Introduction
Lactate, traditionally regarded solely as a
by-product of anaerobic glycolysis and long deemed insignificant in
cellular functions, has undergone a reassessment in light of the
discovery of metabolic reprogramming in the 1920s (1). Metabolic reprogramming is defined
as the adjustment of metabolic pathways to support cellular
functions such as proliferation, differentiation, survival and
stress responses, particularly the transition to aerobic glycolysis
(2). Aerobic glycolysis and the
Warburg effect are essentially the same concept, referring to the
phenomenon where cells primarily use glycolysis to generate energy
even when oxygen is readily available, instead of the more
efficient oxidative phosphorylation pathway. This often occurs
under stress or pathological conditions (3). This shift from mitochondrial
respiration to glycolysis, along with the subsequent production of
lactate in oxygen-sufficient environments, is not only important in
understanding cancer development but also plays a crucial role in
the mechanisms behind inflammatory diseases, indicating a shared
metabolic pathway between the two conditions (3,4).
In the past three decades, numerous studies have revealed a diverse
array of novel and previously undocumented biological functions of
lactate. It is now acknowledged not only as an essential carbon
source for metabolic reprogramming and cellular metabolism but also
as a significant signaling involved in gene expression, immune
activation, energy supply and metabolic regulation (5,6).
In a significant advancement in 2019, Zhang et al (7) uncovered that lactate serves as an
essential substrate in a new form of epigenetic modification known
as histone lysine lactylation. This finding was established through
mass spectrometry analyses of core histones in human and mouse
cells (7). Lactylation is a
ubiquitous and reversible post-translational modification (PTM)
characterized by the addition of a lactyl group to a lysine residue
in the histone or non-histone tail. Lactylation was first
discovered on histones within the nucleosome and this modification
is now understood to significantly impact gene expression by
influencing the initiation of transcription and consequently
affecting important cellular functions. Furthermore, lactylation
has also been observed to occur in non-histone proteins, directly
impacting their functions and corresponding protein functions. In
the last five years, research from various studies has provided
greater insight into the molecular mechanisms underlying histone
and non-histone lactylation, highlighting their significant roles
in a diverse array of diseases, such as cancers, inflammatory
conditions, cardiovascular disorders and certain pulmonary ailments
(8-11). The primary function of the lungs
is to facilitate gas exchange (respiration), however, they also
serve as a significant immune organ. Furthermore, these intricate
metabolic processes occurring in the lungs can be susceptible to
various diseases (12).
Metabolic reprogramming has been identified as a significant factor
in pulmonary diseases, as it influences cellular energy mechanisms
and biosynthetic requirements. This modulation subsequently affects
tumor proliferation in lung cancers and alters immune responses in
inflammatory disorders, including asthma and lung fibrosis
(13,14). The discovery of lysine
lactylation opens new avenues for understanding the interaction of
metabolic reprogramming and epigenetic modification in the context
of pulmonary diseases. In lung cancer, for example, cancer cells
prioritize glycolysis as their primary energy source even when
oxygen is available, leading to the production and accumulation of
lactate within the tumor microenvironment (TME) (15). Elevated lactate subsequently
promotes lysine lactylation, which in turn activates or represses
oncogenes or tumor suppressor genes, contributing to tumorigenesis
and cancer progression (16).
However, a comprehensive review that consolidates and summarizes
its effects on neoplastic and inflammatory pulmonary diseases
remains notably lacking.
In this review, the latest significant advances
concerning the roles of both histone and non-histone lactylation in
neoplastic and inflammatory pulmonary diseases were examined, with
a primary focus on non-small cell lung cancers (NSCLC), malignant
pleural effusion, pulmonary fibrosis, acute lung injury and asthma.
These explorations aim to deepen the understanding of lung
physiology by introducing a novel perspective that could
potentially revolutionize therapeutic approaches through the
targeted modulation of specific lactylation sites. Furthermore, the
study advocates more extensive and detailed future research to
fully elucidate the involvement of PTMs, particularly lactylation,
in the pathogenesis of various pulmonary diseases.
Discovery of lactylation modification
In 2019, a landmark study published in the Nature
Journal was the first to identify 28 lactylation sites on core
histones in mouse bone marrow-derived macrophages and human HeLa
cells. This groundbreaking study revealed that lactate-derived
lactylation of histone lysine residues serves as an epigenetic
modification that directly enhances gene transcription from
chromatin (7). In 2020,
scientific research began to concentrate on lactylation in
non-histone proteins, highlighted by the pioneering study conducted
by Gao et al (17), which
identified the presence of lactylation in the necrotic fungal
pathogen Botrytis cinerea. Their extensive global lysine lactylome
analysis of 166 proteins revealed 273 lactylation sites (17). This period also saw the
identification of lactylation sites in plants and protists, further
expanding the known range of lactylation's biological significance
(18,19). Subsequently, research into
lactylation expanded to encompass various major systemic diseases.
In 2021, significant research findings revealed that patients with
pulmonary fibrosis demonstrated markedly elevated lactate levels in
the lungs. This elevation facilitated lactylation through the
action of acetyltransferase p300 on the promoters of profibrotic
genes, consequently inducing a pro-fibrotic macrophage phenotype.
This discovery marked the first identification of the involvement
of lactylation in pulmonary diseases (11). Furthermore, Yu et al
(8) identified an association
between elevated histone lactylation and poor prognosis in patients
with ocular melanoma. Additionally, scientists elucidated the role
of histone lactylation in uterine remodeling through a proteomic
map of ligand-receptor interactions at the ovine maternal-fetal
interface (20). Furthermore,
Pan et al (21) first
detected that histone lactylation is increased in the brain tissues
of patients with Alzheimer's disease, proposing that the positive
feedback loop glycolysis/lactylation further exacerbated microglial
functional impairment. In 2022, a subsequent exploration indicated
that, in the early stages of post-myocardial infarction, elevated
levels of H3 lysine 18 lactylation (H3K18la) in
monocytes-macrophages promoted the transcription of reparative
genes, suggesting a beneficial role for histone lactylation in
cardiac function improvement following myocardial infarction
(10). A more recent
comprehensive global analysis conducted in 2023 of lactylation in
the human lung under normal physiological conditions identified 724
lactylation sites in 451 proteins, with 141 proteins newly
identified as undergoing lactylation (22). In summary, the discovery of
lactylation modification as an epigenetic modification in 2019 has
significantly expanded the understanding of metabolic and
epigenetic crosstalk in cellular regulation. Lactylation,
originally discovered in histones, was first recognized for its
role in modulating gene transcription. Subsequent studies have
demonstrated its occurrence in non-histone proteins and across a
range of species, thereby expanding its biological significance.
The connection of lactylation to systemic conditions like pulmonary
fibrosis, neoplastic disease, myocardial infarction and Alzheimer's
disease highlights its potential impact on health and disease.
Furthermore, the identification of multiple lactylation sites
across various proteins in the lung represents a vital area of
study with profound implications for future research and
medicine.
Histone lactylation and gene regulation
Histone modifications are pivotal in the field of
epigenetics, serving as crucial regulators of chromatin structure
and gene expression and thereby influencing a vast array of
cellular processes (23). Among
various histone modifications, lactylation is a relatively novel
PTM that has garnered significant attention from researchers.
Lactylation involves the addition of lactyl groups to lysine
residues on histone proteins, forming ε-N-lactyl lysine (24). Lactylation influences the
interaction between histones and DNA, thereby impacting the
compaction of chromatin. This alteration regulates the
accessibility of transcription factors and various epigenetic
regulators to genetic material. Consequently, this indicates that
histone lactylation plays a crucial role in the regulation of gene
activation and repression, as well as in shaping cell fate and
responding to environmental changes (Fig. 1).
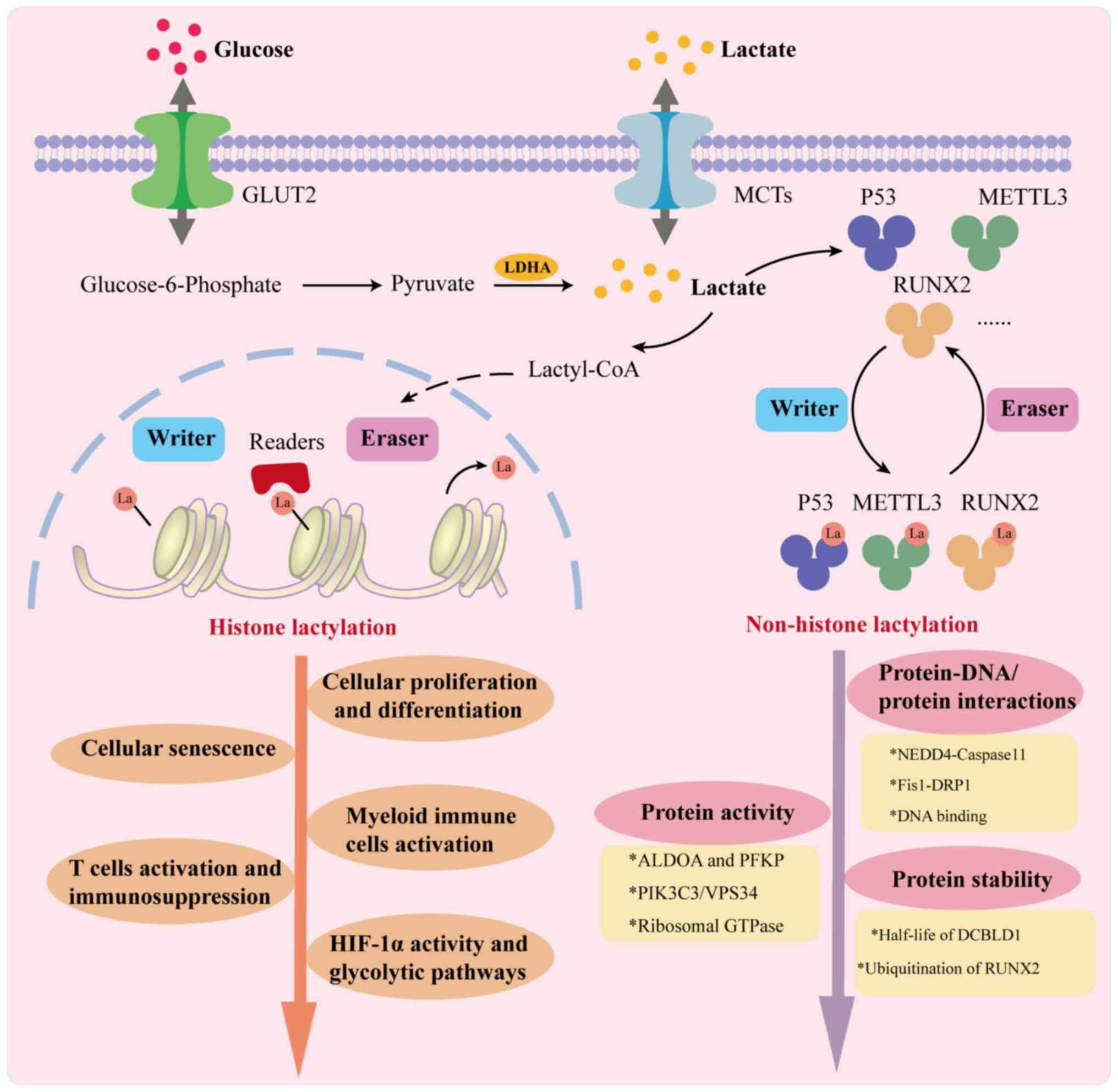 | Figure 1Simplified diagram of histone and
non-histone lactylation. Cells produce lactate through glycolysis
or obtain it from the extracellular environment, with lactate
serving as a substrate for lactylation. Elevated levels of histone
lactylation, particularly at gene promoters, are linked to the
activation of transcription, which plays a crucial role in
regulating cell fate. This includes processes such as cellular
proliferation, differentiation and senescence, as well as the
functions of HIF-1α, glycolysis, tumor immunity and inflammation.
Meanwhile, lactate-induced lysine lactylation of non-histone
proteins such as P53, METTL3 and RUNX2, under the influence of
writers and erasers, can affect protein-DNA/protein interactions,
protein activity and protein stability, ultimately influencing the
pathogenesis and progression of various diseases. ALDOA, aldolase
A; DRP1, dynamin-related protein 1; DCBLD1, discoidin, CUB and LCCL
domain-containing type I; Fis1, fission 1; GTPase, GTP hydrolase;
GLUT1, glucose transporter type 1; HIF-1α, hypoxia-inducible
factor-1α; LDHA, lactate dehydrogenase A; METTL3, methyltransferase
3; MCTs, monocarboxylate Transporters; NEDD4, neuronally expressed
developmentally downregulated 4; NUSAP1, nucleolar and spindle
associated protein 1; PFKP, phosphofructokinase, platelet; PIK3C3,
phosphatidylinositol 3-kinase catalytic subunit type 3; RUNX2,
runt-related transcription factor 2; VPS34, vacuolar protein
sorting 34. |
Cellular proliferation and
differentiation
Recent research has demonstrated that the
lactylation of histones in key genes participates in the
proliferation of tumor cells as well as tumor-associated
macrophages, thereby impacting the progression of various
neoplastic diseases (25,26).
For instance, Li et al (27) discovered that H3K18la was fueled
by tumor-derived lactate in the retinoic acid receptor-γ promoter,
which endowed macrophages with a protumor phenotype and further
activated TRAF6-IL-6-STAT3 signaling in colorectal cancer cells.
Similarly, H3K18la enrichment regulated by lactyltransferases p300
in the promoter of YTH N6-methyladenosine RNA-binding protein 2 in
ocular melanoma cells, as well as c-Myc in breast cancer cells,
facilitated the increase in their transcription and translation
levels. This promoted the proliferation of cancer cells and
contributed to the tumorigenesis of both ocular melanoma and breast
cancer (8,28). In addition to H3K18la, genes with
promoters marked by H4K12la have been found to be enriched in
pathways related to cancer cell proliferation, including connective
tissue growth factor and cyclin E1 (CCNE1) in anaplastic thyroid
cancer cells (29). Histone
lactylation also plays an important role in cell differentiation
and self-renewal. H3K18la has been shown to be enriched on the
proximal promoter of JunB, where it can activate its expression and
subsequently facilitate osteoblast differentiation (30). Previous research indicated that
neuraminidase 2 (Neu2) can induce myoblast differentiation
(31). Dai et al
(32) further established the
prevalence of H3K9la in the promoter region of Neu2 during the
process of cell differentiation which were modulated by p300.
During the Warburg effect in glioblastoma cells, histone
lactylation is increased at the H3K18la site, which in turn leads
to the enhanced transcription of LINC01127; this long non-coding
RNA is linked to the NF-κB signaling pathway and ultimately
promotes the self-renewal capacity of the glioblastoma cells
(33,34). In conclusion, the emerging
evidence highlights the critical role of histone lactylation,
particularly on H3K18la and H4K12la, in regulating both cellular
proliferation and differentiation, underscoring its significant
impact on tumorigenesis and cell fate determination.
Cellular senescence
Cellular senescence is a complex and multifactorial
process marked by an irreversible cessation of cell division,
accompanied by changes in gene expression, chromatin organization
and the secretion of pro-inflammatory factors, collectively known
as the senescence-associated secretory phenotype (SASP) (35,36). Enhanced expression of H3K18la was
reported to result from elevated lactate levels in senescent
microglia, which in turn upregulated SASP components IL-6 and IL-8.
This upregulation occurred through the stimulation of the NF-κB
pathway through increasing binding to the promoter of
Rela(P65) and NF-κB1(P60). These
changes not only accelerated the aging of the brain and the
progression of Alzheimer's disease pathology but also highlighted
the remarkable adaptability of senescent cells to fluctuating
metabolic states. Acetyltransferase p300/CBP and PCAF also worked
as histone lacyltransferases in cells, upregulating H3K18la in
senescent microglia and hippocampus tissues (37). Additionally, senescent cells,
characterized by increased glycolysis and reduced mitochondrial
function, could enhance histone lactylation through metabolic
reprogramming. This process can further strengthen the senescence
program and SASP (38). This
metabolic-epigenetic coupling underscores the adaptability of
senescent cells to their metabolic state. For instance, accumulated
lactate inhibited histone lysine delactylase HDAC3 and promoted
H4K12la. Consequently, enriched H4K12la was assembled in the SASP
promoter, activating SASP transcription, which exacerbated vascular
smooth muscle cell senescence and atherosclerosis, further
demonstrating the far-reaching implications of histone lactylation
beyond individual cellular states (39). As elucidated, the intricate
interplay between histone lactylation and cellular senescence
offers profound insights into the mechanisms driving age-related
pathologies. Furthermore, the irreversible arrest in cell division
that defines senescence is intricately linked to notable changes in
gene expression and chromatin dynamics, notably exemplified through
metabolic reprogramming-mediated histone lactylation
modifications.
Myeloid immune cell activation
Myeloid cells, including macrophages and
neutrophils, serve as the first line of defense against pathogens
and are essential in maintaining homeostasis. Studies have
demonstrated that histone lactylation represents a critical
epigenetic mechanism in myeloid immune cell activation and function
(40,41). For instance, p300-mediated
histone lactylation binding to interleukin (IL)-10 promotor
in monocyte-derived macrophages, promoted IL-10 expression and
enhanced their immunosuppressive activity in glioblastoma (42). This complex interplay between
metabolism and immune response suggests a nuanced role of histone
lactylation in shaping the immune landscape, likely influencing
inflammation and cancer pathogenesis. Lactate-induced H4K12
lactylation and subsequent hypoxia-inducible factor-1α
(Hif-1α) transcription in macrophages further aggravate
metabolic dysregulation and inflammatory infiltration within the
microenvironment in type 2 diabetes (43). Methylsulfonyl-methane increases
lactate levels in peritoneal lavage fluids and subsequently results
in the elevation of histone H3K18la levels. This elevation directly
facilitates the transcription of downstream target genes, including
Arg1, thereby promoting the M2 polarization of peritoneal
macrophages during infection and sepsis (44). Furthermore, elevated H3K18la
levels also upregulate C-X-C motif cytokine ligand 1 (CXCL1) and
CXCL5, promoting neutrophil recruitment, which is crucial for
clearing the infection (45,46). Overall, histone lactylation
functions as a multifaceted regulatory mechanism, influencing
innate immune responses and the activities of myeloid immune cells
across various pathological scenarios, which may have significant
implications for therapeutic interventions in inflammatory
diseases.
T-cell activation and
immunosuppression
Histone lactylation has also emerged as a
significant epigenetic modification influencing T-cell activation
and immunosuppression, critical processes in the TME that affect
tumor progression and immune surveillance (47). Notably, H4K5la enrichment at the
programmed cell death-ligand promoter in leukemic cells attenuates
CD8+ T-cell activation, mediating immunosuppression in
acute myeloid leukemia (48).
Similarly, H3K18la activated CD39, CD73 and CCR8 promoters in
glioma stem cell-co-cultured CD4+ T cells and
macrophages, as well as vascular cellular adhesion molecule 1 in
gastric cancer cells, thereby nurturing an immunosuppressive TME
(49,50). Furthermore, histone lactylation
may equivalently influence regulatory T cells (Tregs), which are
critical mediators of immunosuppression. Improved lactylation may
facilitate the expression of genes that stabilize and increase the
populations of Tregs, thereby strengthening the immunosuppressive
environment. This environment not only inhibits the activity of
effector T cells but also aids in tumor survival by suppressing
anti-tumor immunity (42,51).
These findings underscore the importance of histone lactylation as
a potential target for therapeutic interventions to counteract
tumor-induced immunosuppression, enhance T cell activation and
improve the efficacy of cancer immunotherapies. Further,
understanding the precise mechanisms of histone lactylation in the
TME may pave the way for novel strategies designed to restore
immune surveillance against tumors.
Hif-1α activity and glycolytic
pathways
HIF-1α acts as a central transcription factor that
regulates the metabolic reprogramming process, primarily by
significantly enhancing glycolysis when cells experience low oxygen
conditions (hypoxia), effectively switching the cell's energy
production towards a more efficient anaerobic pathway (52,53). In microglia and uterine stromal
cells, for instance, lactate-dependent H4K12la enrichment at the
HIF-1α promoter upregulates HIF-1α transcription, thereby
forming a feedback loop that enhances glycolysis. This underscores
the influence of histone lactylation in fine-tuning the expression
levels of HIF-1α, which in turn sustains the glycolytic pathway
vital for energy production under hypoxia (21,54). Similarly, the enrichment of
H3K18la at the promoter of ubiquitin-specific peptidase 39 enhanced
its expression, subsequently activating the PI3K/AKT/HIF-1α
signaling pathway. This elucidates the molecular interconnections
in which histone lactylation, metabolic enzymes such as
phosphoglycerate kinase 1, and key signaling pathways converge to
promote glycolytic flux (55).
Furthermore, H3K18la promotes platelet-derived growth factor
receptor β transcription, which leads to an increase of glycolysis,
also forming a glycolytic positive feedback loop in clear cell
renal cell carcinoma (56). Chen
et al (57) also explored
the vital role of histone lactylation in hypoxic pulmonary
hypertension (PH). They revealed that hypoxia environments
specifically promote pulmonary artery smooth muscle cell (PASMC)
proliferation in a mitochondrial reactive oxygen species
(mtROS)-dependent manner, the chemically reactive molecules
containing oxygen and free radicals produced within the
mitochondria. The mtROS-HIF-1α axis triggers the PASMC glycolytic
switch through the HIF-1α/pyruvate dehydrogenase (PDH) kinase 1
(PDK1)&PDK2/p-PDH-E1α axis. This process promotes lactate
secretion and subsequently induces H3K18la and H4K5la of HIF-1α
targets, leading to PASMC proliferation, vascular remodeling and
the occurrence of hypoxic PH (57). Collectively, the crosstalk
between histone lactylation and glycolytic pathways denotes a
feedback mechanism and a vital axis in metabolic adaptation.
Histone lactylation influences gene expression with precision by
modulating HIF-1α activity and enhancing glycolysis, thereby
directly affecting cellular behavior and phenotype in pathological
conditions marked by altered metabolic states, including cancer and
hypoxic diseases. A comprehensive summary of research on histone
lactylation and gene regulation studies is provided in Table I.
 | Table IHistone lactylation and cellular
functions. |
Table I
Histone lactylation and cellular
functions.
| Functions of
histone lactylation | Target | Promoters |
Writers/erasers | Cell type | Specific
functions | (Refs.) |
|---|
| Cellular
proliferation and differentiation | H3K18 | YTHDF2 | p300/CBP | Melanoma cell | Accelerated
tumorigenesis of ocular melanoma | (8) |
| H3K18 | RARγ | / | Macrophage | Endowed macrophages
with tumor-promoting functions | (27) |
| H3K18 | c-Myc | p300/CBP | Breast cancer
cell | Promoted the
proliferation of breast cancer cells | (28) |
| H3K9 | Neu2 | p300/CBP | Myoblast | Affected myoblast
differentiation | (32) |
| H3K18 | LINC01127 | / | Glioma stem
cell | Promoted
self-renewal in glioma stem cells | (33) |
| Cellular
senescence | H3K18 | Rela (P65) and
NF-κB1(P60) | p300/CBP and
PCAF | Microglia | Upregulated SASP
components IL-6 and IL-8, regulates brain aging and Alzheimer's
disease. | (37) |
| H4K12 | SASP | HDAC3 | VSMC | Activated SASP
transcription, exacerbated VSMC senescence and
atherosclerosis. | (39) |
| Myeloid immune cell
activation | Histone | IL-10 | p300/CBP | MDM | Enhanced MDM
immunosuppressive activity in glioblastoma | (42) |
| H3K18 | Arg1 | / | Macrophage | Promoted
anti-inflammatory differentiation of macrophages during infection
and sepsis | (44) |
| H3K18 | CXCL1, CXCL5 | / | Colorectal cancer
cell | Promoted neutrophil
recruitment and modulated the tumor metabolism | (46) |
| T cell activation
and immunosuppression | H4K5 | PD-L1 | / | Leukemic cell | Attenuated CD8+
T-cell activation and mediated immunosuppression | (48) |
| H3K18 | CD39, CD73,
CCR8 | / | Glioma stem
cell | Induced
immunosuppressive TME formation | (49) |
| HIF-1α activity and
glycolytic pathways | H4K12 | Hif-1α, Pkm,
Ldha | / | Microglia | Increased
glycolytic activity and exacerbated microglial dysfunction | (21) |
| H4K12 | Hif-1α | / | Uterine stromal
cell | Formed a
H4K12la-HIF-1α-glycolysis feedback loop to promote
decidualization | (54) |
| H3K18 | USP39 | / | Endometrial
carcinoma cell | Activated the
PI3K/AKT/HIF-1α signaling pathway to stimulate glycolysis | (55) |
| H3K18 | PDGFRβ | / | Clear cell renal
cell | Formed a
H3K18la-PDGFRβ-glycolysis feedback loop and promoted the growth and
metastasis of tumor | (56) |
Non-histone lactylation and protein
modification
Lactylation has also been shown to alter the
functions of non-histone proteins by regulating their activity,
interactions and stability, subsequently influencing cellular
processes, particularly in inflammation and cancer (Fig. 1). Central to these regulatory
mechanisms is the alteration of enzymatic activities through
specific lysine lactylation sites. In cancer metabolism, for
instance, lactylation of glycolytic enzymes such as aldolase A at
K147 and phosphofructokinase at K688 decreases their enzyme
activities. This highlights a pivotal shift in metabolic
reprogramming in colon cancer cells. The decreased activity of
these metabolic enzymes due to lysine lactylation is suggestive of
an adaptive mechanism within the TME that promotes cellular
proliferation and cancer progression. This is not merely an
incidental modification but hints at a broader strategy where
tumors exploit metabolic plasticity conferred by lactylation to
sustain growth and evade regulatory checkpoints (58,59). Furthermore, the lactylation at
K45 leads to the inhibition of glucose-6-phosphate dehydrogenase,
which impairs its capacity to form functional dimers and
consequently disrupts its function in antioxidant defense. This
inhibition is crucial during oxidative stress as it hinders the
cell's capacity to counteract the redox imbalance, potentially
influencing the fate of cells under duress. Likewise, pyruvate
kinase isozyme M2 (PKM2) modulation in macrophages through
lactylation illustrates lactate's role in immune-cell metabolism
and function. The inhibition of tetramer-to-dimer transitions of
PKM2 through K62 lactylation enhances its pyruvate kinase activity
and reorients macrophages towards a reparative phenotype. This
shift not only impacts local inflammatory responses but also
mirrors wider systemic changes that occur during infection and
tumor evolution, highlighting a potential therapeutic angle to
modulate macrophage function (60,61). In the realm of endolysosomal
trafficking, lactate-induced vacuolar protein sorting 34 (VPS34)
K356 and K781 lactylation promoted the interaction of PIK3C3
complex I and II subunits to enhance VPS34 kinase activity and
facilitate sophisticated autophagy. This axis fostered cancer cell
endurance through increased autophagic flux that affected
intracellular signaling and substrate degradation.
Lactylation-induced autophagic modulation appears to be an
attractive target for interventions aiming to disrupt cancer cell
homeostasis and promote cellular death pathways (62). Furthermore, the K408 lactylation
of elongation factor 1α, which intensifies ribosomal GTPase
activity and influences protein synthesis, provides substantial
insight into how cellular machinery is co-opted for oncogenic
purposes. The involvement of lactyltransferases like KAT8 in
modulating this effect suggests a complex interplay between
enzymatic regulation and protein translation that could be
harnessed for therapeutic innovation in tumorigenesis (63). In acute promyelocytic leukemia, a
variety of non-histone protein targets susceptible to lactylation
is revealed during lactylation of methyltransferase 3. The complex
regulation of methyltransferase activity by lactylation highlights
a distinctive regulatory role that may offer insights into cellular
dysregulation in leukemia and possible strategies for therapeutic
intervention. Additionally, the lactylation of methyltransferase 3
significantly influences its enzymatic activity in acute
promyelocytic leukemia cells, emphasizing a unique regulatory role
that could shed light on cellular dysregulation in leukemia and
potential therapeutic pathways (64).
Further, non-histone lactylation also emerges as a
critical modifier of protein interactions and stability,
influencing a variety of cellular processes and pathways with
profound implications for health and disease. The addition of a
lactyl group alters the surface charge of proteins, thereby
modulating their interactions with other partner(s) and stability.
This modification can significantly impact cellular functionality
by either enhancing or inhibiting protein interactions,
consequently affecting pathways ranging from inflammation to tumor
metabolism regulation and transcriptional dynamics (65,66). For instance, lactate-induced
neuronally expressed developmentally downregulated 4 lactylation at
K33 in macrophages hinders its interaction with caspase-11, which
in turn prevents the subsequent ubiquitination of caspase-11,
ultimately promoting macrophage pyroptosis. This process
underscores a crucial regulatory mechanism in inflammation and cell
death pathways, potentially indicating new therapeutic
interventions in inflammatory diseases (67). By contrast, lactylation can also
facilitate interactions, as seen in renal tubular epithelial cells
where mitochondrial fission 1 lactylation at K20 promotes
mitochondrial fission and oxidative stress through enhanced
interaction with dynamin-related protein 1. This finding highlights
lactylation's significant role in mitochondrial dynamics and
emphasizes its potential impact on cellular stress responses
(68). Furthermore, lactylation
has the potential to enhance signaling pathways. Specifically,
lactylation at K72 of the membrane-organizing extension spike
increases its interaction with transforming growth factor β (TGF-β)
receptor I, thereby promoting downstream SMAD3 signaling. This
process strengthens TGF-β signaling in Tregs within the TME,
indicating that lactylation may contribute to tumor progression and
immune evasion. Consequently, it represents a promising target for
cancer immunotherapy (51).
Lactylation's influence also extends to transcription factors,
where it can alter their binding capacity with corresponding DNA
and thus transcriptional activity. For instance, lactylation of p53
at K120 and K139 impairs its liquid-liquid phase separation, DNA
binding and transcriptional activation, correlating with poor
prognosis among cancer patients carrying wild-type p53 (69). This modification can critically
affect tumor suppressor function, providing insights into cancer
biology and potential targets for anti-tumor therapeutic
intervention. Similarly, the lactylation of Yin Yang-1 (YY1)
enhances EGF2 transcription, promoting angiogenesis under hypoxic
conditions. This finding links lactylation with pathological
conditions such as blindness, offering new perspectives on the
regulation of angiogenesis (70). In hepatocellular carcinoma,
lactylation of centromere protein A (CENPA) at K124 accelerates
CENPA binding at YY1 promoter regions to drive oncogene
CCND1 and neuropilins-2 expression (71). Exercise-induced lactylation at
methyl CpG binding protein 2 K271 also binds to the promoter of
epiregulin, leading to the inhibition of its transcription and
facilitating the remission of atherosclerosis (72). These diverse effects on
transcription underscore the broad regulatory potential of
non-histone lactylation in various disease contexts. In addition to
influencing interactions and transcription, lactylation also
affects protein stability by competing with ubiquitination. This
competition can alter protein half-lives, impacting cellular
dynamics and processes. In cervical cancer, for instance, the
lactylation at K172 of discoidin, CUB and LCCL domain-containing
type I enhances its expression while shortening its half-life. This
modification concurrently diminishes its ubiquitination, indicating
a complex role for non-histone lactylation in cancer, as it
influences protein turnover and maintains cellular homeostasis
(73). Similarly, lactylation of
Runt-related transcription factor 2 stabilized the protein by
reducing its ubiquitination, further promoting the osteogenic
differentiation of periodontal ligament stem cells (74). This highlights the positive
regulatory potential of lactylation in tissue regeneration and
healing, illustrating its therapeutic promise beyond disease
mitigation. Overall, non-histone lactylation represents a
multifaceted regulatory mechanism with extensive implications for
cellular function and disease development. By modulating protein
interactions, stability and transcriptional activity, non-histone
lactylation influences a spectrum of biological processes, enhances
the current understanding of cellular regulation and broadens the
horizon for potential therapeutic applications. As research
continues to unveil the complexities of protein modifications,
lactylation stands out as a pivotal modification that could be
harnessed in novel treatment strategies targeting various
pathological conditions, from cancer to inflammatory diseases.
Future studies will benefit from elucidating the precise molecular
mechanisms through which non-histone lactylation exerts its
effects, paving the way for targeted modulation in clinical
settings. Information on additional research on non-histone
lactylation and studies related to protein modification is provided
in Table II.
 | Table IINon-histone lactylation and protein
modification. |
Table II
Non-histone lactylation and protein
modification.
| Functions of
non-histone lactylation | Sites | Target |
Writers/erasers | Cell types | Specific
functions | (Refs.) |
|---|
| Protein
activity | K28 | AK2 | p300/CBP and
HDAC3 | Hepatoma carcinoma
cell | Inhibited the
function of AK2, facilitated the proliferation and metastasis of
hepatoma carcinoma cells | (58) |
| K147 K688 | ALDOA PFKP | / | Colon cancer
cell | Disrupted their
enzyme activity and formed a negative feedback pathway in
glycolysis and lactic acid production | (59) |
| K62 | PKM2 | / | Macrophage | Activated PKM2 and
promoted the transition of pro-inflammatory macrophages towards a
reparative phenotype | (61) |
| K356 and K781 | VPS34 | KAT5/TIP60 | Skeletal muscle
cell | Increased
PIK3C3/VPS34 lipid kinase activity and facilitated autophagy | (62) |
| K408 | eEF1A | KAT8 | Colon cancer
cell | Enhanced ribosomal
GTPase activity | (64) |
| protein-DNA/protein
interactions | K33 | NEDD4 | p300/CBP and
SIRT1 | Macrophage | Inhibited its
binding to caspase-11 and reduced caspase-11 ubiquitination | (67) |
| K20 | Fis1 | SIRT3 | Renal tubular
epithelial cell | Improved Fis1
interaction with DRP1, promoting mitochondrial dysfunction | (68) |
| K72 | MOESIN | / | Regulatory T
cell | Improved the
interaction of MOESIN with TGF-βRI, reinforcing TGF-β
signaling | (51) |
| K120 and K139 | P53 | AARS1 | Tumor cell | Impaired p53
liquid-liquid phase separation, DNA binding and transcriptional
activation | (69) |
| K271 | MECP2 | p300/CBP | Endothelial cell
protein stability | Promoted its
binding to Ereg promoter regions and inhibited its
transcription | (72) |
| K172 | DCBLD1 | / | Cervical cancer
cell | Enhanced the post
transcriptional expression and shortened the half-life of
DCBLD1 | (73) |
| K176 | RUNX2 | / | Periodontal
ligament stem cells | Decreased the
ubiquitination level of RUNX2 and shortened its half-life | (74) |
Writers and erasers
Writers
As with many PTMs, lysine lactylation is
orchestrated by a specific set of enzymes that include
lactyltransferases, or 'writers', which introduce lactate groups to
lysine residues, delactylases (erasers), which remove these
modifications from proteins, and recognition enzymes, often dubbed
'readers', which interpret the presence of lactyl marks to elicit
biological effects (75). The
enzymes responsible for writing the lactyl marks have not been
fully characterized, yet preliminary evidence suggests a likely
overlap with enzyme families known to mediate other forms of PTMs,
such as acetylation and methylation (76). Among these, histone
acetyltransferase families such as p300/CREB binding protein
(p300/CBP), the Moz, Ybf2/Sas3, Sas2 and Tip60 (MYST) proteins and
the GCN5 related N-acetyltransferases (GNAT superfamily) have drawn
particular attention for their potential roles in histone
lactylation (77). A study
conducted by Zhao et al (7) in 2019, for example, identified the
catalytic role of p300 in histone H3 and H4 lactylation and
corresponding transcriptional activation of arginine in a
p53-dependent manner. The p300 enzyme has been demonstrated to
influence the levels of histone lactylation; specifically, its
overexpression resulted in an increase in lactyl marks, whereas its
deletion caused a decrease in this modification. These findings
position p300 not only as a likely writer of histone lactylation
but also as a key player in the regulatory network that links
cellular metabolism with gene expression, particularly in contexts
significant to cancer biology and inflammation. Further insights
come from studies on the GNAT superfamily, specifically the role of
GCN5. This enzyme has emerged as an important writer in
monocyte-macrophage function and reparative gene expression
following myocardial infarction. The silencing of GCN5 in these
cells resulted in significant decreases in H3K18la, underscoring
its role as an upstream regulatory writer in facilitating the
lactylation process. This highlights the broader biological
relevance of lactylation as a modification that might modulate
immune responses and repair mechanisms (10). Beyond these enzymes, the MYST
family member lysine acetyltransferase 7 (KAT7, also known as HBO1)
has been implicated in directing lactylation at specific histone
sites, notably H3K9. Niu et al (78) demonstrated that HBO1, alongside
scaffold proteins like jade family PHD finger 1 and
bromodomain-containing 1, could specifically catalyze the addition
of lysine residues to H3K9. This targeted activity further
underscores the potential specificity and regulatory precision of
histone lactylation, as each writer might differentially influence
various histone sites and subsequent gene expression outcomes
(78). The characterization of
lactylation writers like p300, GCN5 and HBO1 in histone
modification marks a promising frontier in the study of lysine
lactylation, while understanding these 'writers' extends the
possibility for targeted therapeutic interventions.
Erasers and readers
While 'writers' add lactyl groups to proteins,
'erasers' remove them, thereby maintaining the equilibrium of
lactylation and influencing protein function. Just like writers,
the exploration into the specific 'eraser' enzymes involved in
lactylation is under continuous research. However, due to
similarities with the acetylation process, it is plausible to
assume that analogous enzymes to those that erase acetylation, such
as deacetylases, may also serve as 'erasers' for lactylation
(79). Screening studies in
mammals underscore the role of histone deacetylases (HDACs) in this
capacity. Specifically, HDAC1-3 has been identified to possess
delactylase activity, which allows it to effectively remove lactyl
groups from lysine residues on histones. These findings suggest a
conserved mechanism whereby deacetylases not only regulate
acetylation but also participate in modulating lactylation, thereby
influencing gene expression and chromatin dynamics (80,81). Li et al (79) have identified p300 and HDAC2 as
potential 'writer' and 'eraser' enzymes, respectively, involved in
the regulation of H3K18la in pancreatic ductal adenocarcinoma
cells. This discovery highlights the potential cross-talk and
shared pathways between lactylation and other histone
modifications, further explaining the intricate control of gene
expression in cancer cells. The implications of these regulatory
mechanisms extend beyond cancer, as similar processes may be
involved in inflammatory responses and other pathological
conditions of the lung (79).
Furthermore, the involvement of 'readers' in the interpretation of
lactylation signals introduces an extra dimension of enzymatic
regulation. These readers, which identify and attach to lactylated
sites, can influence the functional outcomes of the modifications
made by 'writers' and 'erasers' (8). Proteomic analysis of the H3K18la
immunoprecipitation experiment conducted by Hu et al
(82) revealed the specific
recruitment of BRM/SWI2-related gene 1 (Brg1), a gene that encodes
a protein component of the SWI/SNF chromatin-remodeling complex
which alters the structure of chromatin and modulates access to
DNA, during cellular reprogramming. This study found that both
H3K18la and Brg1 were enriched on the promoters of genes linked to
pluripotency and epithelial junctions, providing insights into how
lactylation can influence cellular identity and transformation
processes (82).
Regulation of non-histone
lactylation
P300/CBP, recognized as a writer of histone
lactylation, has also been identified as a writer of non-histone
lactylation. For instance, p300 catalyzed the lactylation of
nucleolin at K477 in response to hyperglycolytic conditions,
suggesting a direct link between metabolic shifts and PTMs.
Similarly, p300 also acted as a key lactyltransferase responsible
for lactylation at the 128th lysine residue of nicotinamide
nucleotide adenylyltransferase 1 (83,84). The function of p300/CBP extends
into the modulation of key transcription factors such as Snail1, a
well-known TGF-β transcription factor in transformation processes
like endothelial-to-mesenchymal transition (EMT). Lactylation
mediated by p300/CBP was reported to enhance the transcriptional
activity of Snail1, which may contribute to the exacerbation of
conditions such as cardiac fibrosis. This serves as a clear
illustration of how non-histone lactylation connects cellular
metabolism with transcriptional regulation (85). The interaction between PKM2 and
p300 appears to facilitate PKM2 lactylation, consequently
influencing its cellular localization and the cell cycle,
demonstrating the far-reaching effects of lactylation in cell fate
decisions (85,86). In addition to p300/CBP, other
lysine acetyltransferases, including KAT5/TIP60, play a role in
shaping the non-histone lactylation landscape. For instance, the
reduction in KAT5/TIP60 levels results in diminished vacuolar
protein sorting 34 homolog (VPS34) lactylation at lysine-356 and
lysine-781 during intense physical activity, pointing to its role
in muscle adaptation and energy metabolism (62). KAT8, another acetyltransferase,
has been identified as a pan-Kla writer, capable of installing
lactylation across a wide range of substrates. This expands the
spectrum of lactylation beyond individual proteins to encompass
diverse biological processes including translation and signal
transduction (63). In addition,
mitochondrial alanyl-tRNA synthetase 1/2 has recently been
recognized as a non-histone protein lysine lactyltransferase, which
specifically targets metabolic enzymes such as pyruvate
dehydrogenase E1 subunit α1 (PDHA1) and carnitine
palmitoyltransferase 2 (CPT2). This finding introduces an
additional functional role, demonstrating the potential role of
lactylation in the regulation of mitochondrial metabolism and
underscoring possible targets for addressing metabolic disorders
(87,88). Research indicates that sirtuin
(SIRT)3, a class III lysine deacetylase, functions as an eraser of
non-histone lactylation by reversing the lactylation of specific
proteins, including CCNE2, PDHA1 at K336, and CPT2 at K457/8. In
addition, SIRT1 has been identified as the delactylase for α-myosin
heavy chain and canopy FGF signaling regulator 3 (87-91). Collectively, these studies
indicate a sophisticated network of enzymes that facilitate lysine
lactylation, although the landscape remains incompletely mapped.
The enzymatic machinery responsible for lactylation involves
multifaceted roles across different enzyme families, some of which
are shared with other acyl modifications. This points to
evolutionary conservation and potential functional redundancy or
specialization within the realm of PTMs, where similar enzymes may
diversify their substrate preferences and actions under distinct
physiological and pathological conditions. By clarifying the
functions of these regulatory enzymes, knowledge gaps in the
understanding of intricate diseases may be effectively bridged,
thereby enhancing the prognosis for patients afflicted with severe
lung disorders.
NSCLC
Lung cancer remains the leading cause of
cancer-related deaths globally, attributed to its significant
prevalence and poor prognosis, with a 5-year survival rate of
<17% (92). Based on
histological type, therapy and prognosis, lung cancers can be
classified into two major types, SCLC and NSCLC. NSCLC, which
constitutes ~85% of all lung cancer diagnoses, is further
subclassified into three primary types: Lung adenocarcinoma (LUAD),
lung squamous cell carcinoma (LUSC) and large cell carcinoma
(93). The investigation of
lysine lactylation, which involves the modification of lysine
residues by lactate in both histones and non-histone proteins,
provides valuable insights into the significant impact of metabolic
dysregulation on cancer biology (Fig. 2). For instance, Jiang et
al (94) reported the
relevance of lactate metabolism and its aberrations in poor NSCLC
prognosis. In hypoxic tumor environments, lactate accumulates
modulated gene transcription through histone lactylation at
hexokinase 1 (HK-1) and isocitrate dehydrogenase
(IDH3G) promoters, which govern critical metabolic enzymes
involving glycolysis and the tricarboxylic acid cycle. This
metabolic reprogramming supports tumor growth and survival,
creating a robust link between metabolic pathways and cancer
progression (94). Furthermore,
hypoxia-induced lactylation of SRY-related high mobility group box
9 (SOX9), a transcription factor overexpressed in NSCLC,
exemplifies how lactylation modification promotes the stemness,
migration and invasion of NSCLC cells by enhancing glycolysis-a
vital adaptation for tumor growth. This finding highlights the
multifaceted nature of NSCLC pathogenesis where metabolic shifts
dovetail with transcriptional reprogramming (95).
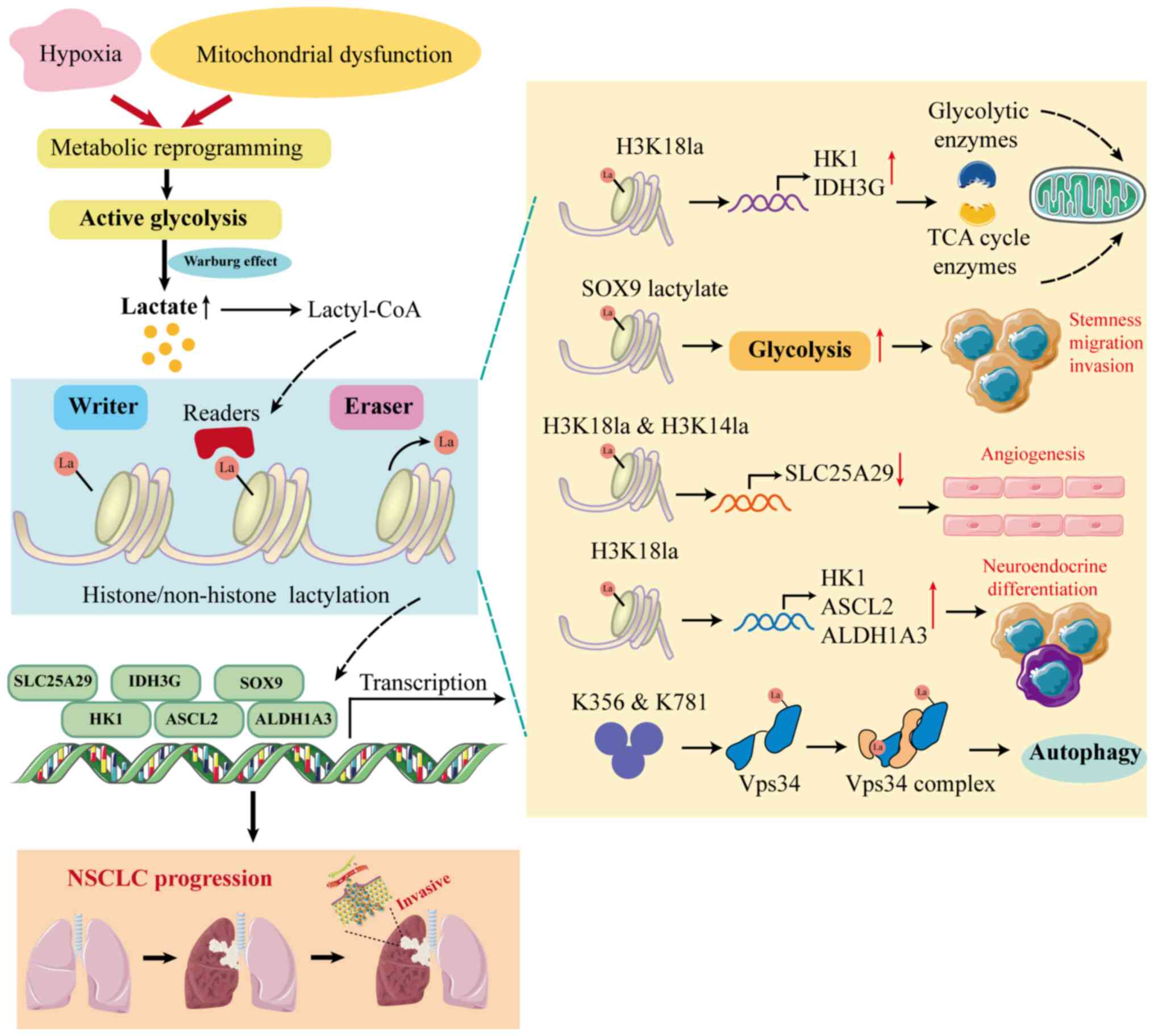 | Figure 2Mechanisms of lactylation that
contribute to NSCLC. In NSCLC, metabolic reprogramming leads to
increased lactate production, which subsequently provides the
substrate lactyl-CoA for lactylation, ultimately contributing to
tumorigenesis and progression. Elevated H3K18la enrichment at
promoters of glycolysis-related genes such as HK-1 and
IDH3G leads to their increased transcription and regulates
cellular metabolism in tumor cells, while its enrichment at
promoters of ASCL2 and ALDH1A3 promotes
neuroendocrine differentiation. H3K18la can also promote the
proliferation, migration and apoptosis of endothelial cells through
its enrichment at the SLC25A29 promoter in non-tumor endothelial
cells. Specifically, lactylation of the non-histone protein SOX9
enhances stemness, migration and invasion in NSCLC cells by
promoting glycolysis, while lactylation of VPS34 at K356 and K781
enhances its lipid kinase activity to facilitate cell autophagy and
endolysosomal degradation. ASCL2, achaete-scute family BHLH
transcription factor 2; ALDH1A3, aldehyde dehydrogenase 3 family
member A1; HK1, hexokinase 1; H3K14la, lactylation of histone 3 on
lysine residue 14; H3K18la, lactylation of histone 3 on lysine
residue 18; IDH3G, isocitrate dehydrogenase 3 non-catalytic subunit
gamma; Lactyl-CoA, lactyl-coenzyme A; NSCLC, non-small cell lung
cancer; SLC25A29, solute carrier family 25 member 29; SOX9 SRY-box
transcription factor 9; VPS34, vacuolar protein sorting 34; TCA,
tricarboxylic acid cycle. |
LUAD
As the most common subtype of NSCLC, LUAD accounts
for a significant proportion of global lung cancer cases (96). Specific lactylation sites have
been implicated in LUAD biology. SLC25A29 belongs to the solute
carrier family and works as a key regulator linked to LUAD staging
(97). Zheng et al
conducted a comprehensive bioinformatics and revealed that lactate
increased the levels of H3K14la and H3K18la at the promoter of
SLC25A29, subsequently enhancing its transcription. This process,
in turn, influenced the proliferation, migration and apoptosis of
endothelial cells, which are critical factors in angiogenesis-a key
characteristic of cancer progression (98). Furthermore, another study focused
on the adenocarcinoma-to-neuroendocrine cell fate transition and
the pivotal role of lactylation in cell energy metabolism. They
discovered that a deficiency in the cell fate determinant
Numb/Parkin pathway in LUAD led to metabolic reprogramming by
upregulated H3K18la and the transcription of
neuroendocrine-associated genes (16). Non-histone lactylation plays a
crucial role in the pathogenesis of LUAD, extending its influence
beyond that of histones. Jia et al (62) highlighted the integral role of
lactylation in autophagy regulation, a pivotal process for cellular
homeostasis in cancer cells. The enhanced activity of the
autophagic mechanism via lactylation, particularly of VPS34 at K356
and K781, emphasizes how metabolic products like lactate
orchestrate cellular survival and proliferation strategies,
exacerbating cancer progression (62).
LUSC
LUSC accounts for ~30% of NSCLC cases; however, it
has been less extensively researched regarding lactylation
(99). However, Hao et al
(100) reported that SLC2A1 may
act as a predictive biomarker for the survival and response to
immunotherapy in LUSC. Tumor tissues with high expression of SLC2A1
were associated with an elevated risk of lactylation, potentially
influencing immune modulation and tumor progression (100). This gap in research presents an
opportunity to investigate the wider implications of lactylation
across different subtypes of NSCLC, particularly given LUSC's
varying response to conventional therapies.
Lung cancer brain metastasis (BM)
BM is a malignant event leading to poor prognosis in
NSCLC (101). Lactylation also
contributes to the challenges of treating BM in LUAD. The enzyme
aldo-keto reductase family 1 member B10 (AKR1B10) is significantly
upregulated in both LUAD cells with high BM propensity and in
patients suffering from brain metastases (102). AKR1B10 functions as a
cyto-detoxification agent within the intestinal system. It
facilitates anaerobic glycolysis and the secretion of lactate,
while also promoting the transcription of the cell cycle-related
gene cyclin B1 through H4K12la. This mechanism may limit the
therapeutic effectiveness for patients with lung cancer BM
(103). This metabolic feedback
loop highlights lactylation as both an indicator of disease
advancement and a promising target for therapeutic strategies.
Conclusively, lysine lactylation emerges as a
central theme in the molecular landscape of NSCLC, particularly
LUAD. Its involvement in modulating genes crucial for metabolism,
angiogenesis and autophagy underlines the profound influence of
metabolic dysregulation in cancer. Future research should explore
the therapeutic possibilities associated with targeting
lactylation, extending beyond its function as a biomarker to
examine its viability as a targeted element within cancer treatment
strategies. Advances in understanding lactylation-specific pathways
could lead to more precise and personalized treatment strategies,
potentially improving outcomes for patients afflicted with this
challenging disease. Given the dynamic nature of cancer biology,
integrating these insights could redefine how to approach NSCLC,
offering new hope for improved prognostic and therapeutic
avenues.
Malignant pleural effusion
The exploration of lysine lactylation's role in
malignant pleural effusion (MPE) further advances the understanding
of the metabolic dynamics and immunological adaptations in
neoplastic environments. MPE, usually caused by the invasion of the
pleura, serves as an independent prognostic marker in NSCLC
(104-106). This phenomenon underscores a
complex interplay of elevated metabolites and versatile cellular
components that harmonize to perpetuate immunosuppression. Within
this milieu, lactate emerges as a particularly influential
metabolite, reshaping the metabolic pathways in the effusion
microenvironment (107,108). Research into lactylation
modifications sheds light on a unique subset of natural killer T
(NKT)-like cells present in MPE. These cells are characterized by
their expression of forkhead box P3 (FOXP3), a transcription factor
typically associated with regulatory T cells and immunosuppression.
The presence of these FOXP3-expressing NKT-like cells signals an
intriguing deviation from their expected antitumor responses,
revealing instead augmented immunomodulatory properties. This shift
is supported by a marked increase in glycolytic activity and
enhanced lactate metabolism, offering insight into how the
metabolic landscape orchestrates immunological roles within the
pleural effusion. In vitro studies revealed that increased
levels of H3K18la lactylation at the promoter region of the
FOXP3 gene correlate with elevated FOXP3 expression.
This suggests that lactylation acts as an epigenetic regulator,
modulating gene expression and function in a metabolically adapted
immune cell population (109).
Consequently, the findings highlight the potential of targeting
lactylation pathways in therapeutic strategies, particularly for
reprogramming immune responses to restore antitumor activity in MPE
environments.
Pulmonary fibrosis
Pulmonary fibrosis, also known as interstitial lung
disease (ILD), is a significant challenge in respiratory medicine,
primarily because it is a chronic, progressive disease with
significant scarring in the lungs that often leads to high
mortality rates due to its irreversible nature and limited
treatment options (110). The
etiology of ILD remains largely elusive. However, since its first
discovery, numerous studies have emerged regarding the lactylation
of profibrotic genes, highlighting the crucial role that
lactylation modification plays in the development of pulmonary
fibrosis (Fig. 3).
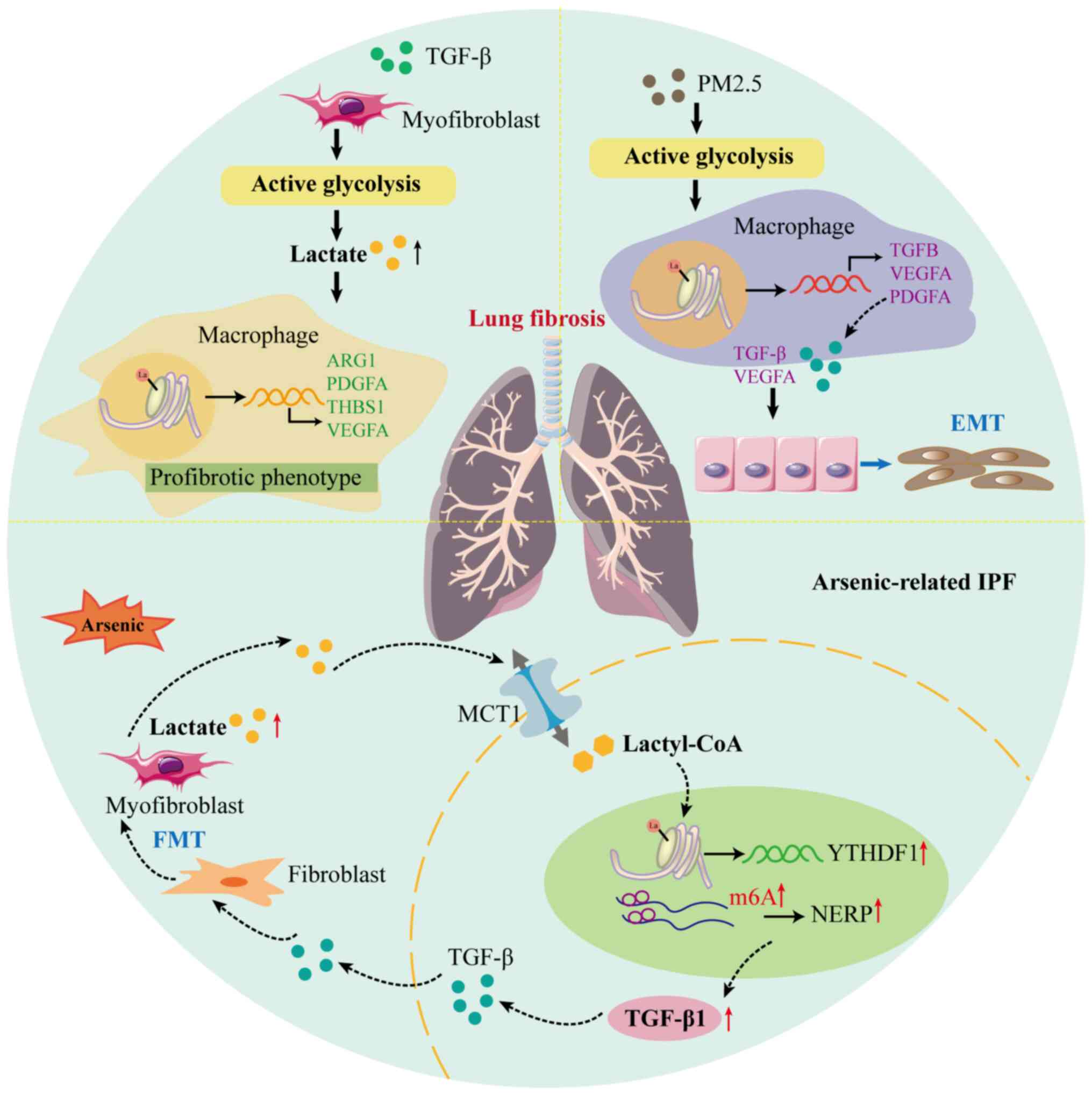 | Figure 3Lactylation plays a significant role
in the progression of lung fibrosis. The abnormally elevated
glycolysis in myofibroblasts, stimulated by TGF-β, causes an
increase in lactate levels that subsequently enhances histone
lactylation at the promoters of pro-fibrotic genes (ARG1,
PDGFA, THBS1 and VEGFA) in macrophages.
Additionally, PM2.5 directly promotes the elevation of macrophage
glycolysis, ultimately leading to histone lactylation at the
promoters of pro-fibrotic genes (TGFB, VEGFA and
PDGFA). This mechanism further promotes EMT by activating
the TGF-β/Smad2/3 and VEGFA/MEK/ERK pathways. Furthermore, in
arsenic-related lung fibrosis, the extracellular lactate secreted
by myofibroblasts is absorbed and converted into lactyl-CoA, which
elevates the H3K18la enriched in the promoter of YTHDF1 in
alveolar epithelial cells. The upregulated YTHDF1 promotes the
translation of Nrep mRNA via recognizing the m6A sites on
the mRNA and activates the secretion of TGF-β1, which further
promotes the FMT. ARG1, arginase 1; EMT, epithelial-to-mesenchymal
transition; FMT, fibroblast-to-myofibroblast transition; IPF,
idiopathic pulmonary fibrosis; Lactyl-CoA, lactyl-coenzyme A; m6A,
N6-methyladenosine; MCT1, monocarboxylate transporter 1; NERP,
neuronal protein 3.1; PDGFA, platelet-derived growth factor
receptor α; TGF-β, transforming growth factor β; THBS1,
thrombospondin-1; VEGFA, vascular endothelial growth factor A;
YTHDF1, YTH N6-methyladenosine RNA-binding protein. |
Idiopathic pulmonary fibrosis (IPF)
As the most severe form of ILD with a median
survival of only 3 to 5 years post-diagnosis, the specific causes
of IPF are unknown, distinguishing it from pulmonary fibrosis
linked to environmental factors, medications or other identifiable
causes (111). Alveolar
macrophages (AMs) are pivotal in pulmonary health, maintaining
homeostasis and modulating immune responses within the alveolar
spaces. In a healthy respiratory environment, these cells typically
manifest with an anti-inflammatory and pro-resolving phenotype.
Yet, in fibrotic lung conditions like IPF, AMs undergo a phenotypic
shift toward a pro-fibrotic state. Pro-fibrotic AMs produce several
soluble factors, such as transforming growth factor-β
(TGF-β), platelet-derived growth factor (PDGF),
vascular endothelial growth factor (VEGF) and thrombospondin
1 (THBS1), which differentiate resident fibroblasts into
myofibroblasts and accelerate the fibrotic processes (112-115). The biochemical environment of
the fibrotic lung is characterized by elevated lactate
concentrations, which serve as a significant indicator of metabolic
reprogramming. A research investigation conducted by Cui et
al (11) revealed increased
levels of lactate in the conditioned media derived from TGF-β1
stimulated lung myofibroblasts, as well as in bronchoalveolar
lavage fluid obtained from fibrotic mouse lungs. This elevation in
lactate was found to play a role in histone lactylation at the
promoters of significant pro-fibrotic genes, such as arginase 1,
PDGFA, THBS1 and VEGFA. This finding not only
underscores the influence of altered metabolic states in pulmonary
fibrosis but also highlights lactylation as a regulatory mechanism
that exacerbates the pro-fibrotic behaviors of AMs. Notably, these
lactylation modifications are facilitated by acetyltransferase
p300, emphasizing a potential target for therapeutic intervention
(11).
Secondary pulmonary fibrosis
Beyond IPF, pulmonary fibrosis can also stem from
secondary environmental influences, such as inhaled small
particulate debris [such as particulate matter of 2.5 microns or
less in diameter (PM2.5)] and microbial agents (116). According to a study by Li et
al (117), exposure to
urban airborne PM2.5 causes lung fibrotic changes and
encourages a metabolic shift toward glycolysis. Mechanically,
PM2.5-induced glycolysis and lactate production promoted
histone lactylation levels at the promoters of the pro-fibrotic
genes TGFB, VEGFA and PDGFA in macrophages.
Furthermore, the upregulated pro-fibrotic genes induced the
excessive secretion of pro-fibrotic cytokines and consequently
triggered EMT through activating TGF-β/Smad2/3 and VEGFA/MEK/ERK
pathways, contributing to fibrosis (117). Arsenic exposure offers another
model for secondary pulmonary fibrosis, with recent research
highlighting elevated pan-lactylation and histone lactylation,
specifically H3K18la, in alveolar epithelial cells among
arsenic-induced fibrotic mice. These modifications foster
fibroblast-to-myofibroblast transition via mechanisms mediating the
m6A methylation site of neuronal regeneration-related protein by
readers like YTH N6-methyladenosine RNA-binding protein F1,
showcasing the intricate epigenetic interplay in environmentally
induced fibrosis (64). Overall,
the exploration of lactylation modifications within pulmonary
fibrosis provides a promising avenue to deepen our understanding of
this complex disease and identify potential therapeutic targets. As
research in this area continues to evolve, it beckons a shift
toward investigating metabolic and epigenetic interventions as
viable strategies to mitigate the progression of pulmonary
fibrosis, ultimately aiming to improve the prognosis of this
devastating illness.
Acute and chronic pulmonary inflammatory
diseases
Pulmonary inflammatory disorders represent a wide
range of conditions that involve inflammation and damage to lung
tissue, which manifest as both acute (short-term) and chronic
(long-term) conditions, including acute lung injury (ALI), asthma
and pulmonary fibrosis (118).
The exploration of lysine lactylation in the context of pulmonary
inflammatory disorders offers a fresh perspective on the underlying
biochemical and molecular changes accompanying these conditions.
Current research has predominantly focused on lactylation in
relation to pulmonary fibrosis; however, its significance in other
pulmonary inflammatory diseases is still relatively underexplored.
This limited focus represents a significant gap considering the
broad spectrum of pulmonary conditions characterized by
inflammation and tissue damage. However, studies have begun to shed
light on the potential significance of lactylation in these
disorders. For instance, the biomimetic anti-inflammation
nanoparticle delivery system developed by Jin et al
(119) demonstrated a promising
approach to treating ALI by targeting inflamed lungs with curcumin
and resveratrol. This approach not only showcased the therapeutic
potential of natural compounds but also highlighted the importance
of inhibiting histone lactylation to modulate macrophage
polarization and reduce lung injury and vascular permeability
(119). Similarly, the
investigation into the effects of corticosteroids on asthma
presents another facet of lactylation's role in pulmonary
inflammation. The study revealed that dexamethasone attenuates the
Hif-1α-glycolysis axis, effectively reducing lactate production and
associated histone lactylation in lung macrophages. These results
underscore the potential of targeting metabolic pathways and
histone modifications to mitigate chronic inflammatory responses in
asthma. Furthermore, ovalbumin stimulation also leads to histone
lactylation in macrophages and the lungs of asthmatic mice, a
process that can be significantly diminished following
dexamethasone treatment; however, the precise mechanism underlying
this phenomenon has yet to be clarified (120). Despite these insights,
significant gaps remain in understanding the precise mechanisms by
which lactylation influences inflammatory processes, warranting
further research. Filling these gaps could unveil novel therapeutic
targets, ultimately improving management strategies for both acute
and chronic pulmonary inflammatory diseases. As Table III indicates, expanding our
knowledge in this area holds great promise for enhancing patient
outcomes in a variety of neoplastic and inflammatory pulmonary
conditions, emphasizing the need for continued exploration in this
emerging field.
 | Table IIILactylation and corresponding targets
in neoplastic and inflammatory pulmonary diseases. |
Table III
Lactylation and corresponding targets
in neoplastic and inflammatory pulmonary diseases.
| Condition | Lactylation
sites | Targets | Biological
functions | (Refs.) |
|---|
| NSCLC | H4K8 | HK1 and IDH3G | Regulated cellular
metabolism reprogramming | (94) |
| / | SOX9 | Promoted glycolysis
in NSCLC cells, accelerating their stemness, migration and
invasion | (95) |
| LUAD | H3K18 and
K3K14 | SLC25A29 | Affected
endothelial cell proliferation, migration and apoptosis associated
with angiogenesis | (98) |
| H3K18 | ASCL2, HK1,
ALDH1A3 | Induced
neuroendocrine differentiation | (16) |
| K356 and K781 | VPS34 | Promoted cancer
cell autophagy to facilitate cancer progression | (62) |
| LUSC | / | / | High SLC2A1
contributed to high protein lactylation levels | (100) |
| BM | H4K12 | CCNB1 | Activated the
transcription of CCNB1 and accelerated the DNA replication and cell
cycle. | (103) |
| MPE | H3K18 | FOXP3 | Maintained
immunosuppressive function in NKT-like cells | (109) |
| Lung fibrosis | Histone | ARG1, PDGFA, THBS1,
VEGFA | Induced the
pro-fibrotic phenotype of macrophages | (11) |
| Histone | TGFB, VEGFA, PDGFA
YTHDF1 | Induced the
expression of pro-fibrotic genes | (117) |
| H3K18 | Mediated m6A
methylation and participated in the fibroblast-to-myofibroblast
transition | (64) |
| Acute lung
injury | Histone | / | Contributed to the
anti-inflammation effects of macrophages | (119) |
| Asthma | / | / | Induced elevated
lactylation levels in lung macrophages | (120) |
Conclusion and future perspectives
The human genome contains ~20,000 genes; however,
the variety of transcripts and post-translational modifications
that create what are known as 'proteoforms' significantly expands
the scale of the proteome. This expansion results in a diverse
protein library capable of engaging in numerous cellular biological
processes (121). As a newly
identified post-translational modification, the lactylation process
has been under investigation and the understanding of it has
significantly advanced in the last five years, revealing its
involvement in numerous diseases including cancers and systemic
diseases. Of note, the various roles and molecular mechanisms of
lactylation in pulmonary diseases ranging from pulmonary fibrosis
to lung cancers are an emerging research frontier. Currently, there
are no comprehensive literature reviews addressing the effects of
lactylation in various pulmonary disorders. This article presents
the findings related to lactylation and provides a thorough
examination of the essential roles that histone and non-histone
lactylation fulfill within cellular and protein contexts,
respectively. Research on histone lactylation has yielded a
significant understanding of how lactate generated during cellular
metabolism can directly alter the chromatin structure, thereby
influencing gene transcription. This novel finding has opened a new
avenue in the field of epigenetics and various cellular functions
including cell fate determination, regulation of immunity and
inflammation, as well as regulation of cellular metabolism. The
specific role of histone lactylation in gene targeting is well
established; however, the function of non-histone lactylation is
still largely unexplored and remains unclear. Preliminary research
suggests that non-histone lactylation also plays an important part
in protein function consisting of protein activity modulation,
influencing protein-DNA/protein interactions and regulation of
protein stability. Nevertheless, further research is required to
comprehend the full spectrum of its functions and mechanisms.
Recent research developments have underscored the
relationship between metabolic reprogramming and pulmonary
diseases. In this context, the accumulation of lactate within
structural and immune cells is not merely a consequence of these
diseases; it also plays a significant role in their progression
(122-124). For instance, in bronchial
epithelial cells and PASMCs, altered lactate metabolism heavily
contributes to their pathological remodeling by stimulating
aberrant inflammatory responses, structural changes and cell
proliferation (125,126). Furthermore, lactate can impair
the bactericidal activity of neutrophils and stimulate macrophages
to produce pro-inflammatory cytokines, exacerbating inflammation
and tissue damage (127).
Although our understanding of the roles and functions of
lactylation in pulmonary diseases is only nascent, several studies
have implicated lactylation in diverse neoplastic and inflammatory
pulmonary diseases, including NSCLC, malignant pleural effusion,
pulmonary fibrosis, acute lung injury and asthma. In general,
histone lactylation, particularly H3K18la and non-histone
lactylation of SOX9/VPS34, have harmful effects on lung tumors by
increasing glycolysis in lung cancer cells, which in turn increases
lactate levels and eventually leads to a vicious cycle (16,62,94,95). Furthermore, histone lactylation
could promote lung fibrosis via modulating the expression of
profibrotic genes in macrophages or mediating m6A methylation of
Nrep mRNA (11,64,117). Research on chronic inflammatory
diseases such as asthma has revealed only a limited correlation
between elevated lactate levels and subsequent lactylation, while
the underlying mechanisms have yet to be thoroughly investigated
(120). In other lung diseases,
research regarding lactylation is currently scarce, yet this field
holds immense potential for future studies. For instance, during
acute exacerbations of chronic obstructive pulmonary disease, an
increase in lactate levels is frequently noted. Elevated lactate
may indicate excessive β2-agonist treatment and warrants further
investigation as a potential biomarker (128). Furthermore, lactate also
exhibits a range of immunomodulatory effects both within the
context of tuberculosis and in other conditions (129). In all cases of plastic
bronchitis in children, lactate dehydrogenase levels increased to
different degrees (130). Given
the potential of lysine lactylation as a biomarker and therapeutic
target, future research is promising. Investigating the reversible
nature of lactylation may unveil novel treatment strategies by
targeting specific molecular pathways. However, a robust
understanding of its clinical mechanism remains imperative. Future
research should explore the functions of lactylation in various
cellular environments and disease conditions, examine its
interactions with other post-translational modifications and assess
the broader implications for pulmonary health. Ultimately,
expanding the knowledge in this domain could revolutionize
therapeutic approaches, offering new hope for targeting pulmonary
diseases at a molecular level. As research progresses, the insights
gained from studying lysine lactylation could have wide-ranging
applications, not just in pulmonary medicine but across the
spectrum of systemic diseases affected by metabolic dysregulation
and epigenetic modification.
Availability of data and materials
Not applicable.
Authors' contributions
SW and HZ wrote and revised the manuscript,
organized the tables and drew the figures. JZ and JX critically
conceptualized and supervised the study and reviewed the
manuscript. All authors have read and approved the final
manuscript. Data authentication is not applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Abbreviations:
|
ALI
|
acute lung injury
|
|
HK-1
|
hexokinase 1
|
|
H3K18la
|
lactylation of histone 3 on lysine
residue 18
|
|
Hif-1α
|
hypoxia-inducible factor-1α
|
|
IL
|
interleukin
|
|
LUAD
|
lung adenocarcinoma
|
|
LUSC
|
lung squamous cell carcinoma
|
|
MPE
|
malignant pleural effusion
|
|
NSCLC
|
non-small cell lung cancer
|
|
p300/CBP
|
p300/CREB binding protein
|
|
PKM2
|
pyruvate kinase isozyme M2
|
|
PTM
|
post-translational modification
|
|
RUNX2
|
runt-related transcription factor
2
|
|
SLC25A29
|
solute carrier family 25 member
29
|
|
TGF-β
|
transforming growth factor β
|
|
VEGFA
|
vascular endothelial growth factor
A
|
|
VPS34
|
vacuolar protein sorting 34
|
|
YTHDF1
|
YTH N6-methyladenosine RNA-binding
protein F1
|
|
YY1
|
Yin Yang-1
|
Acknowledgements
Not applicable.
Funding
This study is supported by the National Natural Science
Foundation of China (grant nos. 82400033, 82070032, 82170049 and
81973986), the Leading Talents of Public Health in Hubei Province
(grant no. 2022SCZ047), the Clinical Collaboration Project of
Traditional Chinese and Western Medicine in the Major Difficult
Diseases in Hubei Province (Respiratory system Diseases), the
Project of Key R&D Program in Hubei Province (grant no.
2023BCB127) and the Major Project of National Science and
Technology (grant no. 2023ZD0506300).
References
|
1
|
Faubert B, Solmonson A and DeBerardinis
RJ: Metabolic reprogramming and cancer progression. Science.
368:eaaw54732020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Xia L, Oyang L, Lin J, Tan S, Han Y, Wu N,
Yi P, Tang L, Pan Q, Rao S, et al: The cancer metabolic
reprogramming and immune response. Mol Cancer. 20:282021.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Brand A, Singer K, Koehl GE, Kolitzus M,
Schoenhammer G, Thiel A, Matos C, Bruss C, Klobuch S, Peter K, et
al: LDHA-associated lactic acid production blunts tumor
immunosurveillance by T and NK cells. Cell Metab. 24:657–671. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu C, Wei W, Huang Y, Fu P, Zhang L and
Zhao Y: Metabolic reprogramming in septic acute kidney injury:
Pathogenesis and therapeutic implications. Metabolism.
158:1559742024. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Certo M, Tsai CH, Pucino V, Ho PC and
Mauro C: Lactate modulation of immune responses in inflammatory
versus tumour microenvironments. Nat Rev Immunol. 21:151–161. 2021.
View Article : Google Scholar
|
|
6
|
Rabinowitz JD and Enerbäck S: Lactate: The
ugly duckling of energy metabolism. Nat Metab. 2:566–571. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang D, Tang Z, Huang H, Zhou G, Cui C,
Weng Y, Liu W, Kim S, Lee S, Perez-Neut M, et al: Metabolic
regulation of gene expression by histone lactylation. Nature.
574:575–580. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yu J, Chai P, Xie M, Ge S, Ruan J, Fan X
and Jia R: Histone lactylation drives oncogenesis by facilitating
m6A reader protein YTHDF2 expression in ocular melanoma.
Genome Biol. 22:852021. View Article : Google Scholar
|
|
9
|
Wang Y, Li H, Jiang S, Fu D, Lu X, Lu M,
Li Y, Luo D, Wu K, Xu Y, et al: The glycolytic enzyme PFKFB3 drives
kidney fibrosis through promoting histone lactylation-mediated
NF-κB family activation. Kidney Int. 106:226–240. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang N, Wang W, Wang X, Mang G, Chen J,
Yan X, Tong Z, Yang Q, Wang M, Chen L, et al: Histone lactylation
boosts reparative gene activation post-myocardial infarction. Circ
Res. 131:893–908. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cui H, Xie N, Banerjee S, Ge J, Jiang D,
Dey T, Matthews QL, Liu RM and Liu G: Lung myofibroblasts promote
macrophage profibrotic activity through lactate-induced histone
lactylation. Am J Respir Cell Mol Biol. 64:115–125. 2021.
View Article : Google Scholar :
|
|
12
|
Schneider JL, Rowe JH, Garcia-de-Alba C,
Kim CF, Sharpe AH and Haigis MC: The aging lung: Physiology,
disease, and immunity. Cell. 184:1990–2019. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bueno M, Calyeca J, Rojas M and Mora AL:
Mitochondria dysfunction and metabolic reprogramming as drivers of
idiopathic pulmonary fibrosis. Redox Biol. 33:1015092020.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lin Z, Li J, Zhang J, Feng W, Lu J, Ma X,
Ding W, Ouyang S, Lu J, Yue P, et al: Metabolic reprogramming
driven by IGF2BP3 promotes acquired resistance to EGFR inhibitors
in non-small cell lung cancer. Cancer Res. 83:2187–2207. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ma Q, Jiang H, Ma L, Zhao G, Xu Q, Guo D,
He N, Liu H, Meng Z, Liu J, et al: The moonlighting function of
glycolytic enzyme enolase-1 promotes choline phospholipid
metabolism and tumor cell proliferation. Proc Natl Acad Sci USA.
120:e22094351202023. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
He Y, Ji Z, Gong Y, Fan L, Xu P, Chen X,
Miao J, Zhang K, Zhang W, Ma P, et al: Numb/Parkin-directed
mitochondrial fitness governs cancer cell fate via metabolic
regulation of histone lactylation. Cell Rep. 42:1120332023.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gao M, Zhang N and Liang W: Systematic
analysis of lysine lactylation in the plant fungal pathogen
botrytis cinerea. Front Microbiol. 11:5947432020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Meng X, Baine JM, Yan T and Wang S:
Comprehensive analysis of lysine lactylation in rice (Oryza sativa)
grains. J Agric Food Chem. 69:8287–8297. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lu Y, Liu L, Pan J, Luo B, Zeng H, Shao Y,
Zhang H, Guan H, Guo D, Zeng C, et al: MFG-E8 regulated by
miR-99b-5p protects against osteoarthritis by targeting chondrocyte
senescence and macrophage reprogramming via the NF-κB pathway. Cell
Death Dis. 12:5332021. View Article : Google Scholar
|
|
20
|
Yang Q, Liu J, Wang Y, Zhao W, Wang W, Cui
J, Yang J, Yue Y, Zhang S, Chu M, et al: A proteomic atlas of
ligand-receptor interactions at the ovine maternal-fetal interface
reveals the role of histone lactylation in uterine remodeling. J
Biol Chem. 298:1014562022. View Article : Google Scholar
|
|
21
|
Pan RY, He L, Zhang J, Liu X, Liao Y, Gao
J, Liao Y, Yan Y, Li Q, Zhou X, et al: Positive feedback regulation
of microglial glucose metabolism by histone H4 lysine 12
lactylation in Alzheimer's disease. Cell Metab. 34:634–648.e6.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yang YH, Wang QC, Kong J, Yang JT and Liu
JF: Global profiling of lysine lactylation in human lungs.
Proteomics. 23:e22004372023. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Neganova ME, Klochkov SG, Aleksandrova YR
and Aliev G: Histone modifications in epigenetic regulation of
cancer: Perspectives and achieved progress. Semin Cancer Biol.
83:452–471. 2022. View Article : Google Scholar
|
|
24
|
Park J, Lee K, Kim K and Yi SJ: The role
of histone modifications: From neurodevelopment to neurodiseases.
Signal Transduct Target Ther. 7:2172022. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Panwar V, Singh A, Bhatt M, Tonk RK,
Azizov S, Raza AS, Sengupta S, Kumar D and Garg M: Multifaceted
role of mTOR (mammalian target of rapamycin) signaling pathway in
human health and disease. Signal Transduct Target Ther. 8:3752023.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lavoie H, Gagnon J and Therrien M: ERK
signalling: A master regulator of cell behaviour, life and fate.
Nat Rev Mol Cell Biol. 21:607–632. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li J, Hou W, Zhao Q, Han W, Cui H, Xiao S,
Zhu L, Qu J, Liu X, Cong W, et al: Lactate regulates major zygotic
genome activation by H3K18 lactylation in mammals. Natl Sci Rev.
11:nwad2952023. View Article : Google Scholar
|
|
28
|
Pandkar MR, Sinha S, Samaiya A and Shukla
S: Oncometabolite lactate enhances breast cancer progression by
orchestrating histone lactylation-dependent c-Myc expression.
Transl Oncol. 37:1017582023. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang X, Ying T, Yuan J, Wang Y, Su X, Chen
S, Zhao Y, Zhao Y, Sheng J, Teng L, et al: BRAFV600E restructures
cellular lactylation to promote anaplastic thyroid cancer
proliferation. Endocr Relat Cancer. 30:e2203442023. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nian F, Qian Y, Xu F, Yang M, Wang H and
Zhang Z: LDHA promotes osteoblast differentiation through histone
lactylation. Biochem Biophys Res Commun. 615:31–35. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fanzani A, Giuliani R, Colombo F, Zizioli
D, Presta M, Preti A and Marchesini S: Overexpression of cytosolic
sialidase Neu2 induces myoblast differentiation in C2C12 cells.
FEBS Lett. 547:183–188. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dai W, Wu G, Liu K, Chen Q, Tao J, Liu H
and Shen M: Lactate promotes myogenesis via activating H3K9
lactylation-dependent up-regulation of Neu2 expression. J Cachexia
Sarcopenia Muscle. 14:2851–2865. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li L, Li Z, Meng X, Wang X, Song D, Liu Y,
Xu T, Qin J, Sun N, Tian K, et al: Histone lactylation-derived
LINC01127 promotes the self-renewal of glioblastoma stem cells via
the cis-regulating the MAP4K4 to activate JNK pathway. Cancer Lett.
579:2164672023. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sabu A, Liu TI, Ng SS, Doong RA, Huang YF
and Chiu HC: Nanomedicines targeting glioma stem cells. ACS Appl
Mater Interfaces. 15:158–181. 2023. View Article : Google Scholar
|
|
35
|
Dou X, Fu Q, Long Q, Liu S, Zou Y, Fu D,
Xu Q, Jiang Z, Ren X, Zhang G, et al: PDK4-dependent
hypercatabolism and lactate production of senescent cells promotes
cancer malignancy. Nat Metab. 5:1887–1910. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wiley CD and Campisi J: From ancient
pathways to aging cells-connecting metabolism and cellular
senescence. Cell Metab. 23:1013–1021. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wei L, Yang X, Wang J, Wang Z, Wang Q,
Ding Y and Yu A: H3K18 lactylation of senescent microglia
potentiates brain aging and Alzheimer's disease through the NFκB
signaling pathway. J Neuroinflammation. 20:2082023. View Article : Google Scholar
|
|
38
|
Jiang X, Yang Y, Li X, Li T, Yu T and Fu
X: Lactylation: An innovative approach to disease control. Aging
Dis. Sep 6–2024.Epub ahead of print.
|
|
39
|
Li X, Chen M, Chen X, He X, Li X, Wei H,
Tan Y, Min J, Azam T, Xue M, et al: TRAP1 drives smooth muscle cell
senescence and promotes atherosclerosis via HDAC3-primed histone H4
lysine 12 lactylation. Eur Heart J. 45:4219–4235. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Greene JT, Brian BF IV, Senevirathne SE
and Freedman TS: Regulation of myeloid-cell activation. Curr Opin
Immunol. 73:34–42. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bassler K, Schulte-Schrepping J,
Warnat-Herresthal S, Aschenbrenner AC and Schultze JL: The myeloid
cell compartment-cell by cell. Annu Rev Immunol. 37:269–293. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
De Leo A, Ugolini A, Yu X, Scirocchi F,
Scocozza D, Peixoto B, Pace A, D'Angelo L, Liu JKC, Etame AB, et
al: Glucose-driven histone lactylation promotes the
immunosuppressive activity of monocyte-derived macrophages in
glioblastoma. Immunity. 57:1105–1123.e8. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ma XM, Geng K, Wang P, Jiang Z, Law BYK
and Xu Y: MCT4-dependent lactate transport: A novel mechanism for
cardiac energy metabolism injury and inflammation in type 2
diabetes mellitus. Cardiovasc Diabetol. 23:962024. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ma W, Ao S, Zhou J, Li J, Liang X, Yang X,
Zhang H, Liu B, Tang W, Liu H, et al: Methylsulfonylmethane
protects against lethal dose MRSA-induced sepsis through promoting
M2 macrophage polarization. Mol Immunol. 146:69–77. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kolaczkowska E and Kubes P: Neutrophil
recruitment and function in health and inflammation. Nat Rev
Immunol. 13:159–175. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhou J, Xu W, Wu Y, Wang M, Zhang N, Wang
L, Feng Y, Zhang T, Wang L and Mao A: GPR37 promotes colorectal
cancer liver metastases by enhancing the glycolysis and histone
lactylation via Hippo pathway. Oncogene. 42:3319–3330. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Tong H, Jiang Z, Song L, Tan K, Yin X, He
C, Huang J, Li X, Jing X, Yun H, et al: Dual impacts of
serine/glycine-free diet in enhancing antitumor immunity and
promoting evasion via PD-L1 lactylation. Cell Metab.
36:2493–2510.e9. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Huang ZW, Zhang XN, Zhang L, Liu LL, Zhang
JW, Sun YX, Xu JQ, Liu Q and Long ZJ: STAT5 promotes PD-L1
expression by facilitating histone lactylation to drive
immunosuppression in acute myeloid leukemia. Signal Transduct
Target Ther. 8:3912023. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Sun T, Liu B, Li Y, Wu J, Cao Y, Yang S,
Tan H, Cai L, Zhang S, Qi X, et al: Oxamate enhances the efficacy
of CAR-T therapy against glioblastoma via suppressing
ectonucleotidases and CCR8 lactylation. J Exp Clin Cancer Res.
42:2532023. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhao Y, Jiang J, Zhou P, Deng K, Liu Z,
Yang M, Yang X, Li J, Li R and Xia J: H3K18 lactylation-mediated
VCAM1 expression promotes gastric cancer progression and metastasis
via AKT-mTOR-CXCL1 axis. Biochem Pharmacol. 222:1161202024.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Gu J, Zhou J, Chen Q, Xu X, Gao J, Li X,
Shao Q, Zhou B, Zhou H, Wei S, et al: Tumor metabolite lactate
promotes tumorigenesis by modulating MOESIN lactylation and
enhancing TGF-β signaling in regulatory T cells. Cell Rep.
39:1109862022. View Article : Google Scholar
|
|
52
|
Li HS, Zhou YN, Li L, Li SF, Long D, Chen
XL, Zhang JB, Feng L and Li YP: HIF-1α protects against oxidative
stress by directly targeting mitochondria. Redox Biol.
25:1011092019. View Article : Google Scholar
|
|
53
|
Zhao Y, Xing C, Deng Y, Ye C and Peng H:
HIF-1α signaling: Essential roles in tumorigenesis and implications
in targeted therapies. Genes Dis. 11:234–251. 2023. View Article : Google Scholar :
|
|
54
|
Zhao W, Wang Y, Liu J, Yang Q, Zhang S, Hu
X, Shi Z, Zhang Z, Tian J, Chu D and An L: Progesterone activates
the histone lactylation-Hif1α-glycolysis feedback loop to promote
decidualization. Endocrinology. 165:bqad1692023. View Article : Google Scholar
|
|
55
|
Wei S, Zhang J, Zhao R, Shi R, An L, Yu Z,
Zhang Q, Zhang J, Yao Y, Li H and Wang H: Histone lactylation
promotes malignant progression by facilitating USP39 expression to
target PI3K/AKT/HIF-1α signal pathway in endometrial carcinoma.
Cell Death Discov. 10:1212024. View Article : Google Scholar
|
|
56
|
Yang J, Luo L, Zhao C, Li X, Wang Z, Zeng
Z, Yang X, Zheng X, Jie H, Kang L, et al: A Positive feedback loop
between inactive VHL-triggered histone lactylation and PDGFRβ
signaling drives clear cell renal cell carcinoma progression. Int J
Biol Sci. 18:3470–3483. 2022. View Article : Google Scholar :
|
|
57
|
Chen J, Zhang M, Liu Y, Zhao S, Wang Y,
Wang M, Niu W, Jin F and Li Z: Histone lactylation driven by
mROS-mediated glycolytic shift promotes hypoxic pulmonary
hypertension. J Mol Cell Biol. 14:mjac0732023. View Article : Google Scholar :
|
|
58
|
Yang Z, Yan C, Ma J, Peng P, Ren X, Cai S,
Shen X, Wu Y, Zhang S, Wang X, et al: Lactylome analysis suggests
lactylation-dependent mechanisms of metabolic adaptation in
hepatocellular carcinoma. Nat Metab. 5:61–79. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Cheng Z, Huang H, Li M and Chen Y:
Proteomic analysis identifies PFKP lactylation in SW480 colon
cancer cells. iScience. 27:1086452023. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Meng Q, Zhang Y, Sun H, Yang X, Hao S, Liu
B, Zhou H, Wang Y and Xu ZX: Human papillomavirus-16 E6 activates
the pentose phosphate pathway to promote cervical cancer cell
proliferation by inhibiting G6PD lactylation. Redox Biol.
71:1031082024. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wang J, Yang P, Yu T, Gao M, Liu D, Zhang
J, Lu C, Chen X, Zhang X and Liu Y: Lactylation of PKM2 suppresses
inflammatory metabolic adaptation in pro-inflammatory macrophages.
Int J Biol Sci. 18:6210–6225. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Jia M, Yue X, Sun W, Zhou Q, Chang C, Gong
W, Feng J, Li X, Zhan R, Mo K, et al: ULK1-mediated metabolic
reprogramming regulates Vps34 lipid kinase activity by its
lactylation. Sci Adv. 9:eadg49932023. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Xie B, Zhang M, Li J, Cui J, Zhang P, Liu
F, Wu Y, Deng W, Ma J, Li X, et al: KAT8-catalyzed lactylation
promotes eEF1A2-mediated protein synthesis and colorectal
carcinogenesis. Proc Natl Acad Sci USA. 121:e23141281212024.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Cheng S, Chen L, Ying J, Wang Y, Jiang W,
Zhang Q, Zhang H, Wang J, Wang C, Wu H, et al: 20(S)-ginsenoside
Rh2 ameliorates ATRA resistance in APL by modulating
lactylation-driven METTL3. J Ginseng Res. 48:298–309. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Narita T, Weinert BT and Choudhary C:
Functions and mechanisms of non-histone protein acetylation. Nat
Rev Mol Cell Biol. 20:156–174. 2019. View Article : Google Scholar
|
|
66
|
Wang S, Osgood AO and Chatterjee A:
Uncovering post-translational modification-associated
protein-protein interactions. Curr Opin Struct Biol. 74:1023522022.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Li Q, Zhang F, Wang H, Tong Y, Fu Y, Wu K,
Li J, Wang C, Wang Z, Jia Y, et al: NEDD4 lactylation promotes APAP
induced liver injury through Caspase11 dependent non-canonical
pyroptosis. Int J Biol Sci. 20:1413–1435. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
An S, Yao Y, Hu H, Wu J, Li J, Li L, Wu J,
Sun M, Deng Z, Zhang Y, et al: PDHA1 hyperacetylation-mediated
lactate overproduction promotes sepsis-induced acute kidney injury
via Fis1 lactylation. Cell Death Dis. 14:4572023. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Zong Z, Xie F, Wang S, Wu X, Zhang Z, Yang
B and Zhou F: Alanyl-tRNA synthetase, AARS1, is a lactate sensor
and lactyltransferase that lactylates p53 and contributes to
tumorigenesis. Cell. 187:2375–2392.e33. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Wang X, Fan W, Li N, Ma Y, Yao M, Wang G,
He S, Li W, Tan J, Lu Q and Hou S: YY1 lactylation in microglia
promotes angiogenesis through transcription activation-mediated
upregulation of FGF2. Genome Biol. 24:872023. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Liao J, Chen Z, Chang R, Yuan T, Li G, Zhu
C, Wen J, Wei Y, Huang Z, Ding Z, et al: CENPA functions as a
transcriptional regulator to promote hepatocellular carcinoma
progression via cooperating with YY1. Int J Biol Sci. 19:5218–5232.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Wang Y, Chen L, Zhang M, Li X, Yang X,
Huang T, Ban Y, Li Y, Li Q, Zheng Y, et al: Exercise-induced
endothelial Mecp2 lactylation suppresses atherosclerosis via the
Ereg/MAPK signalling pathway. Atherosclerosis. 375:45–58. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Meng Q, Sun H, Zhang Y, Yang X, Hao S, Liu
B, Zhou H, Xu ZX and Wang Y: Lactylation stabilizes DCBLD1
activating the pentose phosphate pathway to promote cervical cancer
progression. J Exp Clin Cancer Res. 43:362024. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Wu Y and Gong P: Scopolamine regulates the
osteogenic differentiation of human periodontal ligament stem cells
through lactylation modification of RUNX2 protein. Pharmacol Res
Perspect. 12:e11692024. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Yang Z, Zheng Y and Gao Q: Lysine
lactylation in the regulation of tumor biology. Trends Endocrinol
Metab. 35:720–731. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Chen H, Li Y, Li H, Chen X, Fu H, Mao D,
Chen W, Lan L, Wang C, Hu K, et al: NBS1 lactylation is required
for efficient DNA repair and chemotherapy resistance. Nature.
631:663–669. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Shvedunova M and Akhtar A: Modulation of
cellular processes by histone and non-histone protein acetylation.
Nat Rev Mol Cell Biol. 23:329–349. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Niu Z, Chen C, Wang S, Lu C, Wu Z, Wang A,
Mo J, Zhang J, Han Y, Yuan Y, et al: HBO1 catalyzes lysine
lactylation and mediates histone H3K9la to regulate gene
transcription. Nat Commun. 15:35612024. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Li F, Si W, Xia L, Yin D, Wei T, Tao M,
Cui X, Yang J, Hong T and Wei R: Positive feedback regulation
between glycolysis and histone lactylation drives oncogenesis in
pancreatic ductal adenocarcinoma. Mol Cancer. 23:902024. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Moreno-Yruela C, Zhang D, Wei W, Bæk M,
Liu W, Gao J, Danková D, Nielsen AL, Bolding JE, Yang L, et al:
Class I histone deacetylases (HDAC1-3) are histone lysine
delactylases. Sci Adv. 8:eabi66962022. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Dai SK, Liu PP, Li X, Jiao LF, Teng ZQ and
Liu CM: Dynamic profiling and functional interpretation of histone
lysine crotonylation and lactylation during neural development.
Development. 149:dev2000492022. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Hu X, Huang X, Yang Y, Sun Y, Zhao Y,
Zhang Z, Qiu D, Wu Y, Wu G and Lei L: Dux activates
metabolism-lactylation-MET network during early iPSC reprogramming
with Brg1 as the histone lactylation reader. Nucleic Acids Res.
52:5529–5548. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Yang L, Niu K, Wang J, Shen W, Jiang R,
Liu L, Song W, Wang X, Zhang X, Zhang R, et al: Nucleolin
lactylation contributes to intrahepatic cholangiocarcinoma
pathogenesis via RNA splicing regulation of MADD. J Hepatol.
81:651–666. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Huang H, Wang S, Xia H, Zhao X, Chen K,
Jin G, Zhou S, Lu Z, Chen T, Yu H, et al: Lactate enhances NMNAT1
lactylation to sustain nuclear NAD+ salvage pathway and
promote survival of pancreatic adenocarcinoma cells under
glucose-deprived conditions. Cancer Lett. 588:2168062024.
View Article : Google Scholar
|
|
85
|
Fan M, Yang K, Wang X, Chen L, Gill PS, Ha
T, Liu L, Lewis NH, Williams DL and Li C: Lactate promotes
endothelial-to-mesenchymal transition via Snail1 lactylation after
myocardial infarction. Sci Adv. 9:eadc94652023. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Wang YH, Gao P, Wang YQ, Xu LZ, Zeng KW
and Tu PF: Small-molecule targeting PKM2 provides a molecular basis
of lactylation-dependent fibroblast-like synoviocytes proliferation
inhibition against rheumatoid arthritis. Eur J Pharmacol.
972:1765512024. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Mao Y, Zhang J, Zhou Q, He X, Zheng Z, Wei
Y, Zhou K, Lin Y, Yu H, Zhang H, et al: Hypoxia induces
mitochondrial protein lactylation to limit oxidative
phosphorylation. Cell Res. 34:13–30. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Sun L, Zhang Y, Yang B, Sun S, Zhang P,
Luo Z, Feng T, Cui Z, Zhu T, Li Y, et al: Lactylation of METTL16
promotes cuproptosis via m6A-modification on FDX1 mRNA
in gastric cancer. Nat Commun. 14:65232023. View Article : Google Scholar
|
|
89
|
Jin J, Bai L, Wang D, Ding W, Cao Z, Yan
P, Li Y, Xi L, Wang Y, Zheng X, et al: SIRT3-dependent
delactylation of cyclin E2 prevents hepatocellular carcinoma
growth. EMBO Rep. 24:e560522023. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Zhang XW, Li L, Liao M, Liu D, Rehman A,
Liu Y, Liu ZP, Tu PF and Zeng KW: Thermal proteome profiling
strategy identifies CNPY3 as a cellular target of gambogic acid for
inducing prostate cancer pyroptosis. J Med Chem. 67:10005–10011.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Zhang N, Zhang Y, Xu J, Wang P, Wu B, Lu
S, Lu X, You S, Huang X, Li M, et al: α-myosin heavy chain
lactylation maintains sarcomeric structure and function and
alleviates the development of heart failure. Cell Res. 33:679–698.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Li MY, Liu LZ and Dong M: Progress on
pivotal role and application of exosome in lung cancer
carcinogenesis, diagnosis, therapy and prognosis. Mol Cancer.
20:222021. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Thai AA, Solomon BJ, Sequist LV, Gainor JF
and Heist RS: Lung cancer. Lancet. 398:535–554. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Jiang J, Huang D, Jiang Y, Hou J, Tian M,
Li J, Sun L, Zhang Y, Zhang T, Li Z, et al: Lactate modulates
cellular metabolism through histone lactylation-mediated gene
expression in non-small cell lung cancer. Front Oncol.
11:6475592021. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Yan F, Teng Y, Li X, Zhong Y, Li C, Yan F
and He X: Hypoxia promotes non-small cell lung cancer cell
stemness, migration, and invasion via promoting glycolysis by
lactylation of SOX9. Cancer Biol Ther. 25:23041612024. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Hirsch FR, Scagliotti GV, Mulshine JL,
Kwon R, Curran WJ Jr, Wu YL and Paz-Ares L: Lung cancer: Current
therapies and new targeted treatments. Lancet. 389:299–311. 2017.
View Article : Google Scholar
|
|
97
|
Ruprecht JJ and Kunji ERS: Structural
mechanism of transport of mitochondrial carriers. Annu Rev Biochem.
90:535–558. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Zheng P, Mao Z, Luo M, Zhou L, Wang L, Liu
H, Liu W and Wei S: Comprehensive bioinformatics analysis of the
solute carrier family and preliminary exploration of SLC25A29 in
lung adenocarcinoma. Cancer Cell Int. 23:2222023. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Herbst RS, Morgensztern D and Boshoff C:
The biology and management of non-small cell lung cancer. Nature.
553:446–454. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Hao B, Dong H, Xiong R, Song C, Xu C, Li N
and Geng Q: Identification of SLC2A1 as a predictive biomarker for
survival and response to immunotherapy in lung squamous cell
carcinoma. Comput Biol Med. 171:1081832024. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Steeg PS, Camphausen KA and Smith QR:
Brain metastases as preventive and therapeutic targets. Nat Rev
Cancer. 11:352–363. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Liu W, Song J, Du X, Zhou Y, Li Y, Li R,
Lyu L, He Y, Hao J, Ben J, et al: AKR1B10 (Aldo-keto reductase
family 1 B10) promotes brain metastasis of lung cancer cells in a
multi-organ microfluidic chip model. Acta Biomater. 91:195–208.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Duan W, Liu W, Xia S, Zhou Y, Tang M, Xu
M, Lin M, Li X and Wang Q: Warburg effect enhanced by AKR1B10
promotes acquired resistance to pemetrexed in lung cancer-derived
brain metastasis. J Transl Med. 21:5472023. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Feller-Kopman D and Light R: Pleural
disease. N Engl J Med. 378:740–751. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Morgensztern D, Waqar S, Subramanian J,
Trinkaus K and Govindan R: Prognostic impact of malignant pleural
effusion at presentation in patients with metastatic non-small-cell
lung cancer. J Thorac Oncol. 7:1485–1489. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Zamboni MM, da Silva CT Jr, Baretta R,
Cunha ET and Cardoso GP: Important prognostic factors for survival
in patients with malignant pleural effusion. BMC Pulm Med.
15:292015. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Wang ZH, Peng WB, Zhang P, Yang XP and
Zhou Q: Lactate in the tumour microenvironment: From immune
modulation to therapy. EBioMedicine. 73:1036272021. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Niu Y and Zhou Q: Th17 cells and their
related cytokines: Vital players in progression of malignant
pleural effusion. Cell Mol Life Sci. 79:1942022. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Wang ZH, Zhang P, Peng WB, Ye LL, Xiang X,
Wei XS, Niu YR, Zhang SY, Xue QQ, Wang HL and Zhou Q: Altered
phenotypic and metabolic characteristics of
FOXP3+CD3+CD56+ natural killer T
(NKT)-like cells in human malignant pleural effusion.
Oncoimmunology. 12:21605582022. View Article : Google Scholar
|
|
110
|
Liu GY, Budinger GRS and Dematte JE:
Advances in the management of idiopathic pulmonary fibrosis and
progressive pulmonary fibrosis. BMJ. 377:e0663542022. View Article : Google Scholar
|
|
111
|
Noble PW, Barkauskas CE and Jiang D:
Pulmonary fibrosis: Patterns and perpetrators. J Clin Invest.
122:2756–2762. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Wynn TA and Vannella KM: Macrophages in
tissue repair, regeneration, and fibrosis. Immunity. 44:450–462.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Okabe Y and Medzhitov R: Tissue biology
perspective on macrophages. Nat Immunol. 17:9–17. 2016. View Article : Google Scholar
|
|
114
|
Vannella KM and Wynn TA: Mechanisms of
organ injury and repair by macrophages. Annu Rev Physiol.
79:593–617. 2017. View Article : Google Scholar
|
|
115
|
Byrne AJ, Maher TM and Lloyd CM: Pulmonary
macrophages: A new therapeutic pathway in fibrosing lung disease?
Trends Mol Med. 22:303–316. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Guth AM, Janssen WJ, Bosio CM, Crouch EC,
Henson PM and Dow SW: Lung environment determines unique phenotype
of alveolar macrophages. Am J Physiol Lung Cell Mol Physiol.
296:L936–L946. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Li J, Zeng G, Zhang Z, Wang Y, Shao M, Li
C, Lu Z, Zhao Y, Zhang F and Ding W: Urban airborne
PM2.5 induces pulmonary fibrosis through triggering
glycolysis and subsequent modification of histone lactylation in
macrophages. Ecotoxicol Environ Saf. 273:1161622024. View Article : Google Scholar
|
|
118
|
Kumar M, Jha A, Bharti K, Parmar G and
Mishra B: Advances in lipid-based pulmonary nanomedicine for the
management of inflammatory lung disorders. Nanomedicine (Lond).
17:913–934. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Jin H, Luo R, Li J, Zhao H, Ouyang S, Yao
Y, Chen D, Ling Z, Zhu W, Chen M, et al: Inhaled platelet
vesicle-decoyed biomimetic nanoparticles attenuate inflammatory
lung injury. Front Pharmacol. 13:10502242022. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Chen N, Xie QM, Song SM, Guo SN, Fang Y,
Fei GH and Wu HM: Dexamethasone protects against asthma via
regulating Hif-1α-glycolysis-lactate axis and protein lactylation.
Int Immunopharmacol. 131:1117912024. View Article : Google Scholar
|
|
121
|
Smith LM and Kelleher NL; Consortium for
Top Down Proteomics: Proteoform: A single term describing protein
complexity. Nat Methods. 10:186–187. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Zhao H, Dennery PA and Yao H: Metabolic
reprogramming in the pathogenesis of chronic lung diseases,
including BPD, COPD, and pulmonary fibrosis. Am J Physiol Lung Cell
Mol Physiol. 314:L544–L554. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Faubert B, Li KY, Cai L, Hensley CT, Kim
J, Zacharias LG, Yang C, Do QN, Doucette S, Burguete D, et al:
Lactate metabolism in human lung tumors. Cell. 171:358–371.e9.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Pokharel MD, Marciano DP, Fu P, Franco MC,
Unwalla H, Tieu K, Fineman JR, Wang T and Black SM: Metabolic
reprogramming, oxidative stress, and pulmonary hypertension. Redox
Biol. 64:1027972023. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Babic M, Veljovic K, Popović N, Golic N,
Radojkovic D and Stankovic M: Antioxidant effect of lactic acid
bacteria in human bronchial epithelial cells exposed to cigarette
smoke. J Appl Microbiol. 134:lxad2572023. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Liu X, Wang G, You Z, Qian P, Chen H, Dou
Y, Wei Z, Chen Y, Mao C and Zhang J: Inhibition of hypoxia-induced
proliferation of pulmonary arterial smooth muscle cells by a mTOR
siRNA-loaded cyclodextrin nanovector. Biomaterials. 35:4401–4416.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Colegio OR, Chu NQ, Szabo AL, Chu T,
Rhebergen AM, Jairam V, Cyrus N, Brokowski CE, Eisenbarth SC,
Phillips GM, et al: Functional polarization of tumour-associated
macrophages by tumour-derived lactic acid. Nature. 513:559–563.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
MacDonald MI, Polkinghorne KR, MacDonald
CJ, Leong P, Hamza K, Kathriachchige G, Osadnik CR, King PT and
Bardin PG: Elevated blood lactate in COPD exacerbations associates
with adverse clinical outcomes and signals excessive treatment with
β2-agonists. Respirology. 28:860–868. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Pacl HT, Chinta KC, Reddy VP, Nadeem S,
Sevalkar RR, Nargan K, Lumamba K, Naidoo T, Glasgow JN, Agarwal A
and Steyn AJC: NAD(H) homeostasis underlies host protection
mediated by glycolytic myeloid cells in tuberculosis. Nat Commun.
14:54722023. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Huang JJ, Yang XQ, Zhuo ZQ and Yuan L:
Clinical characteristics of plastic bronchitis in children: A
retrospective analysis of 43 cases. Respir Res. 23:512022.
View Article : Google Scholar : PubMed/NCBI
|

















