Introduction
Lung cancer is the leading cause of cancer-related
mortality worldwide (1) and 85% of
the cases are represented by non-small cell lung cancer (NSCLC),
which is classified into three different histological subtypes:
adenocarcinoma (ADC), squamous cell carcinoma (SCC) and large cell
carcinoma (LCC) (2–4). Despite a better understanding of the
NSCLC pathogenesis and significant improvement in early diagnosis
and treatment, the overall 5-year survival is extremely low (~15%)
and the patients show high recurrence rates even at the early
disease stages (1–7), highlighting the necessity of a deeper
knowledge of NSCLC biology and the identification of more effective
biomarkers.
MicroRNAs (miRNAs) are a highly conserved family of
small (17–22 nucleotides), non-coding, endogenous, single-stranded
RNA molecules that negatively regulate gene expression by binding
to complementary sequences on target messenger RNA (mRNA) (8,9).
Recently, miRNAs have been shown to regulate essential cell
processes, such as cell proliferation, differentiation, apoptosis,
development and metabolism (9–11),
and to play a key role in cancer pathogenesis (12–16).
Moreover, a miRNA prognostic and diagnostic value has been reported
in several malignancies, including lung cancer (14–16).
The first reported miRNA aberrantly expressed in
lung cancer was the Let-7 family (17). A reduced Let-7 expression
has been significantly correlated with a short post-operative
survival in the NSCLC patients (17). Moreover, the ectopic Let-7
expression inhibits cell proliferation in human NSCLC cell lines
(18) and reduces tumor burden in
mouse NSCLC xenografts (19).
Let-7 family members have been demonstrated to behave as
tumor suppressor genes and to functionally inhibit several cell
cycle regulators and oncogenes, such as Ras family,
c-Myc and HMGA2 genes, whose 3′UTRs show multiple
Let-7 binding sites (13,20,21).
Conversely, a role as oncogene has been suggested
for miR-21 that is deregulated in glioblastoma and lung
cancer (22–25). A miR-21 overexpression has
been suggested to be an independent negative prognostic factor for
the overall survival in NSCLC patients (23) and to be related to the lung
carcinogenesis in never smokers (26). Several mRNAs have been identified
as miR-21 targets, including PDCD4, PTEN, TGF-β and
MMP9 genes (22,27).
The aim of this study was to evaluate Let-7g
and miR-21 expression in a series of 80 NSCLC patients to
establish their involvement in the NSCLC pathogenesis and their
potential diagnostic, prognostic and predictive value.
Materials and methods
Patients
Eighty NSCLC patients were retrospectively selected
from patients who had undergone surgery at the Unit of Thoracic
Surgery of the A.O.U.P. between 2005 and 2012. Histological
diagnoses were independently formulated by two pathologists (G.F.
and G.A.) according to the World Health Organization classification
(2–4) and discrepant diagnoses were
re-evaluated and discussed until an agreement was reached.
Clinicopathological characteristics were collected whenever
available for all the patients, while detailed clinical data were
obtained only for 55 patients. The study was approved by the local
Ethics Committee and all the patients gave their informed consent
to the molecular analyses.
DNA and RNA isolation
DNA and RNA were isolated from 10-μm sections of
formalin-fixed and paraffin-embedded (FFPE) tissues or cytological
specimens after manual tumor macrodissection using the QIAamp DNA
Mini kit (Qiagen) and miRNeasy FFPE kit (Qiagen), respectively,
according to the manufacturer's instructions.
MiRNA expression
Quantification of Let-7g, miR-21 and
RNU6B expression was carried out in triplicate into 80 NSCLC
and 27 non-cancer lung tissues using specific TaqMan®
MicroRNA assays (Applied Biosystems) according to the
manufacturer's instructions. Briefly, 10 ng of total RNA were
retro-transcribed by the TaqMan MicroRNA Reverse Transcription (RT)
kit (Applied Biosystems) and 1.3 μl of RT product were analysed by
quantitative real-time PCR (qRT-PCR) on the Rotor-Gene 6000
(Corbett Research). Threshold cycle (Ct) and baselines were
determined by manual settings. MiRNA expression was calculated by
relative quantification and fold expression changes were determined
by the 2−ΔΔCt method using the DataAssist™ software
(Applied Biosystems).
Target prediction and pathway
analysis
Let-7g and miR-21 target genes were
predicted by four different miRNA target prediction algorithms:
miRanda (http://www.microrna.org/microrna), TargetScan
(http://www.targetscan.org), Pictar
(http://www.pictar.org) and miRDB (http://www.mirdb.org). Gene ontology classification
and pathway analysis were performed using the PANTHER software
(http://www.pantherdb.org).
Mutational analysis
K-Ras gene (Reference sequence:
ENSG00000133703) status in codons 12 and 13 was analyzed by
pyrosequencing using the Anti-EGFR MoAb response® kit
(K-Ras status) (Diatech Pharmacogenetics) according to the
manufacturer's instructions.
PCR-single stranded conformation polymorphism
(PCR-SSCP) and sequencing analysis were used for genotyping exons
18–21 of the EGFR gene (Reference sequence:
ENSG00000146648). The primer sequences were as follows: exon 18,
5′-CTCTGTGTTCTTGTCCCCCC-3′ (forward) and 5′-GCCTGTGCCAGGGACCTTAC-3′
(reverse); exon 19, 5′-CATGTGGCACCATCTCACA-3′ (forward) and
5′-CCACACAGCAAAGCAGAAAC-3′ (reverse); exon 20,
5′-CACACTGACGTGCCTCTCC-3′ (forward) and 5′-TATCTCCCCTCCCCGTATCT-3′
(reverse); exon 21, 5′-CCTCACAGCAGGGTCTTCTC-3′ (forward) and
5′-CCTGGTGTCAGGAAAATGCT-3′ (reverse). Briefly, 100 ng of DNA were
amplified by PCR using the FastStart Taq DNA Polymerase (Roche
Diagnostics) on the T3000 Thermocycler 48 (Biometra), as follows: 4
min at 95°C, 40 cycles at 95°C for 30 sec, 58°C for 30 sec and 72°C
for 45 sec and 10 min at 72°C. PCR products were mixed with an
equivalent formamide volume, denatured at 95°C for 5 min and run
onto a non-denaturing 12.5% polyacrylamide gel (GE Healthcare) at
18°C and constant 25 mA for 1 h and 40 min. Denaturated DNA was
visualized by the PlusOne DNA silver staining kit (GE Healthcare)
and samples with altered mobility patterns were sequenced as
previously described (28).
Statistical analysis
One-way analysis of variance and χ2 test
were used to determine the association between miRNA expression and
the different parameters, while survival analysis was performed by
the Kaplan-Meier method. Statistical analyses were performed using
the JMP10 software (SAS) and a two-tailed p<0.05 was considered
statistically significant.
Results
Patient characteristics
This study was conducted in 80 patients with NSCLC,
including 55 ADCs, 21 SCCs, 2 LCCs and 2 undifferentiated NSCLCs.
The median age at diagnosis was 67 years (range 46–85) and the
median follow-up was 32 months (range 7–98). Disease progression
with distant and/or loco-regional recurrence and death from lung
cancer were observed in 34 (61.8%) and 14 (25.5%) of the 55 NSCLC
patients, respectively. The median progression-free survival (PFS)
and overall survival (OS) were 18 months (95% CI, 14–24) and 24
months (95% CI, 18–30), respectively.
Let-7g and miR-21 expression profile
We quantified the mature Let-7g and
miR-21 expression normalized to the RNU6B endogenous
control in 80 NSCLC and 27 non-cancer lung tissues. The
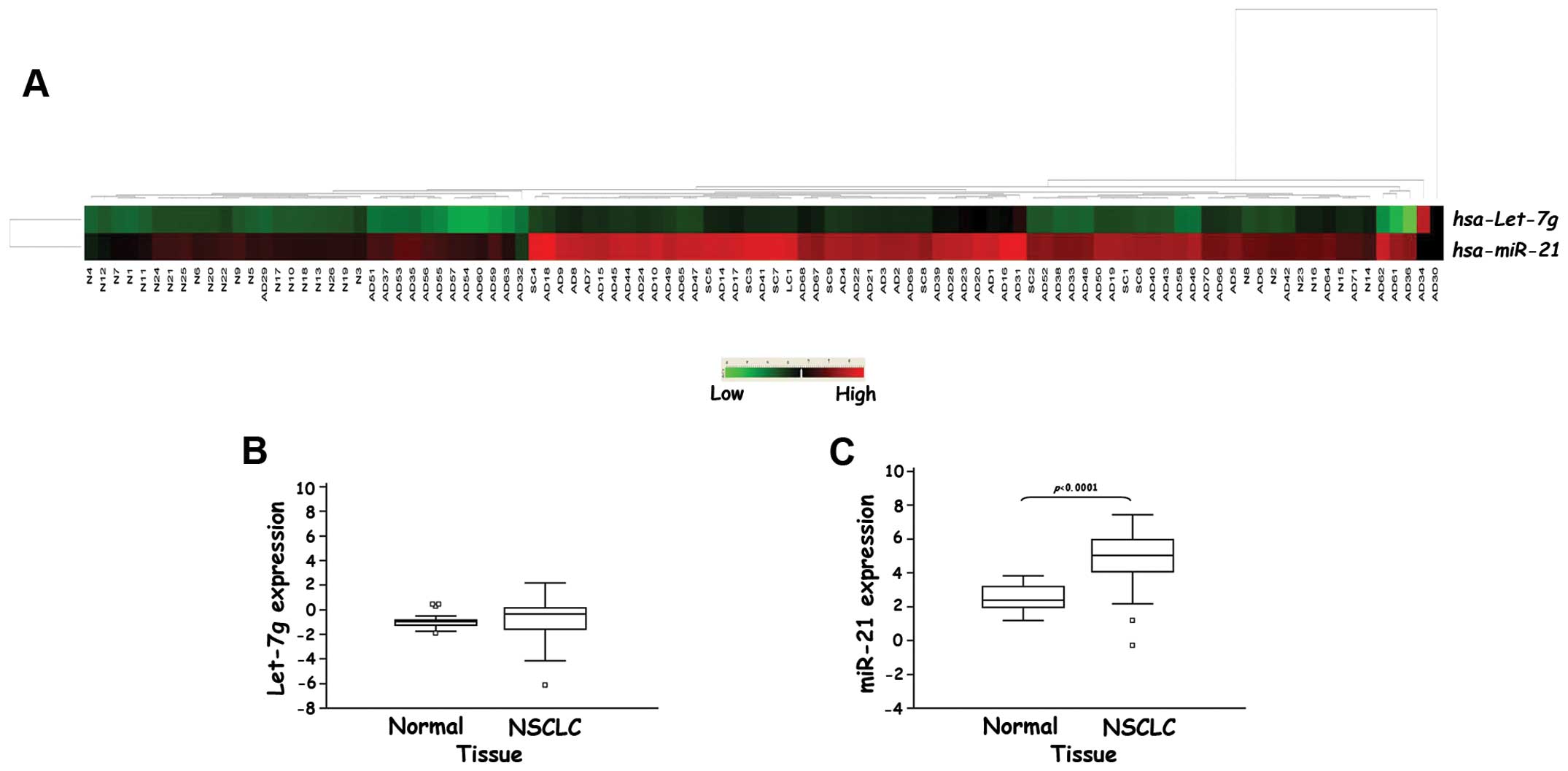
unsupervised hierarchical clustering analysis of miRNA expression
using the Euclidean distance as a similarity measure and average
linkage algorithm revealed two major clusters based on similarities
in miR-21 expression that clearly separated the tumor from
non-cancer tissues. On the contrary, we did not observe a clear
separation between tumor and normal samples based on Let-7g
expression (Fig. 1A).
Let-7g was barely detectable in lung tissues
and we did not observe any significant difference between the NSCLC
and normal samples (−0.897±0.148 vs. −0.709±0.168, p=0.585,
Fig. 1B). Conversely, a highly
significant increase in miR-21 expression was observed in
the NSCLC tissues compared to the non-cancer ones (4.842±0.163 vs.
2.509±0.182, p<0.0001, Fig.
1C).
MiRNA profile and clinicopathological
characteristics
To determine whether miRNA profile was correlated
with the main clinicopathological characteristics, the NSCLC
patients were divided into Let-7g and miR-21 high and
low expression groups based on the median fold-change values
(1.315±0.175 for Let-7g and 6.964±0.759 for miR-21).
Except for a significant association between the low Let-7g
expression and metastatic lymph node presence at diagnosis
(p=0.046), no other statistically significant associations were
observed between the analysed miRNA and the main
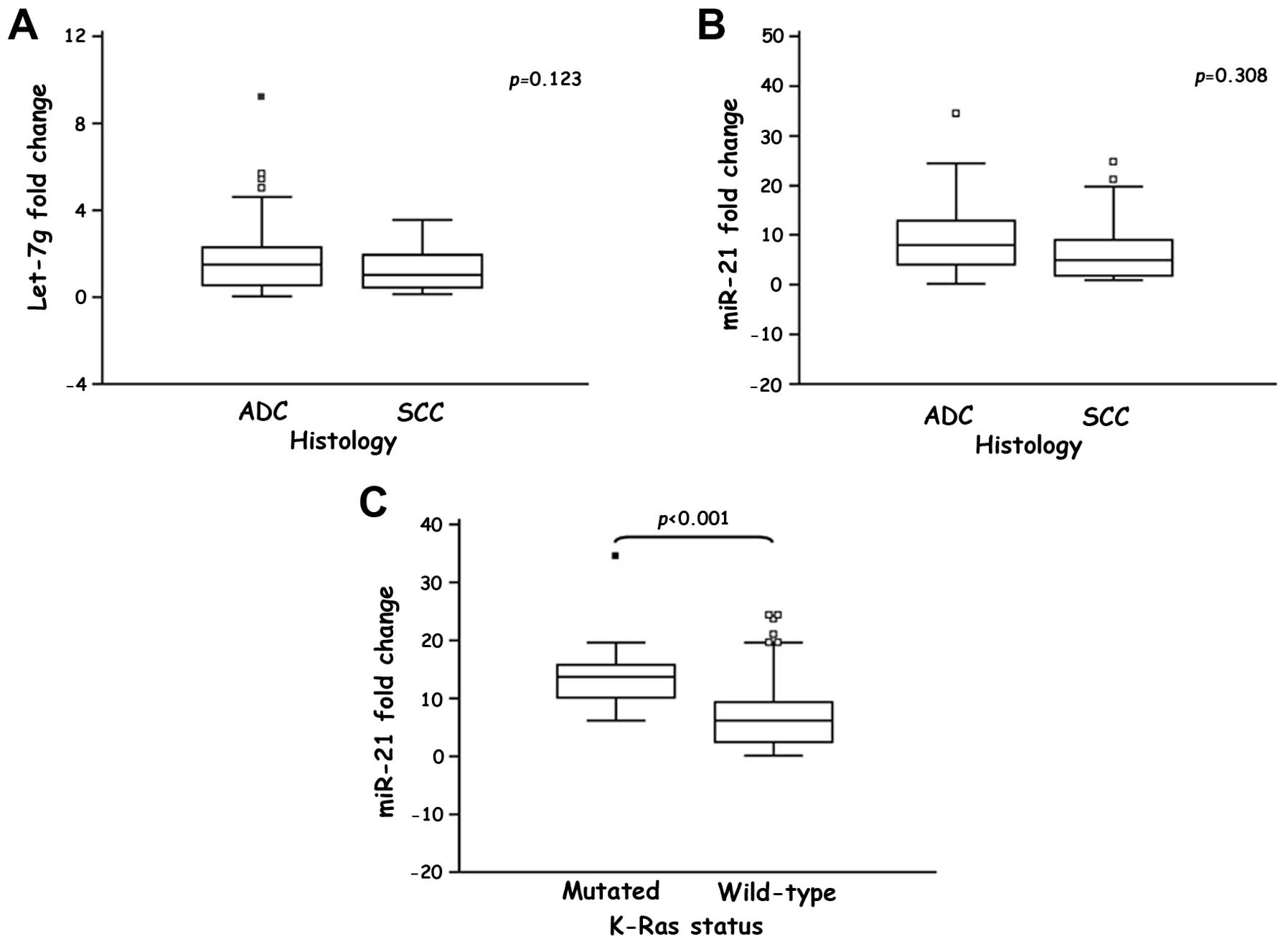
clinicopathological characteristics of the NSCLC patients (Table I). Interestingly, both
Let-7g and miR-21 were upregulated in ADCs compared
to SCCs, although these relationships were not statistically
significant (Fig. 2A and B).
 | Table ICorrelations between the
Let-7g and miR-21 expression and the main
clinicopathological characteristics of the NSCLC patients. |
Table I
Correlations between the
Let-7g and miR-21 expression and the main
clinicopathological characteristics of the NSCLC patients.
| Let-7g
expressiona | | miR-21
expressiona | |
|---|
|
| |
| |
|---|
| Characteristic | Low | High | p-valueb | Low | High | p-valueb |
|---|
| Age |
| ≤67 years | 22 (57.9) | 16 (42.1) | 0.263 | 19 (50) | 19 (50) | 0.823 |
| >67 years | 18 (42.8) | 24 (57.2) | | 21 (50) | 21 (50) | |
| Gender |
| Males | 28 (50.9) | 27 (49.1) | 0.809 | 27 (49.1) | 28 (50.9) | 0.809 |
| Females | 12 (48) | 13 (52) | | 13 (52) | 12 (48) | |
| Histology |
| ADC | 27 (49.1) | 28 (50.9) | 0.157 | 24 (43.6) | 31 (56.4) | 0.065 |
| SCC | 13 (61.9) | 8 (38.1) | | 14 (66.7) | 7 (33.3) | |
| LCC | 0 (0) | 2 (100) | | 2 (100) | 0 (0) | |
| Others | 0 (0) | 2 (100) | | 0 (0) | 2 (100) | |
| Tumor stage |
| T1 (T1a-T1b) | 6 (50) | 6 (50) | 0.111 | 5 (41.7) | 7 (58.3) | 0.793 |
| T2 (T2a-T2b) | 16 (51.6) | 15 (48.4) | | 13 (41.9) | 18 (58.1) | |
| T3 | 2 (14.3) | 12 (85.7) | | 8 (57.2) | 6 (42.8) | |
| T4 | 3 (50) | 3 (50) | | 3 (50) | 3 (50) | |
| Lymph node
status |
| Negative | 4 (21.1) | 15 (78.9) | 0.046 | 9 (47.4) | 10 (52.6) | 0.633 |
| Positive | 20 (52.6) | 18 (47.4) | | 14 (36.8) | 24 (63.2) | |
| Smoking |
| Never smoking | 7 (53.8) | 6 (46.2) | 0.425 | 7 (53.8) | 6 (46.2) | 0.872 |
| Former
smoking | 17 (65.4) | 9 (34.6) | | 16 (61.5) | 10 (38.5) | |
| Current
smoking | 2 (100) | 0 (0) | | 1 (50) | 1 (50) | |
| Performance
status |
| ECOG 0 | 5 (45.5) | 6 (54.5) | 0.396 | 5 (45.5) | 6 (54.5) | 0.149 |
| ECOG 1 | 20 (69) | 9 (31) | | 18 (62.1) | 11 (37.9) | |
| ECOG 2 | 1 (50) | 1 (50) | | 2 (100) | 0 (0) | |
| TKI response |
| Complete
response | 1 (100) | 0 (0) | 0.181 | 1 (100) | 0 (0) | 0.218 |
| Partial
response | 13 (72.2) | 5 (27.8) | | 10 (55.6) | 8 (44.4) | |
| Stable
disease | 7 (70) | 3 (30) | | 6 (60) | 4 (40) | |
| Progressive
disease | 5 (55.6) | 4 (44.4) | | 7 (77.8) | 2 (22.2) | |
| Disease
recurrence |
| NED | 10 (47.6) | 11 (52.4) | 0.072 | 8 (38.1) | 13 (61.9) | 0.189 |
| Recurrence | 20 (58.8) | 14 (41.2) | | 21 (61.8) | 13 (38.2) | |
MiRNA profile and K-Ras and EGFR
status
To investigate whether Let-7g and
miR-21 expression was correlated to K-Ras and
EGFR mutational status, we performed genotyping in the 80
NSCLC patients, except 2, due to insufficient tissue. K-Ras
and EGFR mutations were observed in 16 (20.5%) and 23
(29.5%) of the 78 NSCLC patients, respectively (Table II).
 | Table IIEGFR and K-Ras
mutational status in the NSCLC patients. |
Table II
EGFR and K-Ras
mutational status in the NSCLC patients.
| Gene | Exon | ID sample | Nucleotide
substitution | Amino acid
substitution |
|---|
| EGFR | 19 | AD20, AD23, AD24,
AD26, AD32, AD39 | c.2235_2249del | p.E746_A750 |
| EGFR | 19 | AD28, AD29, AD35,
AD40 | c.2236_2250del | p.E746_A750 |
| EGFR | 19 | AD16, AD21,
AD37 |
c.2237_2255delinsT |
p.E746_S752delinsV |
| EGFR | 19 | AD22 |
c.2239_2264delinsGCCAA |
p.L747_A755delinsAN |
| EGFR | 19 | AD25 | c.2240_2257del |
p.L747_P753delinsS |
| EGFR | 19+20 | AD27 |
c.2235_2249del+c.2369C>T |
p.E746_A750+p.T790M |
| EGFR | 20 | AD41 |
c.2311_2312insGCGTGGACA |
p.D770_N771insSVD |
| EGFR | 20 | AD36 | c.2353A>C | p.T785P |
| EGFR | 21 | AD7 | c.2570G>A | p.G857E |
| EGFR | 21 | AD31, AD33, AD38,
AD42 | c.2573T>G | p.L858R |
| K-Ras | 2 | AD9, AD10, AD15,
AD17, AD18, AD44 | c.34G>T | p.G12C |
| K-Ras | 2 | AD65 |
c.34_35GG>TT | p.G12F |
| K-Ras | 2 | LC1, AD19, AD45,
AD46 | c.35G>T | p.G12V |
| K-Ras | 2 | AD1, AD14,
AD62 | c.35G>C | p.G12A |
| K-Ras | 2 | AD61 | c.35G>A | p.G12D |
| K-Ras | 2 | AD43 |
c.37_38GG>CC | p.G13P |
K-Ras and EGFR mutations were mutually
exclusive, observed only in the NSCLC patients with ADC and
associated with gender (Table
III). As is shown in Table
III, EGFR status was also significantly associated with
the smoking habit (p=0.0086), performance status (p=0.0008) and
response to the treatment with EGFR tyrosine kinase
inhibitors (TKIs) (p=0.0076).
 | Table IIIEGFR and K-Ras status
in relation to the main clinicopathological and biological
characteristics of the NSCLC patients. |
Table III
EGFR and K-Ras status
in relation to the main clinicopathological and biological
characteristics of the NSCLC patients.
| EGFR
statusa | | K-Ras
statusa | |
|---|
|
| |
| |
|---|
| Characteristic | Wild-type | Mutated | p-valueb | Wild-type | Mutated | p-valueb |
|---|
| EGFR
status |
| Wild-type | | | | 39 (70.9) | 16 (29.1) | 0.0095 |
| Mutated | | | | 23 (100) | 0 (0) | |
| Age |
| ≤67 years | 24 (43.6) | 12 (52.2) | 0.6595 | 25 (40.3) | 11 (68.8) | 0.0797 |
| >67 years | 31 (56.4) | 11 (47.8) | | 37 (59.7) | 5 (31.2) | |
| Gender |
| Males | 47 (85.5) | 7 (30.4) |
<0.0001 | 39 (62.9) | 15 (93.8) | 0.0376 |
| Females | 8 (14.5) | 16 (69.6) | | 23 (37.1) | 1 (6.2) | |
| Histology |
| ADC | 32 (58.2) | 22 (95.7) | 0.0049 | 39 (62.9) | 15 (93.8) | 0.0369 |
| SCC | 20 (36.4) | 0 (0) | | 20 (32.3) | 0 (0) | |
| LCC | 2 (3.6) | 0 (0) | | 2 (3.2) | 0 (0) | |
| Others | 1 (1.8) | 1 (4.3) | | 1 (1.6) | 1 (6.2) | |
| Tumor stage |
| T1 (T1a-T1b) | 9 (20.0) | 3 (16.7) | 0.1511 | 9 (18.4) | 3 (21.4) | 0.3189 |
| T2 (T2a-T2b) | 20 (44.4) | 11 (61.1) | | 27 (55.1) | 4 (28.6) | |
| T3 | 13 (28.9) | 1 (5.5) | | 9 (18.4) | 5 (35.7) | |
| T4 | 3 (6.7) | 3 (16.7) | | 4 (8.1) | 2 (14.3) | |
| Lymph node
status |
| Negative | 16 (39) | 3 (18.8) | 0.2516 | 15 (34.1) | 4 (30.8) | 0.9111 |
| Positive | 25 (61) | 13 (81.2) | | 29 (65.9) | 9 (69.2) | |
| Smoking |
| Never smoking | 4 (15.4) | 9 (60) | 0.0086 | 12 (35.3) | 1 (14.3) | 0.2975 |
| Former
smoking | 21 (80.8) | 5 (33.3) | | 21 (61.8) | 5 (71.4) | |
| Current
smoking | 1 (3.8) | 1 (6.7) | | 1 (2.9) | 1 (14.3) | |
| Performance
status |
| ECOG 0 | 2 (18.2) | 9 (81.8) | 0.0008 | 10 (90.9) | 1 (9.1) | 0.5509 |
| ECOG 1 | 23 (79.3) | 6 (20.7) | | 23 (79.3) | 6 (20.7) | |
| ECOG 2 | 2 (100) | 0 (0) | | 2 (100) | 0 (0) | |
| TKI response |
| Complete
response | 0 (0) | 1 (100) | 0.0076 | 1 (100) | 0 (0) | 0.5417 |
| Partial
response | 7 (38.9) | 11 (61.1) | | 16 (88.9) | 2 (11.1) | |
| Stable
disease | 9 (90) | 1 (10) | | 7 (70) | 3 (30) | |
| Progressive
disease | 8 (88.9) | 1 (11.1) | | 8 (88.9) | 1 (11.1) | |
| Disease
recurrence |
| NED | 13 (35.1) | 7 (41.2) | 0.9016 | 14 (34.1) | 6 (46.2) | 0.6515 |
| Recurrence | 24 (64.9) | 10 (58.8) | | 27 (65.9) | 7 (53.8) | |
| Let-7g
expression |
| Low | 27 (49.1) | 11 (47.8) | 0.8835 | 32 (51.6) | 6 (37.5) | 0.4676 |
| High | 28 (50.9) | 12 (52.2) | | 30 (48.4) | 10 (62.5) | |
| miR-21
expression |
| Low | 27 (49.1) | 12 (52.2) | 0.8039 | 38 (61.3) | 1 (6.2) | 0.0003 |
| High | 28 (50.9) | 11 (47.8) | | 24 (38.7) | 15 (93.8) | |
Statistical analysis did not show any significant
association between EGFR mutations and Let-7g or
miR-21 expression, while we found a highly significant
association between K-Ras status and miR-21
expression (p=0.0003, Table
III). Noteworthy, a significantly higher miR-21
expression was observed in the NSCLC patients with
K-Ras-mutated tumors (14.237±1.638, p<0.001) compared to
the patients with K-Ras-wild-type tumors (7.316±0.792,
Fig. 2C).
miRNA target prediction and pathway
analysis
Let-7g and miR-21 target gene analysis
by miRanda, TargetScan, Pictar and miRDB prediction algorithms led
to the identification of a plethora of putative target genes for
these miRNAs. In order to minimize the number of false positives,
we recorded a gene as a putative target gene of the analysed miRNAs
only if it was predicted by at least two prediction algorithms with
a high confidence score. According to these stringent criteria, we
identified 24 putative target genes for Let-7g, including
HMGA2, ERCC6 and MAP3K3 genes and 26 putative target
genes for miR-21, including PDCD4, MSH2 and
SPRY1/SPRY2 genes (Table
IV).
 | Table IVPutative target genes of the
dysregulated Let-7g and miR-21 in the NSCLC
patients. |
Table IV
Putative target genes of the
dysregulated Let-7g and miR-21 in the NSCLC
patients.
| miRNA | Locus | Pathway | Target genes |
|---|
| Let-7g | 3p21.1 | Cell cycle | HMGA2, E2F5,
COIL, DNA2, CCNJ, CCND2, CDC25A, LIN28B, BACH1 |
| |
Transcription/transduction | BZW1,
HIC2 |
| | DNA repair | ERCC6, SMARCAD1,
BACH1 |
| | Apoptosis | N-MYC, CASP3,
MAP4K3 |
| | MAPK/ERK
pathway | N-RAS, MAP3K3,
MAP4K3, MAPK6 |
| | Insulin/TGFβ
pathway | FOXP2, IGF1R,
IGF2BP2 |
| | PI3K/Akt
pathway | N-RAS, FOXP2,
CCND2 |
| | Wnt pathway | END1, END2,
N-MYC |
| miR-21 | 17q23.2 | Cell cycle | STAG2,
KIF6 |
| | DNA repair | MSH2, FANCC,
CHD7 |
| | Apoptosis | PDCD4, APAF1,
STAT3, MALT1, SGK3 |
| | Angiogenesis | SOS2, JAG1,
MAP3K1, STAT3 |
| | Proteolysis | WWP1 |
| | Cell adhesion | CCL1, MATN2,
TGFBI, VCL |
| | MAPK/ERK
pathway | MAP3K1, STAT3,
SOS2, NKIRAS1, SPRY1, SPRY2 |
| | TGFβ pathway | BMPR2,
SMAD7 |
| | G-protein
pathway | SOS2, TIAM2,
GPR64, KRIT1 |
We further investigated the biological consequences
of Let-7g and miR-21 aberrant expression grouping the
predicted target genes by gene ontology terms. This analysis
revealed that most of cell processes regulated by these miRNAs play
a key role in cancer pathogenesis and are mainly involved in cell
proliferation, apoptosis, DNA repair, cell adhesion and signal
transduction pathways (Table
IV).
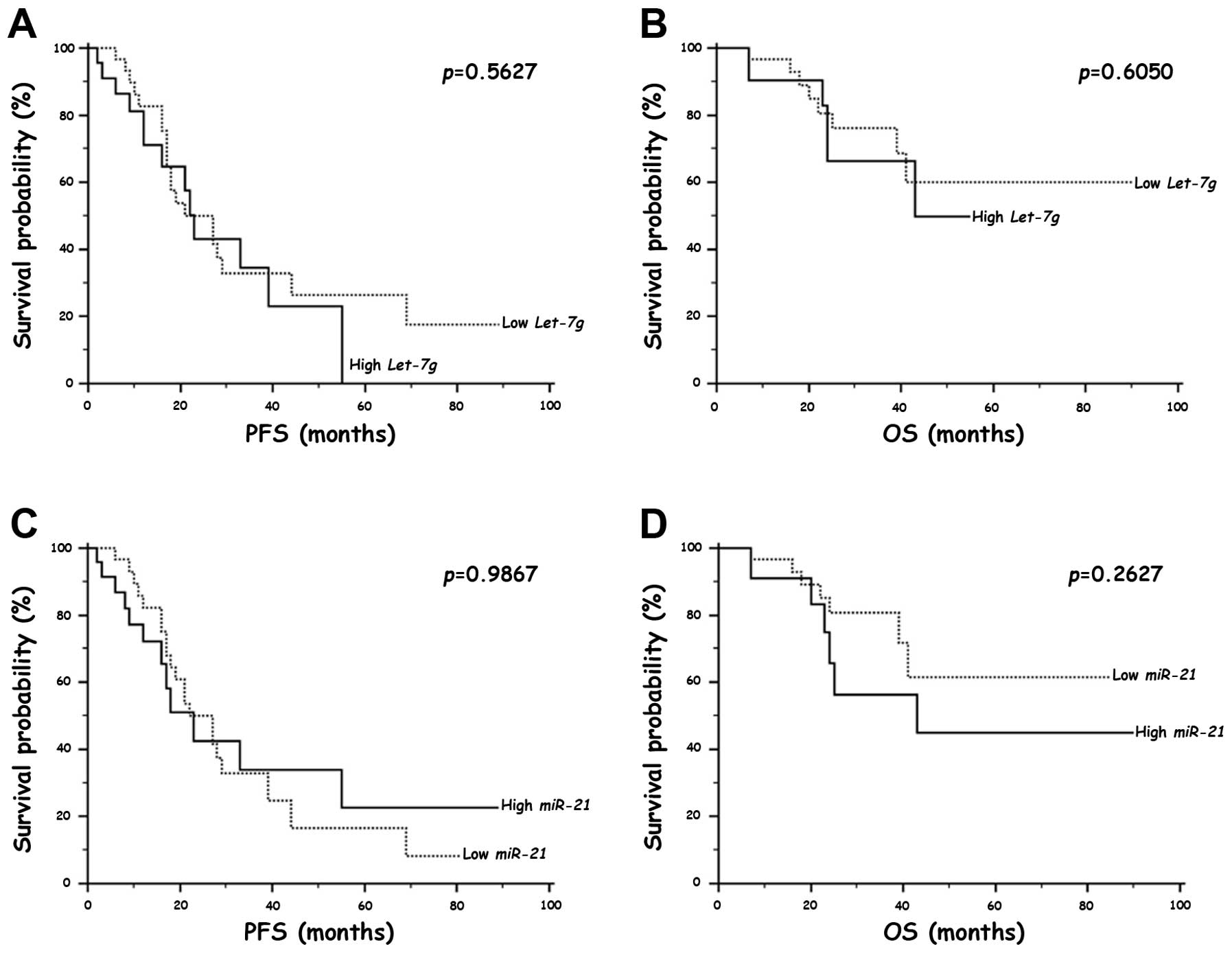
Survival analysis
To evaluate the relationships of Let-7g and
miR-21 expression with the prognosis of the NSCLC patients,
we performed a survival analysis by the Kaplan-Meier method using
the disease recurrence and the overall post-operative survival as
end-points. We did not observe any significant difference in PFS
and OS of the NSCLC patients with a high Let-7g or
miR-21 expression compared to the patients with a low
expression of these miRNAs (Fig.
3). However, we further investigated Let-7g and
miR-21 as prognostic indicators by restricting our analysis
to the first 30 months of the follow-up to verify a possible
short-term prognostic value of Let-7g and miR-21
evaluation. Interestingly, we found that the NSCLC patients with a
high Let-7g or miR-21 expression showed a
significantly shorter mean PFS and OS compared to the patients with
a low expression of these miRNAs (Table V).
 | Table VShort-term correlations between the
prognosis of the NSCLC patients and the Let-7 and
miR-21 expression. |
Table V
Short-term correlations between the
prognosis of the NSCLC patients and the Let-7 and
miR-21 expression.
| Characteristic | PFS
(months)a | p-valueb | OS (months)a | p-valueb |
|---|
| Let-7g
expression | | | | |
| Low | 18 (15–22) | 0.01 | 20 (16–23) | 0.023 |
| High | 12 (8–16) | | 13 (9–17) | |
| miR-21
expression | | | | |
| Low | 19 (16–23) | 0.0003 | 21 (17–25) | 0.0045 |
| High | 11 (8–14) | | 13 (9–17) | |
Discussion
Lung cancer is the first cause of death for cancer
worldwide and >80% of the cases are NSCLC (1–4).
Although early diagnosis and patient care have greatly improved in
recent years, most of the NSCLC patients show locally advanced or
metastatic disease at diagnosis and their prognosis remains
extremely poor (1–7). Currently, no appropriate diagnostic
biomarker exists for NSCLC, highlighting the need of a better
knowledge of its biology to improve prevention, diagnosis and
treatment.
MiRNAs are a highly conserved family of small
non-coding RNA molecules that negatively regulate gene expression
(8,9) and their aberrant expression has been
found to play a key role in pathogenesis of several malignancies,
including NSCLC (13–16). This study was aimed to evaluate
Let-7g and miR-21 expression profile in the NSCLC
patients in order to establish their role in NSCLC pathogenesis and
their potential diagnostic, prognostic and predictive
significance.
We demonstrated that miR-21 expression
strongly differentiates the NSCLC from non-cancer lung tissues,
while we did not observe any Let-7g discriminative value. In
our study, a highly significant increase was found in miR-21
expression in NSCLC tissues compared to the non-cancer ones, in
agreement with previous results that demonstrated a miR-21
overexpression in tumor tissues from several human malignancies
(23,29–31).
Conversely, we observed a reduced Let-7g expression that was
expressed at comparable levels in NSCLC and non-cancer lung
tissues. Let-7g downregulation in NSCLC tissues has been
previously reported by several authors, who have also demonstrated
that the aberrant expression of Let-7 family represents an
early event during NSCLC carcinogenesis and is more common in SCCs
compared to ADCs (12,19,32,33).
In our study, Let-7g and miR-21 are downregulated in
SCCs compared to ADCs, but their evaluation has not been shown to
have a significant diagnostic value in discriminating between these
two different histotypes, as previously reported in larger miRNA
profiling studies (29,30). Landi et al(30) reported that Let-7g and
miR-21 differential expression allows to discriminate
between ADC and SCC in the early-stage tumors (stage I), but not in
the advanced stage (stage II–IV), suggesting that miRNA expression
loses its histology-specificity in the more advanced and less
differentiated tumors. Therefore, the lack of a statistical
significance we observed between the altered Let-7g and
miR-21 expression and NSCLC histology could be explained by
the fact that most of the enrolled patients were diagnosed in an
advanced stage, where miRNA histology-related expression may be not
specific.
Concerning the other analysed clinicopathological
characteristics, we did not observe any significant correlation
between the Let-7g and miR-21 dysregulated expression
and the clinicopathological features, including age, tumor stage,
therapy response and smoking habit. In particular, our results
concerning the relationship between the miR-21 expression
and the smoking habit are in disagreement with data reported by
Seike et al(26), who
demonstrated that miR-21 expression is significantly higher
in tumors from smokers than from never smokers; however, this
discrepancy could be due to the small number of patients for whom
we had smoking data. Interestingly, we found that Let-7g
expression is significantly associated with lymph nodal status. We
showed that most of the NSCLC patients with a low Let-7g
expression present metastatic lymph nodes at diagnosis, while no
substantial differences were observed for the patients with a high
Let-7g expression. This result suggests an important role of
Let-7g in NSCLC tumor progression and acquisition of
metastatic potential and is supported by in vivo studies
showing that ectopic Let-7g expression in NSCLC xenografts
induce a significant decrease in tumor growth and spread (17,18).
Since the importance of EGFR and K-Ras
mutation detection in current management of the NSCLC patients, we
explored the relationship between their mutational status and
Let-7g and miR-21 expression profile. In our study,
the frequency of K-Ras mutation (20.5%) was in agreement
with previously reported data (34,35),
whereas EGFR mutation incidence was slightly higher (29.5%)
than that reported in the literature for lung cancer (15–20%)
(6,36–38),
because many of the recruited patients belonged to a larger study
designed to evaluate the EGFR TKI response. According to previously
reported data (20,21,30,39),
Let-7g expression was not correlated with EGFR or
K-Ras mutational status. However, several studies have
demonstrated that Let-7g acts as a K-Ras negative
regulator by binding to multiple sites of their 3′UTRs (40) and that lung cancer tissues with
reduced Let-7g levels have significantly higher K-Ras levels
compared to their corresponding normal tissues (18,20).
Therefore, it is possible that the aberrant expression of
Let-7g and K-Ras mutations are mutually exclusive in
NSCLC carcinogenesis with a more predominant effect of
Let-7g dysregulation in SCCs, which show a low expression of
this miRNA compared to the other NSCLC histotypes and a more
prominent role of K-Ras mutations in ADC carcinogenesis
(32,33).
Furthermore, we first demonstrated a strong and
highly significant correlation between the high miR-21
expression and the presence of mutations in the codons 12 and 13 of
K-Ras gene, suggesting a synergistic interplay between
miR-21 and K-Ras oncogenes that supports neoplastic
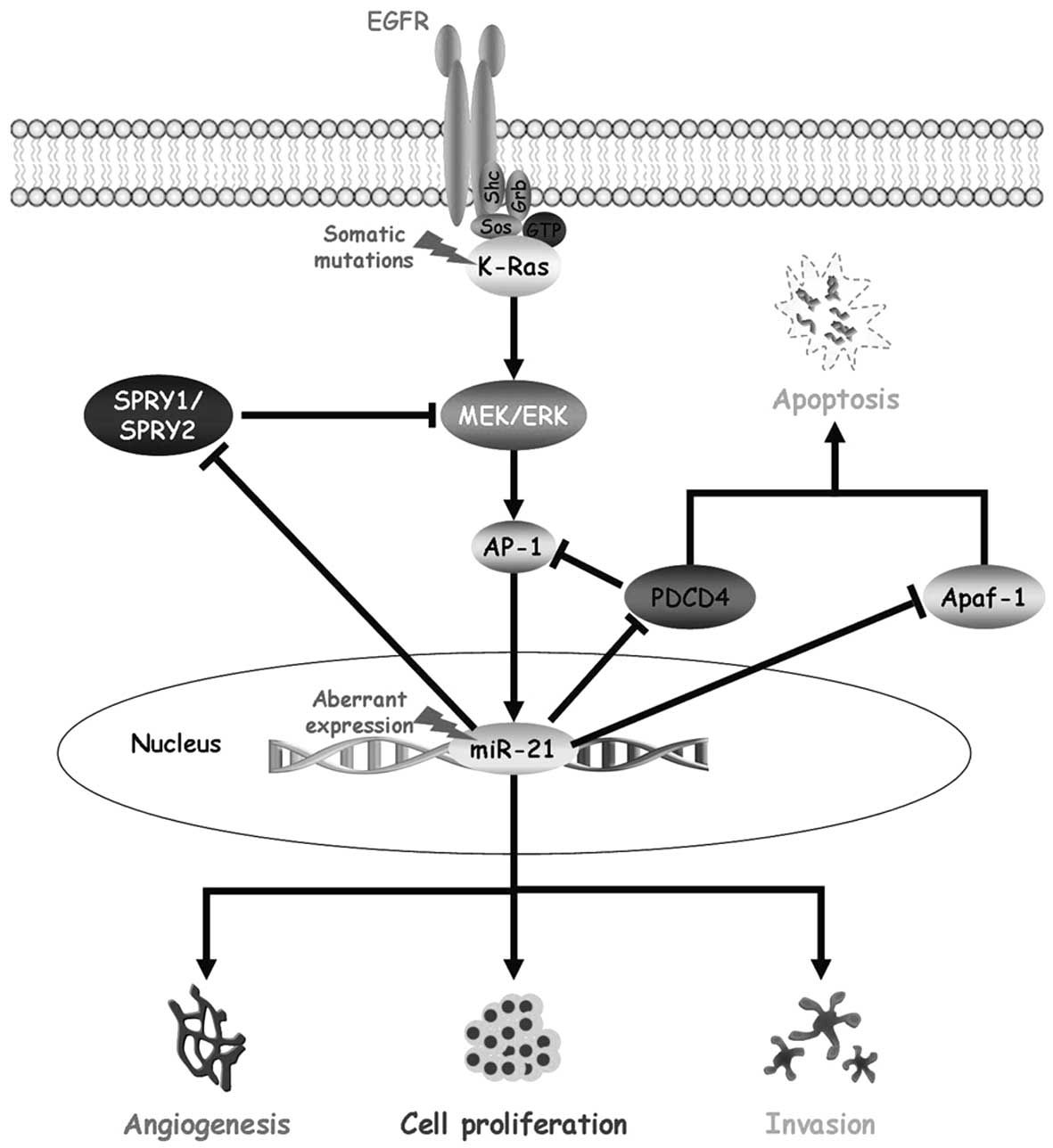
phenotype in NSCLC. Based on miR-21 expression and target
gene prediction results, we might hypothesize an auto-regulatory
loop between oncogenic K-Ras and miR-21 mediated by
the MAPK/ERK signalling pathway, SPRY1/SPRY2 and
PDCD4 (Fig. 4).
K-Ras mutations determine a constitutive protein activation
with a consequent activation of the MAPK/ERK signalling pathway,
which plays an important role in lung carcinogenesis by inhibiting
apoptosis and promoting cell proliferation, cell growth,
angiogenesis, invasion and metastasis (41). On the other hand, miR-21
modulates several components critical to the NSCLC pathogenesis by
targeting apoptotic effectors and antagonists of the MAPK/ERK
signalling pathway (22,25,27).
The high miR-21 expression observed in our NSCLC patients
might cause a decrease in SPRY1/SPRY2 expression that has
been demonstrated to negatively regulate the MAPK/ERK signalling
pathway and to enhance cell migration (42). In addition, the negative regulation
of PDCD4 and Apaf-1 genes by miR-21 leads to
apoptosis inhibition (22,27,43),
as well as to the removal of the PDCD4 inhibitory effect on AP-1,
which is downstream the MAPK/ERK signalling pathway and promotes
miR-21 expression (44,45).
This complex and auto-regulatory circuit might justify the high
levels of miR-21 expression observed in our study in the
NSCLC patients harbouring K-Ras mutations and might have a
final stimulation effect on the processes that promote tumor
progression and therapy resistance (Fig. 4).
We investigated the relationship between the
differential Let-7g and miR-21 expression and
prognosis of the NSCLC patients without observing any statistically
significant correlation. These results are in disagreement with
data reported by other authors that support a negative prognostic
role for Let-7g, whose downregulation has been associated
with a reduced overall post-operative survival in NSCLC patients
(18–21), and miR-21, whose
overexpression has been associated with a poor prognosis
irrespective of the TNM stage and lymph nodal status (23,26,46).
However, these discrepant results could be due to the small number
of patients with available clinical data. Interestingly, by
restricting our analysis to the first 30 months of the follow-up
observation, we demonstrated that the NSCLC patients with a high
expression of either Let-7g or miR-21 show a highly
significant shorter PFS and OS compared to the patients with a low
expression of both these miRNAs, suggesting a possible negative
short-term prognostic value of the evaluation of Let-7g and
miR-21 expression.
In conclusion, our data show that Let-7g and
miR-21 are aberrantly expressed in the NSCLC patients and
that there is a close interplay among K-Ras, miR-21
and Let-7g in NSCLC, suggesting that their systematic
evaluation could represent a useful biomarker in the molecular
characterization and management of NSCLC patients.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar
|
|
2
|
Travis WD, Brambilla E, Muller-Hemerlink
HK and Harris CC; World Health Organization Classification of
Tumours. Pathology and Genetics of Tumours of the Lung, Pleura,
Thymus and Heart. IARC Press; Lyon: 2004
|
|
3
|
Travis WD, Brambilla E, Noguchi M,
Nicholson AG, Geisinger KR, Yatabe Y, Beer DG, Powell CA, Riely GJ,
Van Schil PE, Garg K, Austin JH, Asamura H, Rusch VW, Hirsch FR,
Scagliotti G, Mitsudomi T, Huber RM, Ishikawa Y, Jett J,
Sanchez-Cespedes M, Sculier JP, Takahashi T, Tsuboi M,
Vansteenkiste J, Wistuba I, Yang PC, Aberle D, Brambilla C, Flieder
D, Franklin W, Gazdar A, Gould M, Hasleton P, Henderson D, Johnson
B, Johnson D, Kerr K, Kuriyama K, Lee JS, Miller VA, Petersen I,
Roggli V, Rosell R, Saijo N, Thunnissen E, Tsao M and Yankelewitz
D: International association for the study of lung cancer/American
thoracic society/European respiratory society international
multidisciplinary classification of lung adenocarcinoma. J Thorac
Oncol. 6:244–285. 2011. View Article : Google Scholar
|
|
4
|
Travis WD, Brambilla E, Noguchi M,
Nicholson A, Geisinger K, Yatabe Y, Ishikawa Y, Wistuba I, Flieder
DB, Franklin W, Gazdar A, Hasleton PS, Henderson DW, Kerr KM,
Petersen I, Roggli V, Thunnissen E and Tsao M: Diagnosis of lung
cancer in small biopsies and cytology: implications of the 2011
International Association for the Study of Lung Cancer/American
Thoracic Society/European Respiratory Society Classification. Arch
Pathol Lab Med. 137:668–684. 2012. View Article : Google Scholar
|
|
5
|
Bronte G, Rizzo S, La Paglia L, Adamo V,
Siragusa S, Ficorella C, Santini D, Bazan V, Colucci G, Gebbia N
and Russo A: Driver mutations and differential sensitivity to
targeted therapies: a new approach to the treatment of lung
adenocarcinoma. Cancer Treat Rev. 36:S21–S29. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pao W and Girard N: New driver mutations
in non-small-cell lung cancer. Lancet Oncol. 12:175–180. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Saintigny P and Burger JA: Recent advances
in non-small cell lung cancer biology and clinical management.
Discov Med. 13:287–297. 2012.PubMed/NCBI
|
|
8
|
He L and Hannon GJ: MicroRNAs: small RNAs
with big role in gene regulation. Nat Rev Genet. 5:522–531. 2002.
View Article : Google Scholar
|
|
9
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Schmittgen TD: Regulation of microRNA
processing in development, differentiation and cancer. J Cell Mol
Med. 12:1811–1819. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Huang Y, Shen XJ, Zou Q, Wang SP, Tang SM
and Zhang GZ: Biological functions of microRNAs: a review. J
Physiol Biochem. 67:129–139. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Calin GA, Sevignani C, Dumitru CD, Hyslop
T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M
and Croce CM: Human microRNA genes are frequently located at
fragile sites and genomic regions involved in cancers. Proc Natl
Acad Sci USA. 101:2999–3004. 2004. View Article : Google Scholar
|
|
13
|
Zhang B, Pan X, Cobb GP and Anderson TA:
microRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bartels CL and Tsongalis GJ: MicroRNAs:
novel biomarkers for human cancer. Clin Chem. 55:623–631. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Farazi TA, Spitzer JI, Morozov P and
Tuschl T: miRNAs in human cancer. J Pathol. 223:102–115. 2011.
View Article : Google Scholar
|
|
16
|
Iorio MV and Croce CM: MicroRNA
dysregulation in cancer: diagnostics, monitoring and therapeutics.
A comprehensive review. EMBO Mol Med. 4:143–159. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Takamizawa J, Konishi H, Yanagisawa K,
Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y,
Mitsudomi T and Takahashi T: Reduced expression of the let-7
microRNAs in human lung cancers in association with shortened
postoperative survival. Cancer Res. 64:3753–3756. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Johnson CD, Esquela-Kerscher A, Stefani G,
Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J,
Shingara J, Chin L, Brown D and Slack FJ: The let-7 microRNA
represses cell proliferation pathways in human cells. Cancer Res.
67:7713–7722. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kumar MS, Erkeland SJ, Pester RE, Chen CY,
Ebert MS, Sharp PA and Jacks T: Suppression of non-small cell lung
tumor development by the let-7 microRNA family. Proc Natl Acad Sci
USA. 105:3903–3908. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Johnson SM, Grosshans H, Shingara J, Byrom
M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D and Slack
FJ: RAS is regulated by the let-7 microRNA family. Cell.
120:635–647. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Roush S and Slack FJ: The let-7 family of
microRNAs. Trends Cell Biol. 18:505–516. 2008. View Article : Google Scholar
|
|
22
|
Krichevsky AM and Gabriely G: miR-21: a
small multi-faceted RNA. J Cell Mol Med. 13:39–53. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gao W, Yu Y, Cao H, Shen H, Li X, Pan S
and Shu Y: Deregulated expression of miR-21, miR-143 and miR-181a
in non small cell lung cancer is related to clinicopathologic
characteristics or patient prognosis. Biomed Pharmacother.
64:399–408. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Moore LM and Zhang W: Targeting miR-21 in
glioma: a small RNA with big potential. Expert Opin Ther Targets.
14:1247–1257. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pan X, Wang ZX and Wang R: MicroRNA-21: a
novel therapeutic target in human cancer. Cancer Biol Ther.
10:1224–1232. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Seike M, Goto A, Okano T, Bowman ED,
Schetter AJ, Horikawa I, Mathe EA, Jen J, Yang P, Sugimura H, Gemma
A, Kudoh S, Croce CM and Harris CC: MiR-21 is an EGFR-regulated
anti-apoptotic factor in lung cancer in never-smokers. Proc Natl
Acad Sci USA. 106:12085–12090. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Selcuklu SD, Donoghue MT and Spillane C:
miR-21 as a key regulator of oncogenic processes. Biochem Soc
Trans. 37:918–925. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Capodanno A, Boldrini L, Alì G,
Pelliccioni S, Mussi A and Fontanini G:
Phosphatidylinositol-3-kinase α catalytic subunit gene somatic
mutations in bronchopulmonary neuroendocrine tumours. Oncol Rep.
28:1559–1566. 2012.
|
|
29
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M,
Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M,
Harris CC and Croce CM: A microRNA expression signature of human
solid tumors defines cancer gene targets. Proc Natl Acad Sci USA.
103:2257–2261. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Landi MT, Zhao Y, Rotunno M, Koshiol J,
Liu H, Bergen AW, Rubagotti M, Goldstein AM, Linnoila I, Marincola
FM, Tucker MA, Bertazzi PA, Pesatori AC, Caporaso NE, McShane LM
and Wang E: MicroRNA expression differentiates histology and
predicts survival of lung cancer. Clin Cancer Res. 16:430–441.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Meng F, Henson R, Wehbe-Janek H, Ghoshal
K, Jacob ST and Patel T: MicroRNA-21 regulates expression of the
PTEN tumor suppressor gene in human hepatocellular cancer.
Gastroenterology. 133:647–658. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mitsudomi T, Oyama T, Nishida K, Ogami A,
Osaki T, Sugio K, Yasumoto K, Sugimachi K and Gazdar AF: Loss of
heterozygosity at 3p in non-small cell lung cancer and its
prognostic implication. Clin Cancer Res. 2:1185–1189.
1996.PubMed/NCBI
|
|
33
|
Zabarovsky ER, Lerman MI and Minna JD:
Tumor suppressor genes on chromosome 3p involved in the
pathogenesis of lung and other cancers. Oncogene. 21:6915–6935.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Riely GJ, Marks J and Pao W: KRAS
mutations in non-small cell lung cancer. Proc Am Thorac Soc.
6:201–205. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mao C, Qiu LX, Liao RY, Du FB, Ding H,
Yang WC, Li J and Chen Q: KRAS mutations and resistance to
EGFR-TKIs treatment in patients with non-small cell lung cancer: a
meta-analysis of 22 studies. Lung Cancer. 69:272–278. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yatabe Y and Mitsudomi T: Epidermal growth
factor receptor mutations in lung cancers. Pathol Int. 57:233–244.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ladanyi M and Pao W: Lung adenocarcinoma:
guiding EGFR-targeted therapy and beyond. Mod Pathol. 21:S16–S22.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Dacic S: Molecular diagnostics of lung
carcinomas. Arch Pathol Lab Med. 135:622–629. 2011.PubMed/NCBI
|
|
39
|
Dacic S, Kelly L, Shuai Y and Nikiforova
MN: miRNA expression profiling of lung adenocarcinomas: correlation
with mutational status. Mod Pathol. 23:1577–1582. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chin LJ, Ratner E, Leng S, Zhai R, Nallur
S, Babar I, Muller RU, Straka E, Su L, Burki EA, Crowell RE, Patel
R, Kulkarni T, Homer R, Zelterman D, Kidd KK, Zhu Y, Christiani DC,
Belinsky SA, Slack FJ and Weidhaas JB: A SNP in a let-7 microRNA
complementary site in the KRAS 3′ untranslated region increases
non-small cell lung cancer risk. Cancer Res. 68:8535–8540.
2008.PubMed/NCBI
|
|
41
|
Boutros T, Chevet E and Metrakos P:
Mitogen-activated protein (MAP) kinase/MAP kinase phosphatase
regulation: roles in cell growth, death and cancer. Pharmacol Rev.
60:261–310. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hatley ME, Patrick DM, Garcia MR,
Richardson JA, Bassel-Duby R, van Rooij E and Olson EN: Modulation
of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell.
18:282–293. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lu Z, Liu M, Stribinskis V, Klinge CM,
Ramos KS, Colburn NH and Li Y: MicroRNA-21 promotes cell
transformation by targeting the programmed cell death 4 gene.
Oncogene. 27:4373–4379. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hwang SK, Jin H, Kwon JT, Chang SH, Kim
TH, Cho CS, Lee KH, Young MR, Colburn NH, Beck GR Jr, Yang HS and
Cho MH: Aerosol-delivered programmed cell death 4 enhanced
apoptosis, controlled cell cycle and suppressed AP-1 activity in
the lungs of AP-1 luciferase reporter mice. Gene Ther.
14:1353–1361. 2007.PubMed/NCBI
|
|
45
|
Talotta F, Cimmino A, Matarazzo MR,
Casalino L, De Vita G, D'Esposito M, Di Lauro R and Verde P: An
autoregulatory loop mediated by miR-21 and PDCD4 controls the AP-1
activity in RAS transformation. Oncogene. 28:73–84. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Markou A, Tsaroucha EG, Kaklamanis L,
Fotinou M, Georgoulias V and Lianidou ES: Prognostic value of
mature microRNA-21 and microRNA-205 overexpression in non-small
cell lung cancer by quantitative real-time RT-PCR. Clin Chem.
54:1696–1704. 2008. View Article : Google Scholar : PubMed/NCBI
|