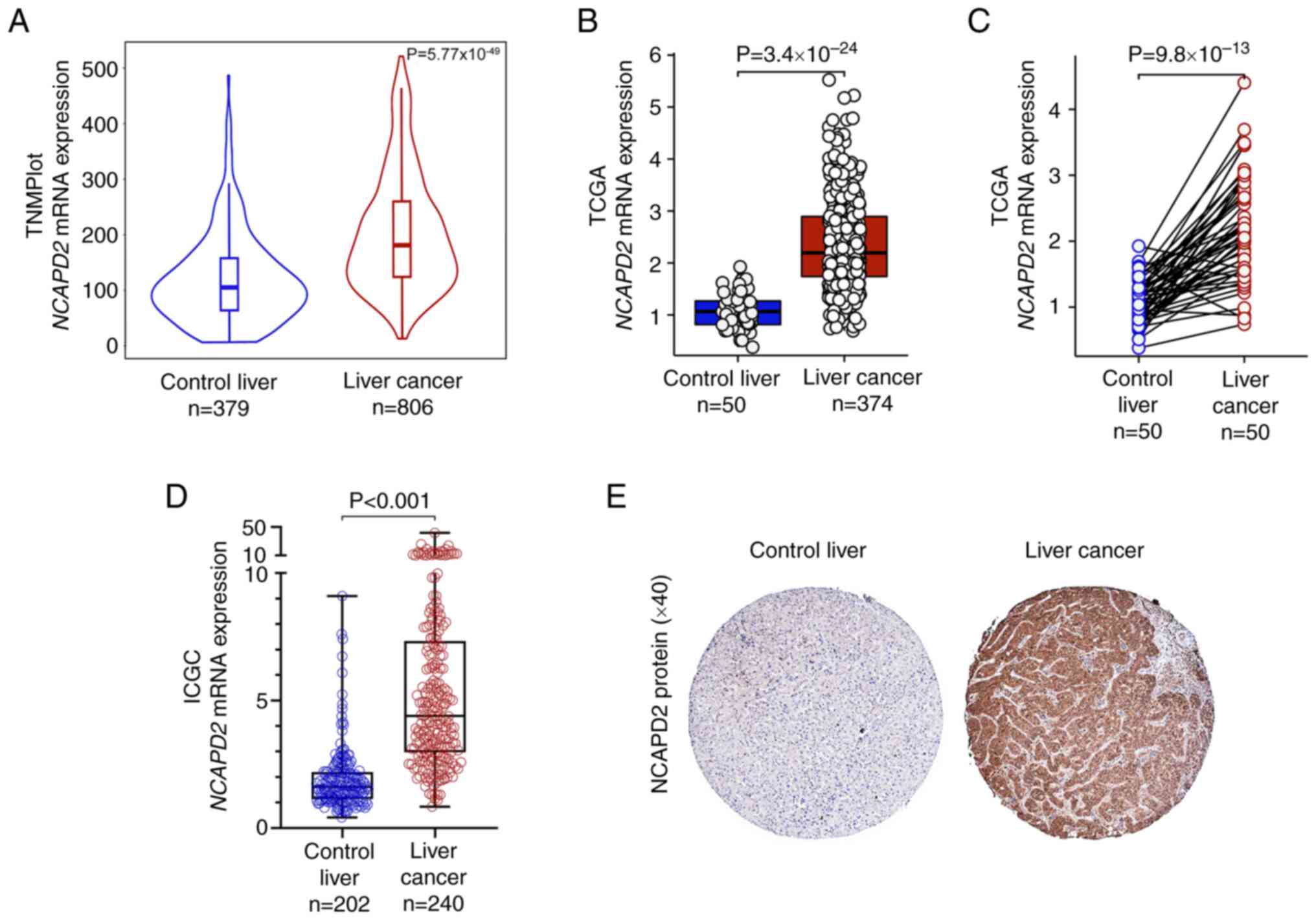
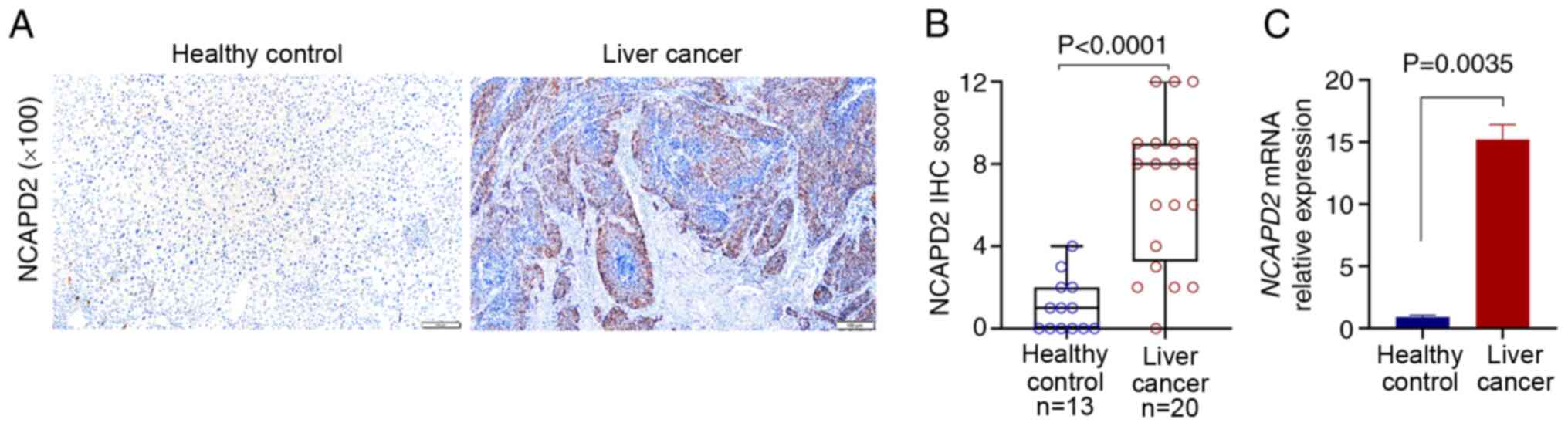
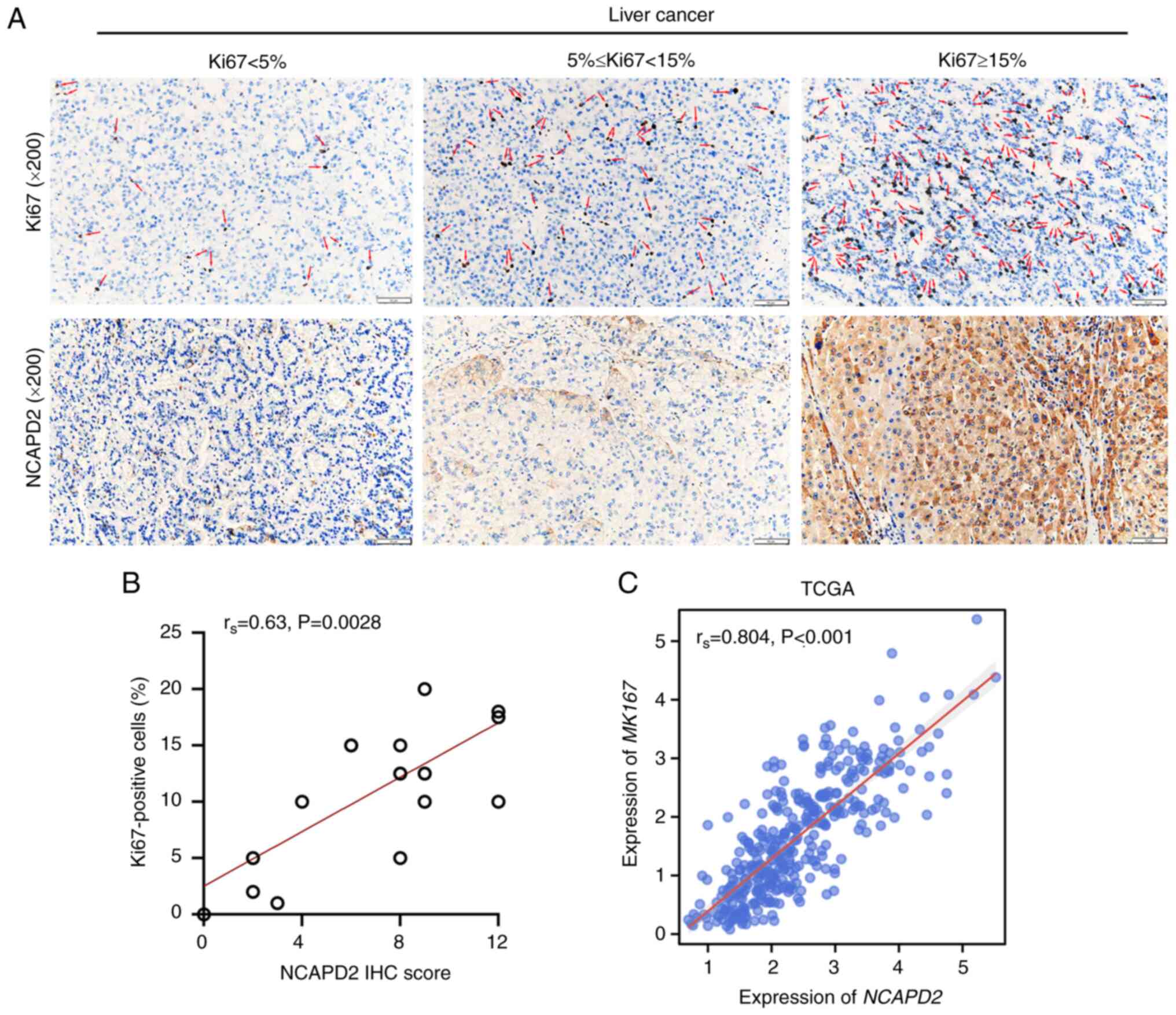
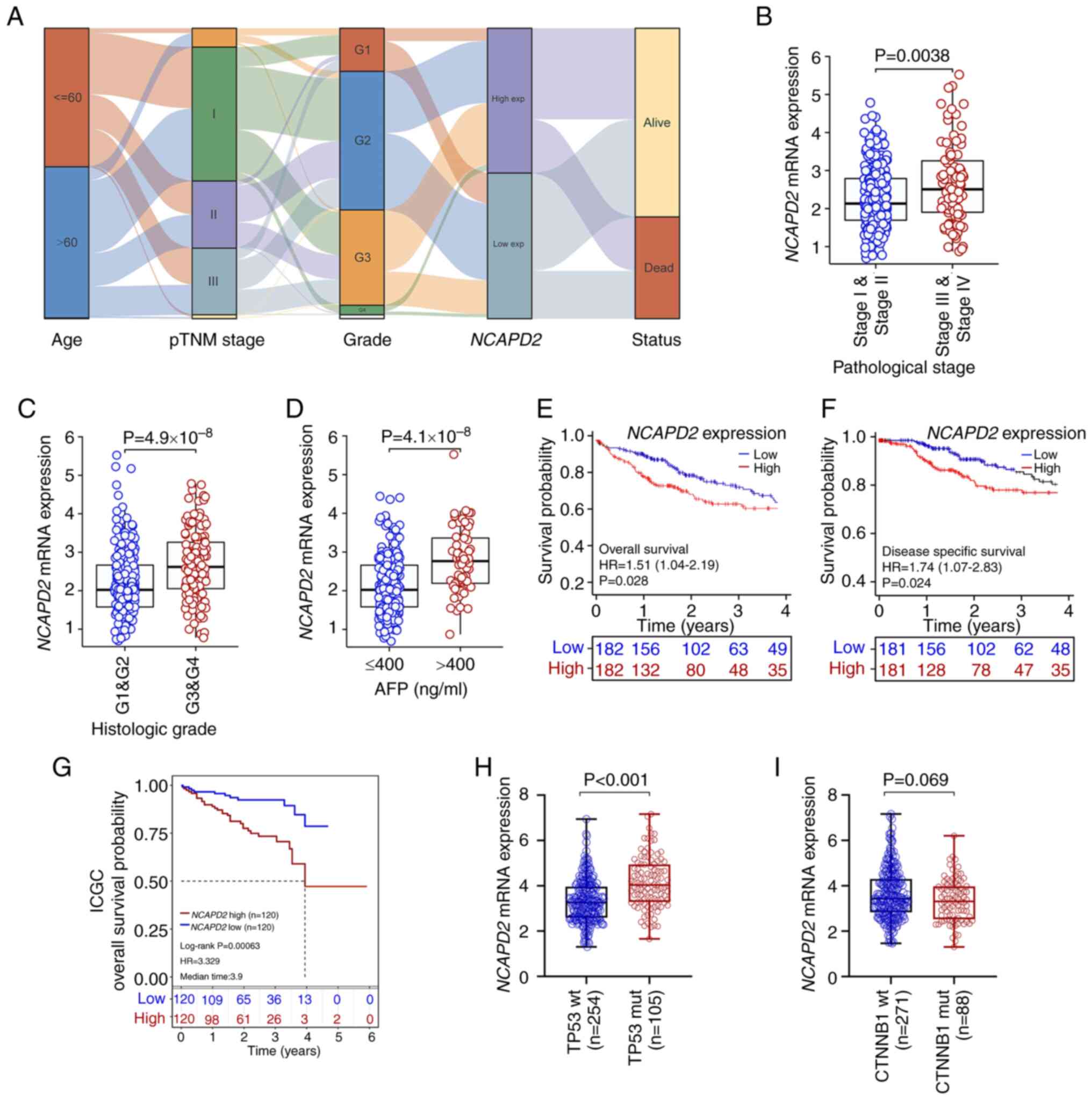
|
1
|
Forner A, Reig M and Bruix J:
Hepatocellular carcinoma. Lancet. 391:1301–1314. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ganesan P and Kulik LM: Hepatocellular
carcinoma: New developments. Clin Liver Dis. 27:85–102. 2023.
View Article : Google Scholar
|
|
3
|
Llovet JM, Kelley RK, Villanueva A, Singal
AG, Pikarsky E, Roayaie S, Lencioni R, Koike K, Zucman-Rossi J and
Finn RS: Hepatocellular carcinoma. Nat Rev Dis Primers. 7:62021.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Vogel A, Meyer T, Sapisochin G, Salem R
and Saborowski A: Hepatocellular carcinoma. Lancet. 400:1345–1362.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nagtegaal ID, Odze RD, Klimstra D, Paradis
V, Rugge M, Schirmacher P, Washington KM, Carneiro F and Cree IA;
WHO Classification of Tumours Editorial Board: The 2019 WHO
classification of tumours of the digestive system. Histopathology.
76:182–188. 2020. View Article : Google Scholar :
|
|
6
|
Zheng H, Pomyen Y, Hernandez MO, Li C,
Livak F, Tang W, Dang H, Greten TF, Davis JL, Zhao Y, et al:
Single-cell analysis reveals cancer stem cell heterogeneity in
hepatocellular carcinoma. Hepatology. 68:127–140. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Friemel J, Rechsteiner M, Frick L, Böhm F,
Struckmann K, Egger M, Moch H, Heikenwalder M and Weber A:
Intratumor heterogeneity in hepatocellular carcinoma. Clin Cancer
Res. 21:1951–1961. 2015. View Article : Google Scholar
|
|
8
|
Ball AR Jr, Schmiesing JA, Zhou C, Gregson
HC, Okada Y, Doi T and Yokomori K: Identification of a
chromosome-targeting domain in the human condensin subunit
CNAP1/hCAP-D2/Eg7. Mol Cell Biol. 22:5769–5781. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Savvidou E, Cobbe N, Steffensen S,
Cotterill S and Heck MM: Drosophila CAP-D2 is required for
condensin complex stability and resolution of sister chromatids. J
Cell Sci. 118:2529–2543. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Watrin E and Legagneux V: Contribution of
hCAP-D2, a non-SMC subunit of condensin I, to chromosome and
chromosomal protein dynamics during mitosis. Mol Cell Biol.
25:740–750. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jing Z, He X, Jia Z, Sa Y, Yang B and Liu
P: NCAPD2 inhibits autophagy by regulating
Ca(2+)/CAMKK2/AMPK/mTORC1 pathway and PARP-1/SIRT1 axis to promote
colorectal cancer. Cancer Lett. 520:26–37. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
He J, Gao R, Yang J, Li F, Fu Y, Cui J,
Liu X, Huang K, Guo Q, Zhou Z and Wei W: NCAPD2 promotes breast
cancer progression through E2F1 transcriptional regulation of CDK1.
Cancer Sci. 114:896–907. 2023. View Article : Google Scholar :
|
|
13
|
Zhang Y, Liu F, Zhang C, Ren M, Kuang M,
Xiao T, Di X, Feng L, Fu L and Cheng S: Non-SMC condensin I complex
subunit D2 is a prognostic factor in triple-negative breast cancer
for the ability to promote cell cycle and enhance invasion. Am J
Pathol. 190:37–47. 2020. View Article : Google Scholar
|
|
14
|
Dong X, Liu T, Li Z and Zhai Y: Non-SMC
condensin I complex subunit D2 (NCAPD2) reveals its prognostic and
immunologic features in human cancers. Aging (Albany NY).
15:7237–7257. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bartha Á and Győrffy B: TNMplot.com: A web
tool for the comparison of gene expression in normal, tumor and
metastatic tissues. Int J Mol Sci. 22:26222021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
17
|
Zeng D, Li M, Zhou R, Zhang J, Sun H, Shi
M, Bin J, Liao Y, Rao J and Liao W: Tumor microenvironment
characterization in gastric cancer identifies prognostic and
immunotherapeutically relevant gene signatures. Cancer Immunol Res.
7:737–750. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yoshihara K, Shahmoradgoli M, Martínez E,
Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW,
Levine DA, et al: Inferring tumour purity and stromal and immune
cell admixture from expression data. Nat Commun. 4:26122013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bindea G, Mlecnik B, Tosolini M,
Kirilovsky A, Waldner M, Obenauf AC, Angell H, Fredriksen T,
Lafontaine L, Berger A, et al: Spatiotemporal dynamics of
intratumoral immune cells reveal the immune landscape in human
cancer. Immunity. 39:782–795. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xiao Z, Dai Z and Locasale JW: Metabolic
landscape of the tumor microenvironment at single cell resolution.
Nat Commun. 10:37632019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wei J, Huang K, Chen Z, Hu M, Bai Y, Lin S
and Du H: Characterization of glycolysis-associated molecules in
the tumor microenvironment revealed by pan-cancer tissues and lung
cancer single cell data. Cancers (Basel). 12:17882020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Camps J, Noël F, Liechti R, Massenet-Regad
L, Rigade S, Götz L, Hoffmann C, Amblard E, Saichi M, Ibrahim MM,
et al: Meta-analysis of human cancer single-cell RNA-seq datasets
using the IMMUcan database. Cancer Res. 83:363–373. 2023.
View Article : Google Scholar :
|
|
23
|
Zhang Q, He Y, Luo N, Patel SJ, Han Y, Gao
R, Modak M, Carotta S, Haslinger C, Kind D, et al: Landscape and
dynamics of single immune cells in hepatocellular carcinoma. Cell.
179:829–845.e20. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Losic B, Craig AJ, Villacorta-Martin C,
Martins-Filho SN, Akers N, Chen X, Ahsen ME, von Felden J, Labgaa
I, D-Avola D, et al: Intratumoral heterogeneity and clonal
evolution in liver cancer. Nat Commun. 11:2912020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Verloh N, Probst U, Utpatel K, Zeman F,
Brennfleck F, Werner JM, Fellner C, Stroszczynski C, Evert M,
Wiggermann P and Haimerl M: Influence of hepatic fibrosis and
inflammation: Correlation between histopathological changes and
Gd-EOB-DTPA-enhanced MR imaging. PLoS One. 14:e02157522019.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Seipold S, Priller FC, Goldsmith P, Harris
WA, Baier H and Abdelilah-Seyfried S: Non-SMC condensin I complex
proteins control chromosome segregation and survival of
proliferating cells in the zebrafish neural retina. BMC Dev Biol.
9:402009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang P, Liu L, Huang J, Shao L, Wang H,
Xiong N and Wang T: Non-SMC condensin I complex, subunit D2 gene
polymorphisms are associated with Parkinson's disease: A Han
Chinese study. Genome. 57:253–257. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lin Y, Zeng C, Lu Z, Lin R and Liu L: A
novel homozygous splice-site variant of NCAPD2 gene identified in
two siblings with primary microcephaly: The second case report.
Clin Genet. 96:98–101. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yuan CW, Sun XL, Qiao LC, Xu HX, Zhu P,
Chen HJ and Yang BL: Non-SMC condensin I complex subunit D2 and
non-SMC condensin II complex subunit D3 induces inflammation via
the IKK/NF-κB pathway in ulcerative colitis. World J Gastroenterol.
25:6813–6822. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li Z, Zheng Y, Wu Z, Zhuo T, Zhu Y, Dai L,
Wang Y and Chen M: NCAPD2 is a novel marker for the poor prognosis
of lung adenocarcinoma and is associated with immune infiltration
and tumor mutational burden. Medicine (Baltimore). 102:e326862023.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhao Q, Zhang Y, Shao S, Sun Y and Lin Z:
Identification of hub genes and biological pathways in
hepatocellular carcinoma by integrated bioinformatics analysis.
PeerJ. 9:e105942021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Thompson EB: The many roles of c-Myc in
apoptosis. Annu Rev Physiol. 60:575–600. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Meškytė EM, Keskas S and Ciribilli Y: MYC
as a multifaceted regulator of tumor microenvironment leading to
metastasis. Int J Mol Sci. 21:77102020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Dang CV: MYC on the path to cancer. Cell.
149:22–35. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Dhanasekaran R, Deutzmann A,
Mahauad-Fernandez WD, Hansen AS, Gouw AM and Felsher DW: The MYC
oncogene-the grand orchestrator of cancer growth and immune
evasion. Nat Rev Clin Oncol. 19:23–36. 2022. View Article : Google Scholar
|
|
36
|
Gao FY, Li XT, Xu K, Wang RT and Guan XX:
c-MYC mediates the crosstalk between breast cancer cells and tumor
microenvironment. Cell Commun Signal. 21:282023. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Janku F, Yap TA and Meric-Bernstam F:
Targeting the PI3K pathway in cancer: Are we making headway? Nat
Rev Clin Oncol. 15:273–291. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Alzahrani AS: PI3K/Akt/mTOR inhibitors in
cancer: At the bench and bedside. Semin Cancer Biol. 59:125–132.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Peng Y, Wang Y, Zhou C, Mei W and Zeng C:
PI3K/Akt/mTOR pathway and its role in cancer therapeutics: Are we
making headway? Front Oncol. 12:8191282022. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Guo C, He J, Song X, Tan L, Wang M, Jiang
P, Li Y, Cao Z and Peng C: Pharmacological properties and
derivatives of shikonin-A review in recent years. Pharmacol Res.
149:1044632019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Rebello RJ, Pearson RB, Hannan RD and
Furic L: Therapeutic approaches targeting MYC-driven prostate
cancer. Genes (Basel). 8:712017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Nussinov R, Tsai CJ, Jang H, Korcsmáros T
and Csermely P: Oncogenic KRAS signaling and YAP1/β-catenin:
Similar cell cycle control in tumor initiation. Semin Cell Dev
Biol. 58:79–85. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Beck M, Covino R, Hänelt I and
Müller-McNicoll M: Understanding the cell: Future views of
structural biology. Cell. 187:545–562. 2024. View Article : Google Scholar : PubMed/NCBI
|