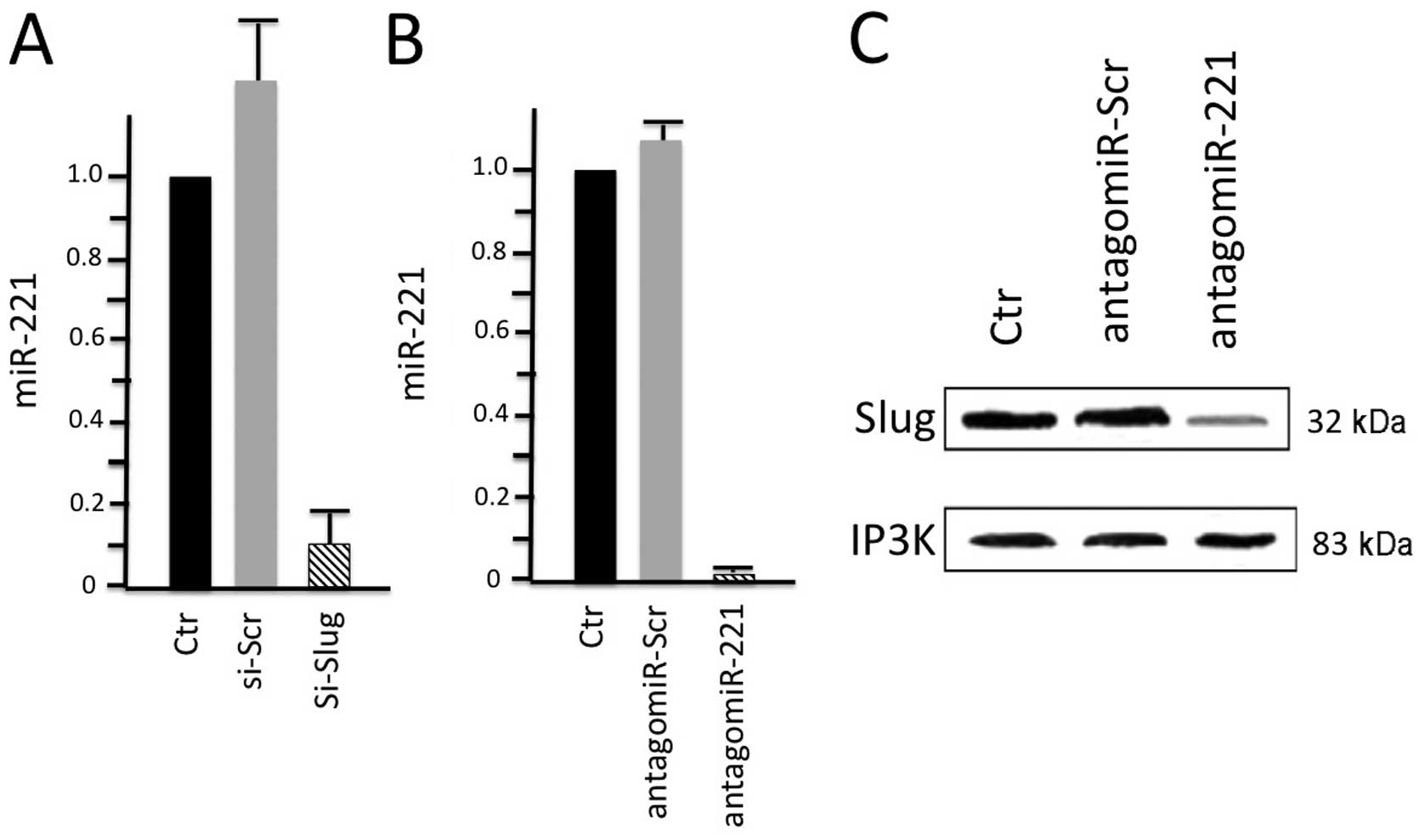
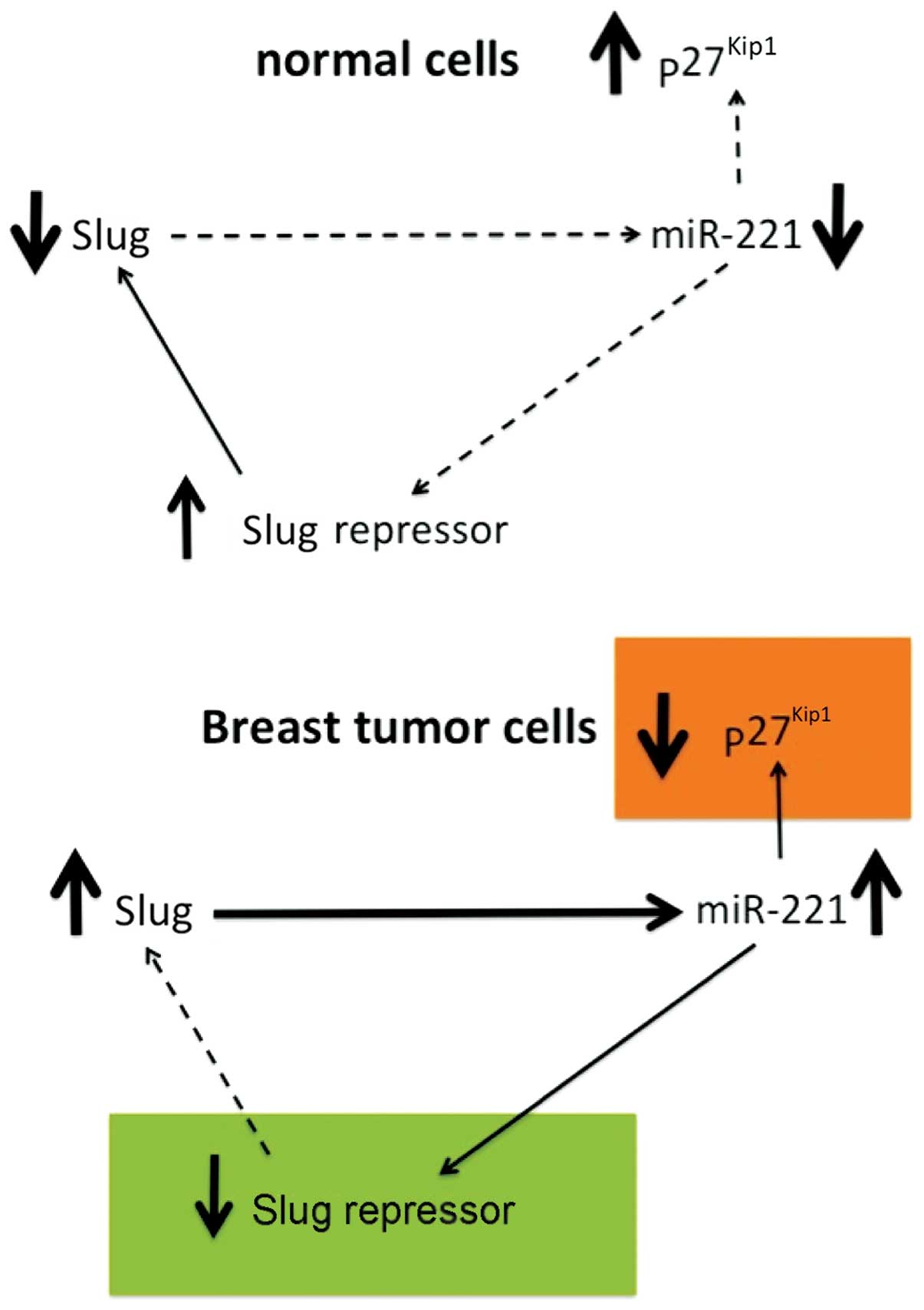
|
1.
|
Filipowicz W, Jaskiewicz L, Kolb FA and
Pillai RS: Post-transcriptional gene silencing by siRNAs and
miRNAs. Curr Opin Struc Biol. 15:331–341. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
He L and Hannon GJ: MicroRNAs: small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Kozomara A and Griffiths-Jones S: miRBase:
integrating microRNA annotation and deep-sequencing data. Nucleic
Acids Res. 39:D152–D157. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Krol J, Loedige I and Filipowicz W: The
widespread regulation of microRNA biogenesis, function and decay.
Nat Rev Genet. 11:597–610. 2010.PubMed/NCBI
|
|
5.
|
Sontheimer EJ and Carthew RW: Silence from
within: endogenous siRNAs and miRNAs. Cell. 122:9–12. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Taccioli C, Fabbri E, Visone R, Volinia S,
Calin GA, Fong LY, et al: UCbase and miRfunc: a database of
ultracon-served sequences and microRNA function. Nucleic Acids Res.
37:D41–D48. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Griffiths-Jones S: miRBase: the microRNA
sequence database. Methods Mol Biol. 342:129–138. 2006.PubMed/NCBI
|
|
8.
|
Witwer KW: Data submission and quality in
microarray-based microRNA profiling. Clin Chem. 59:392–400. 2013.
View Article : Google Scholar
|
|
9.
|
Sablok G, Milev I, Minkov G, Minkov I,
Varotto C, Yahubyan G and Baev V: isomiRex: Web-based
identification of microRNAs, isomiR variations and differential
expression using next-generation sequencing datasets. FEBS Lett.
Jul 4–2013.(Epub ahead of print).
|
|
10.
|
Russo F, Di Bella S, Nigita G, Macca V,
Laganà A, Giugno R, Pulvirenti A and Ferro A: miRandola:
extracellular circulating microRNAs database. PLoS One.
7:e477862012. View Article : Google Scholar
|
|
11.
|
Krützfeldt J, Kuwajima S, Braich R, Rajeev
KG, Pena J, Tuschl T, Manoharan M and Stoffel M: Specificity,
duplex degradation and subcellular localization of antagomirs.
Nucleic Acids Res. 35:2885–2892. 2007.PubMed/NCBI
|
|
12.
|
Dalmay T: Mechanism of miRNA-mediated
repression of mRNA translation. Essays Biochem. 54:29–38. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Jiang Q, Wang Y, Hao Y, Juan L, Teng M,
Zhang X, Li M, Wang G and Liu Y: miR2Disease: a manually curated
database for microRNA deregulation in human disease. Nucleic Acids
Res. 37:D98–D104. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Subramanian S and Steer CJ: MicroRNAs as
gatekeepers of apoptosis. J Cell Physiology. 223:89–98. 2010.
|
|
15.
|
Wang YM and Blelloch R: Cell cycle
regulation by MicroRNAs in embryonic stem cells. Cancer Res.
69:4093–4096. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Alvarez-Garcia I and Miska EA: MicroRNA
functions in animal development and human disease. Development.
132:4653–4662. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Tsai LM and Yu D: MicroRNAs in common
diseases and potential therapeutic applications. Clin Exp Pharmacol
Physiol. 7:102–107. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Hemida MG, Ye X, Thair S and Yang D:
Exploiting the therapeutic potential of microRNAs in viral
diseases: expectations and limitations. Mol Diagn Ther. 14:271–282.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Kota SK and Balasubramanian S: Cancer
therapy via modulation of micro RNA levels: a promising future.
Drug Discov Today. 15:733–740. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Bader AG, Brown D and Winkler M: The
promise of microRNA replacement therapy. Cancer Res. 70:7027–7030.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Sibley CR, Seow Y and Wood MJ: Novel
RNA-based strategies for therapeutic gene silencing. Mol Ther.
18:466–476. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Ge YF, Sun J, Jin CJ, Cao BQ, Jiang ZF and
Shao JF: AntagomiR-27a targets FOXO3a in glioblastoma and
suppresses U87 cell growth in vitro and in vivo. Asian Pac J Cancer
Prev. 14:963–968. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Rather MI, Nagashri MN, Swamy SS, Gopinath
KS and Kumar A: Oncogenic microRNA-down-regulates tumor suppressor
CDC73 and promotes oral squamous cell carcinoma cell proliferation:
implications for cancer therapeutics. J Biol Chem. 288:608–618.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Shu M, Zheng X, Wu S, Lu H, Leng T, Zhu W,
Zhou Y, Ou Y, Lin X, Lin Y, Xu D, Zhou Y and Yan G: Targeting
oncogenic miR-335 inhibits growth and invasion of malignant
astrocytoma cells. Mol Cancer. 10:592011. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Haug BH, Henriksen JR, Buechner J, Geerts
D, Tømte E, Kogner P, Martinsson T, Flægstad T, Sveinbjørnsson B
and Einvik C: MYCN-regulated miRNA-92 inhibits secretion of the
tumor suppressor DICKKOPF-3 (DKK3) in neuroblastoma.
Carcinogenesis. 32:1005–1012. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Tang H, Liu X, Wang Z, She X, Zeng X, Deng
M, Liao Q, Guo X, Wang R, Li X, Zeng F, Wu M and Li G: Interaction
of hsa-miR-381 and glioma suppressor LRRC4 is involved in glioma
growth. Brain Res. 1390:21–32. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Ma L, Reinhardt F, Pan E, Soutschek J,
Bhat B, Marcusson EG, Teruya-Feldstein J, Bell GW and Weinberg RA:
Therapeutic silencing of miR-10b inhibits metastasis in a mouse
mammary tumor model. Nat Biotechnol. 28:341–347. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Mercatelli N, Coppola V, Bonci D, Miele F,
Costantini A, Guadagnoli M, Bonanno E, Muto G, Frajese GV, De Maria
R, Spagnoli LG, Farace MG and Ciafrè SA: The inhibition of the
highly expressed miR-221 and miR-222 impairs the growth of prostate
carcinoma xenografts in mice. PLoS One. 3:e40292008. View Article : Google Scholar
|
|
29.
|
Scheibner KA, Teaboldt B, Hauer MC, Chen
X, Cherukuri S, Guo Y, Kelley SM, Liu Z, Baer MR, Heimfeld S and
Civin CI: MiR-27a functions as a tumor suppressor in acute leukemia
by regulating 14-3-3θ. PLoS One. 7:e508952012.PubMed/NCBI
|
|
30.
|
Endo H, Muramatsu T, Furuta M, Uzawa N,
Pimkhaokham A, Amagasa T, Inazawa J and Kozaki K: Potential of
tumor-suppressive miR-596 targeting LGALS3BP as a therapeutic agent
in oral cancer. Carcinogenesis. 34:560–569. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Liang Z, Ahn J, Guo D, Votaw JR and Shim
H: MicroRNA-302 replacement therapy sensitizes breast cancer cells
to ionizing radiation. Pharm Res. 30:1008–1016. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Thomas M, Lange-Grünweller K, Weirauch U,
Gutsch D, Aigner A, Grünweller A and Hartmann RK: The
proto-oncogene Pim-1 is a target of miR-33a. Oncogene. 31:918–928.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Ibrahim AF, Weirauch U, Thomas M,
Grünweller A, Hartmann RK and Aigner A: MicroRNA replacement
therapy for miR-145 and miR-33a is efficacious in a model of colon
carcinoma. Cancer Res. 71:5214–5224. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Wiggins JF, Ruffino L, Kelnar K, Omotola
M, Patrawala L, Brown D and Bader AG: Development of a lung cancer
therapeutic based on the tumor suppressor microRNA-34. Cancer Res.
70:5923–5930. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Trang P, Wiggins JF, Daige CL, Cho C,
Omotola M, Brown D, Weidhaas JB, Bader AG and Slack FJ: Systemic
delivery of tumor suppressor microRNA mimics using a neutral lipid
emulsion inhibits lung tumors in mice. Mol Ther. 19:1116–1122.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Wu Y, Crawford M, Mao Y, Lee RJ, Davis IC,
Elton TS, Lee LJ and Nana-Sinkam SP: Therapeutic delivery of
microRNA-29b by cationic lipoplexes for lung cancer. Mol Ther
Nucleic Acids. 2:e842013. View Article : Google Scholar : PubMed/NCBI
|
|
37.
|
Huang X, Schwind S, Yu B, Santhanam R,
Wang H, Hoellerbauer P, Mims A, Klisovic R, Walker AR, Chan KK,
Blum W, Perrotti D, Byrd JC, Bloomfield CD, Caligiuri MA, Lee RJ,
Garzon R, Muthusamy N, Lee LJ and Marcucci G: Targeted delivery of
microRNA-29b by transferrin-conjugated anionic lipopolyplex
nanoparticles: a novel therapeutic strategy in acute myeloid
leukemia. Clin Cancer Res. 19:2355–2367. 2013.
|
|
38.
|
Voorhoeve PM, le Sage C, Schrier M, Gillis
AJ, Stoop H, Nagel R, Liu YP, van Duijse J, Drost J, Griekspoor A,
Zlotorynski E, Yabuta N, De Vita G, Nojima H, Looijenga LH and
Agami R: A genetic screen implicates miRNA-372 and miRNA-373 as
oncogenes in testicular germ cell tumors. Cell. 124:1169–1181.
2006. View Article : Google Scholar
|
|
39.
|
Voorhoeve PM, le Sage C, Schrier M, Gillis
AJ, Stoop H, Nagel R, Liu YP, van Duijse J, Drost J, Griekspoor A,
Zlotorynski E, Yabuta N, De Vita G, Nojima H, Looijenga LH and
Agami R: A genetic screen implicates miRNA-372 and miRNA-373 as
oncogenes in testicular germ cell tumors. Adv Exp Med Biol.
604:17–46. 2007. View Article : Google Scholar
|
|
40.
|
Huang Q, Gumireddy K, Schrier M, le Sage
C, Nagel R, Nair S, Egan DA, Li A, Huang G, Klein-Szanto AJ,
Gimotty PA, Katsaros D, Coukos G, Zhang L, Puré E and Agami R: The
microRNAs miR-373 and miR-520c promote tumour invasion and
metastasis. Nat Cell Biol. 10:202–210. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41.
|
Galardi S, Mercatelli N, Giorda E,
Massalini S, Frajese GV, Ciafrè SA and Farace MG: miR-221 and
miR-222 expression affects the proliferation potential of human
prostate carcinoma cell lines by targeting p27Kip1. J
Biol Chem. 282:23716–23724. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
42.
|
Valastyan S, Reinhardt F, Benaich N,
Calogrias D, Szász AM, Wang ZC, Brock JE, Richardson AL and
Weinberg RA: A pleiotropically acting microRNA, miR-31, inhibits
breast cancer metastasis. Cell. 137:1032–1046. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43.
|
Hurst DR, Edmonds MD and Welch DR:
Metastamir: the field of metastasis-regulatory microRNA is
spreading. Cancer Res. 69:7495–7498. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44.
|
Wotschofsky Z, Liep J, Meyer HA, Jung M,
Wagner I, Disch AC, Schaser KD, Melcher I, Kilic E, Busch J,
Weikert S, Miller K, Erbersdobler A, Mollenkopf HJ and Jung K:
Identification of metastamirs as metastasis-associated microRNAs in
clear cell renal cell carcinomas. Int J Biol Sci. 8:1363–1374.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
45.
|
Taylor MA, Sossey-Alaoui K, Thompson CL,
Danielpour D and Schiemann WP: TGF-β upregulates miR-181a
expression to promote breast cancer metastasis. J Clin Invest.
123:150–163. 2013.
|
|
46.
|
Welch DR and Hurst DR: Unraveling the
‘TGF-β paradox’ one metastamir at a time. Breast Cancer Res.
15:3052013.
|
|
47.
|
Moldovan L, Batte K, Wang Y, Wisler J and
Piper M: Analyzing the circulating microRNAs in
exosomes/extracellular vesicles from serum or plasma by qRT-PCR.
Methods Mol Biol. 1024:129–145. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
48.
|
Chen X, Liang H, Zhang J, Zen K and Zhang
CY: Horizontal transfer of microRNAs: molecular mechanisms and
clinical applications. Protein Cell. 3:28–37. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
49.
|
Kosaka N and Ochiya T: Unraveling the
mystery of cancer by secretory microRNA: horizontal microRNA
transfer between living cells. Front Genet. 2:972011.PubMed/NCBI
|
|
50.
|
Chen X, Liang H, Zhang J, Zen K and Zhang
CY: Secreted microRNAs: a new form of intercellular communication.
Trends Cell Biol. 22:125–132. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
51.
|
Ramachandran S and Palanisamy V:
Horizontal transfer of RNAs: exosomes as mediators of intercellular
communication. Wiley Interdiscip Rev RNA. 3:286–293. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
52.
|
Muralidharan-Chari V, Clancy JW, Sedgwick
A and D’Souza-Schorey C: Microvesicles: mediators of extracellular
communication during cancer progression. J Cell Sci1. 23:1603–1611.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
53.
|
Piovan C, Palmieri D, Di Leva G, Braccioli
L, Casalini P, Nuovo G, Tortoreto M, Sasso M, Plantamura I, Triulzi
T, Taccioli C, Tagliabue E, Iorio MV and Croce CM: Oncosuppressive
role of p53-induced miR-205 in triple negative breast cancer. Mol
Oncol. 6:458–472. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
54.
|
Lee YM, Lee JY, Ho CC, Hong QS, Yu SL,
Tzeng CR, Yang PC and Chen HW: miRNA-34b as a tumor suppressor in
estrogen-dependent growth of breast cancer cells. Breast Cancer
Res. 13:R1162011. View Article : Google Scholar : PubMed/NCBI
|
|
55.
|
Iorio MV and Croce CM: Causes and
consequences of microRNA dysregulation. Cancer J. 18:215–222. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
56.
|
Xu X, Chen H, Lin Y, Hu Z, Mao Y, Wu J, Xu
X, Zhu Y, Li S, Zheng X and Xie L: MicroRNA-409-3p inhibits
migration and invasion of bladder cancer cells via targeting c-Met.
Mol Cells. 36:62–68. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
57.
|
He J, Deng Y, Yang G and Xie W:
MicroRNA-203 down-regulation is associated with unfavorable
prognosis in human glioma. J Surg Oncol. 108:121–125. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
58.
|
Iorio MV and Croce CM: MicroRNAs in
cancer: small molecules with a huge impact. J Clin Oncol.
27:5848–5856. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
59.
|
Volinia S, Galasso M, Sana ME, Wise TF,
Palatini J, Huebner K and Croce CM: Breast cancer signatures for
invasiveness and prognosis defined by deep sequencing of microRNA.
Proc Natl Acad Sci USA. 109:3024–3029. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
60.
|
Lu Y, Roy S, Nuovo G, Ramaswamy B, Miller
T, Shapiro C, Jacob ST and Majumder S: Anti-microRNA-222
(anti-miR-222) and -181B suppress growth of tamoxifen-resistant
xenografts in mouse by targeting TIMP3 protein and modulating
mitogenic signal. J Biol Chem. 286:42292–42302. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
61.
|
Shah MY and Calin GA: MicroRNAs miR-221
and miR-222: a new level of regulation in aggressive breast cancer.
Genome Med. 3:562011. View
Article : Google Scholar : PubMed/NCBI
|
|
62.
|
Stinson S, Lackner MR, Adai AT, Yu N, Kim
HJ, O’Brien C, Spoerke J, Jhunjhunwala S, Boyd Z, Januario T,
Newman RJ, Yue P, Bourgon R, Modrusan Z, Stern HM, Warming S, de
Sauvage FJ, Amler L, Yeh RF and Dornan D: miR-221/222 targeting of
trichorhinophalangeal 1 (TRPS1) promotes epithelial-to-mesenchymal
transition in breast cancer. Sci Signal. 4(pt5)2011.PubMed/NCBI
|
|
63.
|
Cochrane DR, Cittelly DM, Howe EN,
Spoelstra NS, McKinsey EL, LaPara K, Elias A, Yee D and Richer JK:
MicroRNAs link estrogen receptor alpha status and Dicer levels in
breast cancer. Horm Cancer. 1:306–319. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
64.
|
Yoshimoto N, Toyama T, Takahashi S,
Sugiura H, Endo Y, Iwasa M, Fujii Y and Yamashita H: Distinct
expressions of microRNAs that directly target estrogen receptor α
in human breast cancer. Breast Cancer Res Treat. 130:331–339.
2011.PubMed/NCBI
|
|
65.
|
Stinson S, Lackner MR, Adai AT, Yu N, Kim
HJ, O’Brien C, Spoerke J, Jhunjhunwala S, Boyd Z, Januario T,
Newman RJ, Yue P, Bourgon R, Modrusan Z, Stern HM, Warming S, de
Sauvage FJ, Amler L, Yeh RF and Dornan D: TRPS1 targeting by
miR-221/222 promotes the epithelial-to-mesenchymal transition in
breast cancer. Sci Signal. 4:ra412011.PubMed/NCBI
|
|
66.
|
Guttilla IK, Phoenix KN, Hong X, Tirnauer
JS, Claffey KP and White BA: Prolonged mammosphere culture of MCF-7
cells induces an EMT and repression of the estrogen receptor by
microRNAs. Breast Cancer Res Treat. 132:75–85. 2012. View Article : Google Scholar
|
|
67.
|
Gordanpour A, Stanimirovic A, Nam RK,
Moreno CS, Sherman C, Sugar L and Seth A: miR-221 is down-regulated
in TMPRSS2: ERG fusion-positive prostate cancer. Anticancer Res.
31:403–410. 2011.PubMed/NCBI
|
|
68.
|
Radojicic J, Zaravinos A, Vrekoussis T,
Kafousi M, Spandidos DA and Stathopoulos EN: MicroRNA expression
analysis in triple-negative (ER, PR and Her2/neu) breast cancer.
Cell Cycle. 10:507–517. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
69.
|
Pelletier C, Speed WC, Paranjape T, Keane
K, Blitzblau R, Hollestelle A, Safavi K, van den Ouweland A,
Zelterman D, Slack FJ, Kidd KK and Weidhaas JB: Rare BRCA1
haplotypes including 3’UTR SNPs associated with breast cancer risk.
Cell Cycle. 10:90–99. 2011.
|
|
70.
|
Rao X, Di Leva G, Li M, Fang F, Devlin C,
Hartman-Frey C, Burow ME, Ivan M, Croce CM and Nephew KP:
MicroRNA-221/222 confers breast cancer fulvestrant resistance by
regulating multiple signaling pathways. Oncogene. 30:1082–1097.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
71.
|
Zhou M, Liu Z, Zhao Y, Ding Y, Liu H, Xi
Y, Xiong W, Li G, Lu J, Fodstad O, Riker AI and Tan M:
MicroRNA-125b confers the resistance of breast cancer cells to
paclitaxel through suppression of pro-apoptotic Bcl-2 antagonist
killer 1 (Bak1) expression. J Biol Chem. 285:21496–21507. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
72.
|
Di Leva G, Gasparini P, Piovan C, Ngankeu
A, Garofalo M, Taccioli C, Iorio MV, Li M, Volinia S, Alder H,
Nakamura T, Nuovo G, Liu Y, Nephew KP and Croce CM: MicroRNA
cluster 221–222 and estrogen receptor alpha interactions in breast
cancer. J Natl Cancer Inst. 102:706–721. 2010.
|
|
73.
|
Pogribny IP, Filkowski JN, Tryndyak VP,
Golubov A, Shpyleva SI and Kovalchuk O: Alterations of microRNAs
and their targets are associated with acquired resistance of MCF-7
breast cancer cells to cisplatin. Int J Cancer. 127:1785–1794.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
74.
|
Lambertini E, Lolli A, Vezzali F,
Penolazzi L, Gambari R and Piva R: Correlation between Slug
transcription factor and miR-221 in MDA-MB-231 breast cancer cells.
BMC Cancer. 12:4452012. View Article : Google Scholar : PubMed/NCBI
|
|
75.
|
Zhao R, Wu J, Jia W, Gong C, Yu F, Ren Z,
Chen K, He J and Su F: Plasma miR-221 as a predictive biomarker for
chemoresistance in breast cancer patients who previously received
neoadjuvant chemotherapy. Onkologie. 34:675–680. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
76.
|
Velu CS and Grimes HL: Utilizing antagomiR
(antisense microRNA) to knock down microRNA in murine bone marrow
cells. Methods Mol Biol. 928:185–195. 2012.PubMed/NCBI
|
|
77.
|
Poltronieri P, D’Urso PI, Mezzolla V and
D’Urso OF: Potential of anti-cancer therapy based on anti-miR-155
oligonucleotides in glioma and brain tumours. Chem Biol Drug Des.
81:79–84. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
78.
|
Ma D, Tao X, Gao F, Fan C and Wu D:
miR-224 functions as an onco-miRNA in hepatocellular carcinoma
cells by activating AKT signaling. Oncol Lett. 4:483–488.
2012.PubMed/NCBI
|
|
79.
|
Nielsen PE, Egholm M, Berg RH and Buchardt
O: Sequence-selective recognition of DNA by strand displacement
with a thymine-substituted polyamide. Science. 254:1497–1500. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
80.
|
Demidov VV and Frank-Kamenetskii MD:
Sequence-specific targeting of duplex DNA by peptide nucleic acids
via triplex strand invasion. Methods. 23:108–122. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
81.
|
Gambari R: Peptide-nucleic acids (PNAs): a
tool for the development of gene expression modifiers. Curr Pharm
Des. 7:1839–1862. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
82.
|
Karkare S and Bhatnagar D: Promising
nucleic acid analogs and mimics: characteristic features and
applications of PNA, LNA, and morpholino. Appl Microbiol
Biotechnol. 71:575–586. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
83.
|
Nielsen PE: Antisense peptide nucleic
acids. Curr Opin Mol Ther. 2:282–287. 2002.
|
|
84.
|
Soomets U, Hällbrink M and Langel U:
Antisense properties of peptide nucleic acids. Front Biosci.
4:D782–D786. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
85.
|
Ray A and Nordén B: Peptide nucleic acid
(PNA): its medical and biotechnical applications and promise for
the future. FASEB J. 14:1041–1060. 2000.PubMed/NCBI
|
|
86.
|
Nielsen PE: Targeting double stranded DNA
with peptide nucleic acid (PNA). Curr Med Chem. 8:545–550. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
87.
|
Gambari R: Biological activity and
delivery of peptide nucleic acids (PNA)-DNA chimeras for
transcription factor decoy (TFD) pharmacotherapy. Curr Med Chem.
11:1253–1263. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
88.
|
Corradini R, Sforza S, Tedeschi T,
Totsingan F and Marchelli R: Peptide nucleic acids with a
structurally biased backbone: effects of conformational constraints
and stereochemistry. Curr Top Med Chem. 7:681–694. 2007. View Article : Google Scholar
|
|
89.
|
Sforza S, Tedeschi T, Calabretta A,
Corradini R, Camerin C, Tonelli R, Pession A and Marchelli R: A
peptide nucleic acid embedding a pseudopeptide nuclear localization
sequence in the backbone behaves as a peptide mimic. Eur J Org
Chem. 13:2441–2444. 2010. View Article : Google Scholar
|
|
90.
|
Sforza S, Corradini R, Ghirardi S, Dossena
A and Marchelli R: DNA binding of a D-Lysine-based chiral PNA:
direction control and mismatch recognition. Eur J Org Chem.
16:2905–2913. 2000. View Article : Google Scholar
|
|
91.
|
Sforza S, Tedeschi T, Corradini R and
Marchelli R: Induction of helical handedness and DNA binding
properties of peptide nucleic acids (PNAs) with two stereogenic
centres. Eur J Org Chem. 35:5879–5885. 2007. View Article : Google Scholar
|
|
92.
|
Tedeschi T, Sforza S, Corradini R and
Marchelli R: Synthesis of new chiral PNAs bearing a dipeptide-mimic
monomer with two lysine-derived stereogenic centres. Tetrahedron
Lett. 46:8395–8399. 2005. View Article : Google Scholar
|
|
93.
|
Dragulescu-Andrasi A, Zhou P, He G and Ly
DH: Cell-permeable GPNA with appropriate backbone stereochemistry
and spacing binds sequence-specifically to RNA. Chem Commun.
3:244–246. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
94.
|
Brognara E, Fabbri E, Aimi F, Manicardi A,
Bianchi N, Finotti A, Breveglieri G, Borgatti M, Corradini R,
Marchelli R and Gambari R: Peptide nucleic acids targeting miR-221
modulate p27Kip1 expression in breast cancer MDA-MB-231
cells. Int J Oncol. 41:2119–2127. 2012.PubMed/NCBI
|
|
95.
|
Gambari R, Fabbri E, Borgatti M, Lampronti
I, Finotti A, Brognara E, Bianchi N, Manicardi A, Marchelli R and
Corradini R: Targeting microRNAs involved in human diseases: a
novel approach for modification of gene expression and drug
development. Biochem Pharmacol. 82:1416–1429. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
96.
|
Fabani MM and Gait MJ: miR-122 targeting
with LNA/2′-O-methyloligonucleotide mixmers, peptide nucleic acids
(PNA), and PNA-peptide conjugates. RNA. 14:336–346. 2008.
|
|
97.
|
Fabani MM, Abreu-Goodger C, Williams D,
Lyons PA, Torres AG, Smith KGC, et al: Efficient inhibition of
miR-155 function in vivo by peptide nucleic acids. Nucleic Acids
Res. 38:4466–4475. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
98.
|
Fabbri E, Manicardi A, Tedeschi T, Sforza
S, Bianchi N, Brognara E, Finotti A, Breveglieri G, Borgatti M,
Corradini R, Marchelli R and Gambari R: Modulation of the
biological activity of microRNA-210 with peptide nucleic acids
(PNAs). Chem Med Chem. 6:2192–2202. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
99.
|
Fabbri E, Brognara E, Borgatti M,
Lampronti I, Finotti A, Bianchi N, Sforza S, Tedeschi T, Manicardi
A, Marchelli R, Corradini R and Gambari R: miRNA therapeutics:
delivery and biological activity of peptide nucleic acids targeting
miRNAs. Epigenomics. 3:733–745. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
100.
|
Manicardi A, Fabbri E, Tedeschi T, Sforza
S, Bianchi N, Brognara E, Gambari R, Marchelli R and Corradini R:
Cellular uptakes, biostabilities and anti-miR-210 activities of
chiral arginine-PNAs in leukaemic K562 cells. Chembiochem.
13:1327–1337
|
|
101.
|
Yan LX, Wu QN, Zhang Y, Li YY, Liao DZ,
Hou JH, Fu J, Zeng MS, Yun JP, Wu QL, Zeng YX and Shao JY:
Knockdown of miR-21 in human breast cancer cell lines inhibits
proliferation, in vitro migration and in vivo tumor growth. Breast
Cancer Res. 13:R22011. View Article : Google Scholar : PubMed/NCBI
|