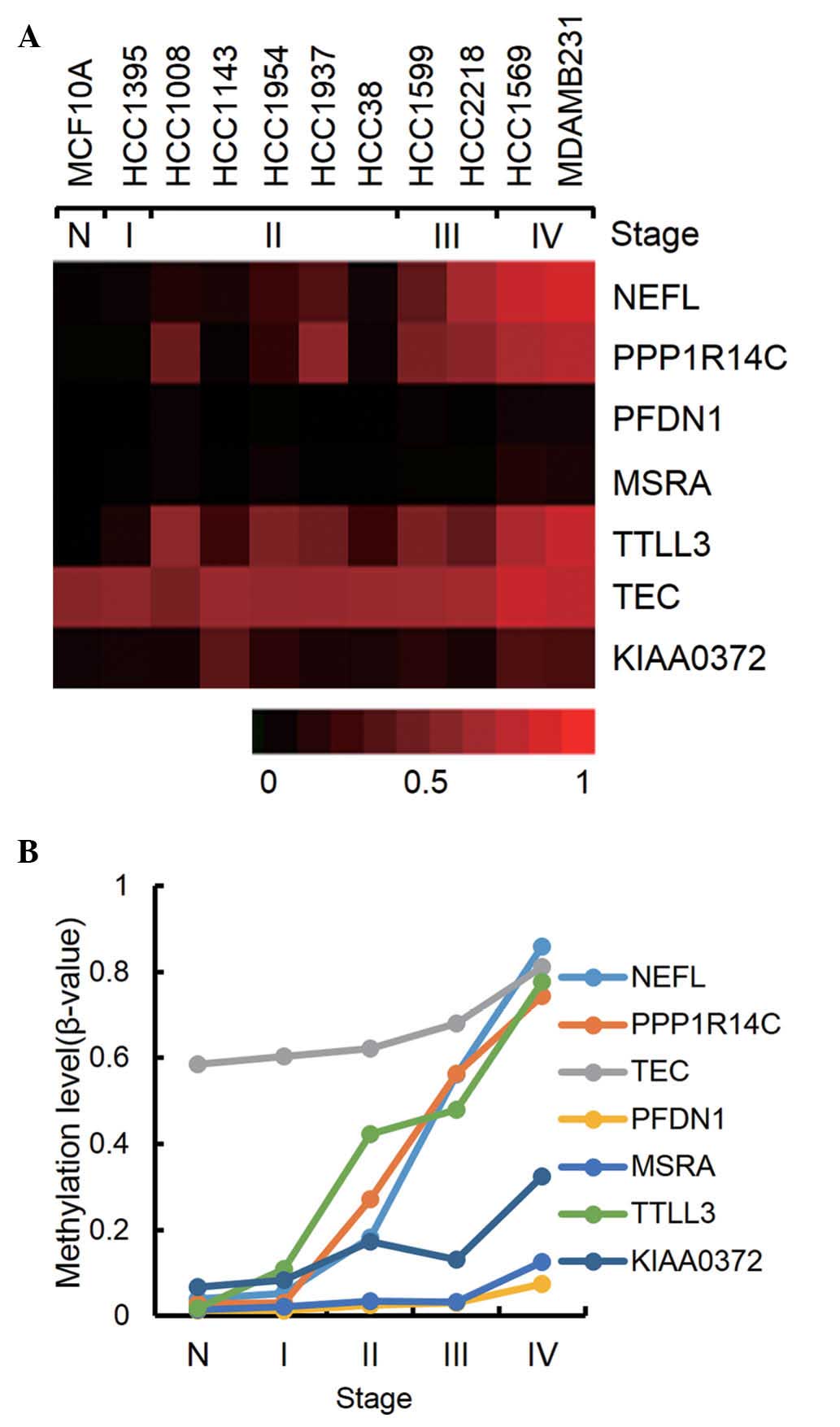
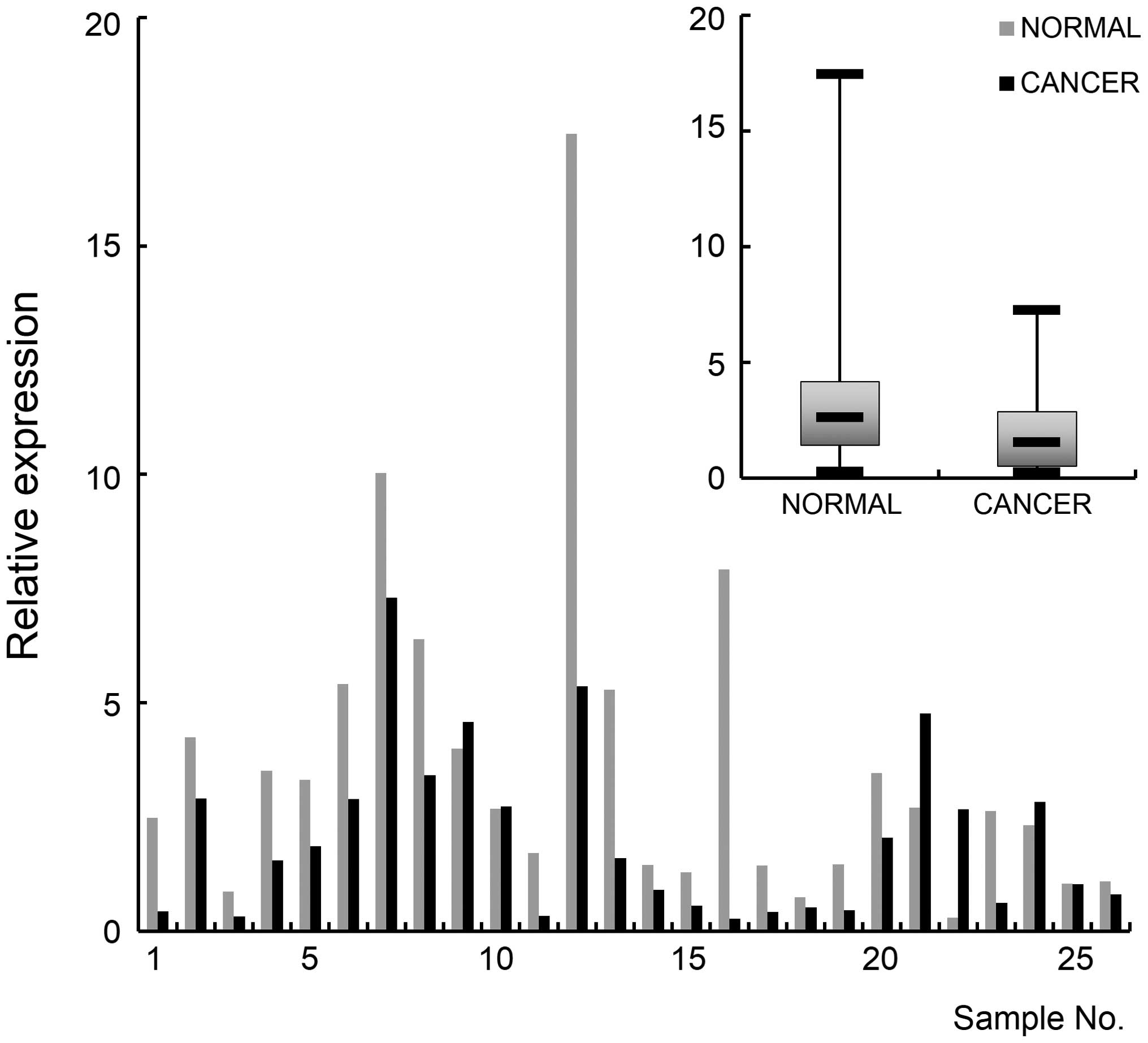
|
1.
|
Pare R, Yang T, Shin JS and Lee CS: The
significance of the senescence pathway in breast cancer
progression. J Clin Pathol. 66:491–495. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Harburg GC and Hinck L: Navigating breast
cancer: axon guidance molecules as breast cancer tumor suppressors
and oncogenes. J Mammary Gland Biol Neoplasia. 16:257–270. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Shi L, Sun L, Li Q, et al: Histone
demethylase JMJD2B coordinates H3K4/H3K9 methylation and promotes
hormonally responsive breast carcinogenesis. Proc Natl Acad Sci
USA. 108:7541–7546. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Krell J, Frampton AE, Jacob J, Castellano
L and Stebbing J: miRNAs in breast cancer: ready for real time?
Pharmacogenomics. 13:709–719. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Kang JH, Kim SJ, Noh DY, et al:
Methylation in the p53 promoter is a supplementary route to breast
carcinogenesis: correlation between CpG methylation in the p53
promoter and the mutation of the p53 gene in the progression from
ductal carcinoma in situ to invasive ductal carcinoma. Lab Invest.
81:573–579. 2001. View Article : Google Scholar
|
|
6.
|
Yan PS, Shi H, Rahmatpanah F, et al:
Differential distribution of DNA methylation within the RASSF1A CpG
island in breast cancer. Cancer Res. 63:6178–6186. 2003.PubMed/NCBI
|
|
7.
|
Matros E, Wang ZC, Lodeiro G, Miron A,
Iglehart JD and Richardson AL: BRCA1 promoter methylation in
sporadic breast tumors: relationship to gene expression profiles.
Breast Cancer Res Treat. 91:179–186. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Faryna M, Konermann C, Aulmann S, et al:
Genome-wide methylation screen in low-grade breast cancer
identifies novel epigenetically altered genes as potential
biomarkers for tumor diagnosis. FASEB J. 26:4937–4950. 2012.
View Article : Google Scholar
|
|
9.
|
Van Hoesel AQ, Sato Y, Elashoff DA, et al:
Assessment of DNA methylation status in early stages of breast
cancer development. Br J Cancer. 108:2033–2038. 2013.PubMed/NCBI
|
|
10.
|
Miyamoto K, Fukutomi T, Akashi-Tanaka S,
et al: Identification of 20 genes aberrantly methylated in human
breast cancers. Int J Cancer. 116:407–414. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Chen ST, Lin SY, Yeh KT, et al:
Mutational, epigenetic and expressional analyses of caveolin-1 gene
in breast cancers. Int J Mol Med. 14:577–582. 2004.PubMed/NCBI
|
|
12.
|
Hu M, Yao J, Cai L, et al: Distinct
epigenetic changes in the stromal cells of breast cancers. Nat
Genet. 37:899–905. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Sebova K, Zmetakova I, Bella V, et al:
RASSF1A and CDH1 hypermethylation as potential epimarkers in breast
cancer. Cancer Biomark. 10:13–26. 2011.PubMed/NCBI
|
|
14.
|
Mohammad J, Zeerak A and Hjerten S:
Dye-ligand affinity chromatography on continuous beds. Biomed
Chromatogr. 9:80–84. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Jeschke J, Van Neste L, Glockner SC, et
al: Biomarkers for detection and prognosis of breast cancer
identified by a functional hypermethylome screen. Epigenetics.
7:701–709. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Fernandez SV, Snider KE, Wu YZ, Russo IH,
Plass C and Russo J: DNA methylation changes in a human cell model
of breast cancer progression. Mutat Res. 688:28–35. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Loss LA, Sadanandam A, Durinck S, et al:
Prediction of epigenetically regulated genes in breast cancer cell
lines. BMC Bioinformatics. 11:3052010. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Kim JH, Kang HS, Kim TW and Kim SJ:
Differential methylation hybridization profiling identifies
involvement of STAT1-mediated pathways in breast cancer. Int J
Oncol. 39:955–963. 2011.PubMed/NCBI
|
|
19.
|
Wee EJ, Peters K, Nair SS, et al: Mapping
the regulatory sequences controlling 93 breast cancer-associated
miRNA genes leads to the identification of two functional promoters
of the Hsa-mir-200b cluster, methylation of which is associated
with metastasis or hormone receptor status in advanced breast
cancer. Oncogene. 31:4182–4195. 2012.
|
|
20.
|
Fackler MJ, Umbricht CB, Williams D, et
al: Genome-wide methylation analysis identifies genes specific to
breast cancer hormone receptor status and risk of recurrence.
Cancer Res. 71:6195–6207. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Bonazzi VF, Irwin D and Hayward NK:
Identification of candidate tumor suppressor genes inactivated by
promoter methylation in melanoma. Genes Chromosomes Cancer.
48:10–21. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Wloga D, Webster DM, Rogowski K, et al:
TTLL3 is a tubulin glycine ligase that regulates the assembly of
cilia. Dev Cell. 16:867–876. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Abe A, Numakura C, Saito K, et al:
Neurofilament light chain polypeptide gene mutations in
Charcot-Marie-Tooth disease: nonsense mutation probably causes a
recessive phenotype. J Hum Genet. 54:94–97. 2009. View Article : Google Scholar
|
|
24.
|
Li XQ, Li L, Xiao CH and Feng YM: NEFL
mRNA expression level is a prognostic factor for early-stage breast
cancer patients. PLoS One. 7:e311462012. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Shen L, Kondo Y, Guo Y, et al: Genome-wide
profiling of DNA methylation reveals a class of normally methylated
CpG island promoters. PLoS Genet. 3:2023–2036. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Sincic N and Herceg Z: DNA methylation and
cancer: ghosts and angels above the genes. Curr Opin Oncol.
23:69–76. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Carmona FJ and Esteller M: DNA methylation
in early neoplasia. Cancer Biomark. 9:101–111. 2010.PubMed/NCBI
|
|
28.
|
Seitz S, Werner S, Fischer J, Nothnagel A,
Schlag PM and Scherneck S: Refined deletion mapping in sporadic
breast cancer at chromosomal region 8p12-p21 and association with
clinicopathological parameters. Eur J Cancer. 36:1507–1513. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Chen B, Chen J, House MG, Cullen KJ,
Nephew KP and Guo Z: Role of neurofilament light polypeptide in
head and neck cancer chemoresistance. Mol Cancer Res. 10:305–315.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Fumagalli C, Pruneri G, Possanzini P, et
al: Methylation of O6-methylguanine-DNA methyltransferase (MGMT)
promoter gene in triple-negative breast cancer patients. Breast
Cancer Res Treat. 134:131–137. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Li L, Lee KM, Han W, et al: Estrogen and
progesterone receptor status affect genome-wide DNA methylation
profile in breast cancer. Hum Mol Genet. 19:4273–4277. 2010.
View Article : Google Scholar : PubMed/NCBI
|