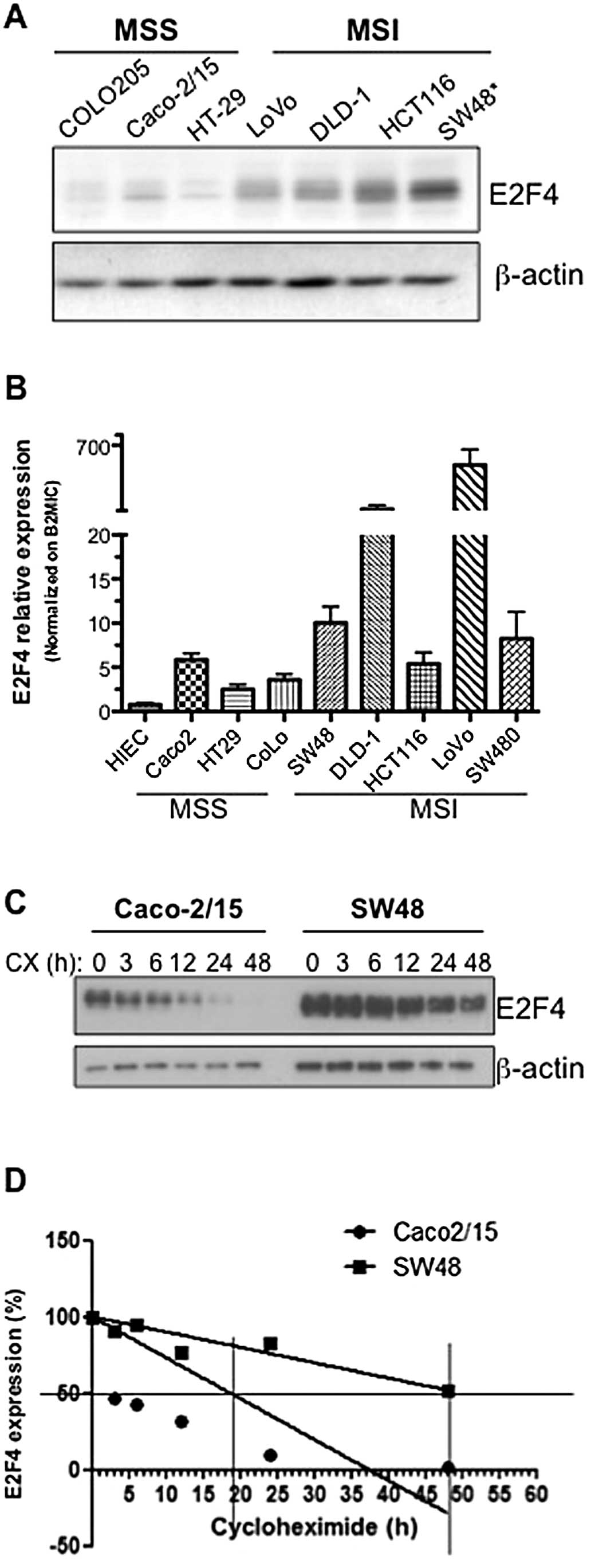
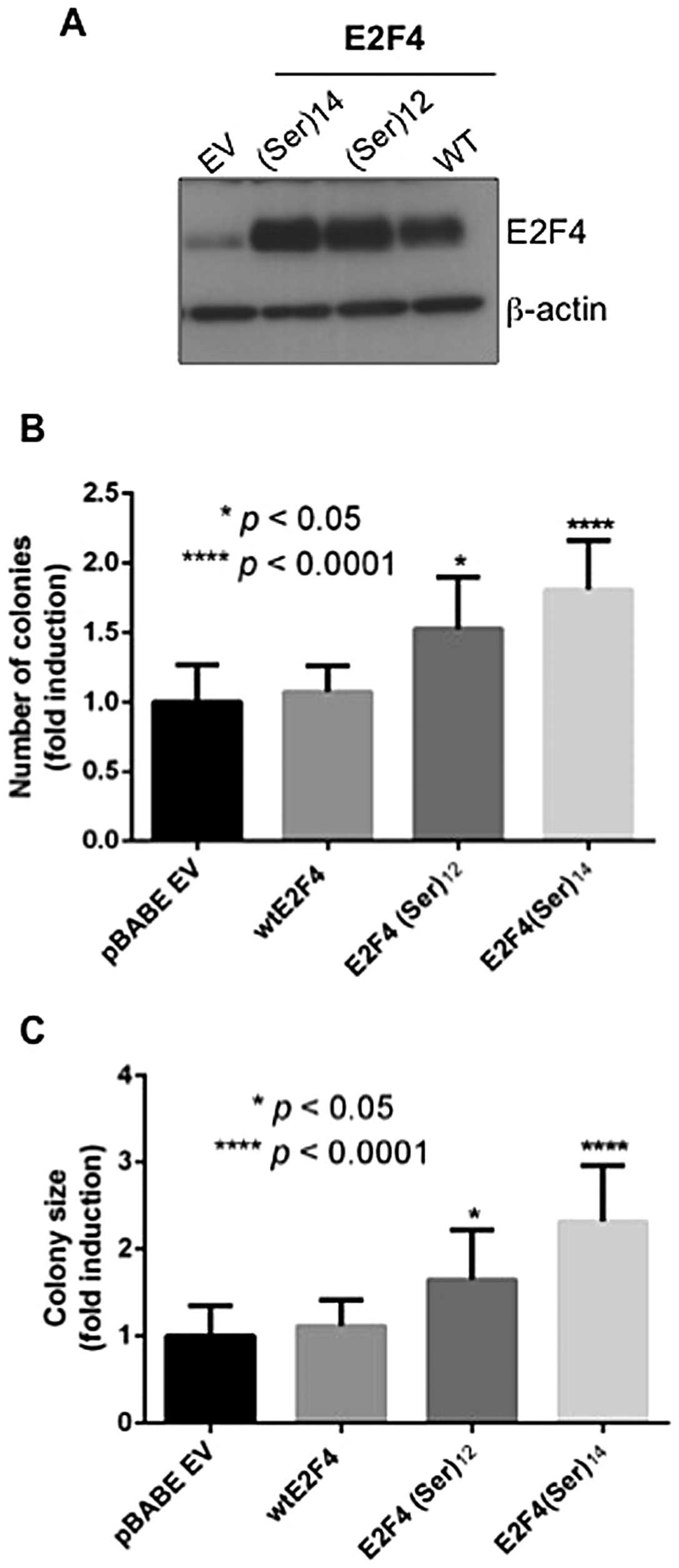
|
1.
|
Yang VW, Lewis J, Wang TC and Rustgi AK:
Colon cancer: an update and future directions. Gastroenterology.
138:2027–2028. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
van der Flier LG and Clevers H: Stem
cells, self-renewal, and differentiation in the intestinal
epithelium. Annu Rev Physiol. 71:241–260. 2009.
|
|
3.
|
Worthley DL and Leggett BA: Colorectal
cancer: molecular features and clinical opportunities. Clin Biochem
Rev. 31:31–38. 2010.PubMed/NCBI
|
|
4.
|
Boland CR and Goel A: Microsatellite
instability in colorectal cancer. Gastroenterology.
138:2073–2087.e3. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Tsantoulis PK and Gorgoulis VG:
Involvement of E2F transcription factor family in cancer. Eur J
Cancer. 41:2403–2414. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Chen HZ, Tsai SY and Leone G: Emerging
roles of E2Fs in cancer: an exit from cell cycle control. Nat Rev
Cancer. 9:785–797. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Gaubatz S, Lindeman GJ, Ishida S, Jakoi L,
Nevins JR, Livingston DM and Rempel RE: E2F4 and E2F5 play an
essential role in pocket protein-mediated G1 control. Mol Cell.
6:729–735. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Garneau H, Paquin MC, Carrier JC and
Rivard N: E2F4 expression is required for cell cycle progression of
normal intestinal crypt cells and colorectal cancer cells. J Cell
Physiol. 221:350–358. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Deschenes C, Alvarez L, Lizotte ME, Vezina
A and Rivard N: The nucleocytoplasmic shuttling of E2F4 is involved
in the regulation of human intestinal epithelial cell proliferation
and differentiation. J Cell Physiol. 199:262–273. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Yoshitaka T, Matsubara N, Ikeda M, Tanino
M, Hanafusa H, Tanaka N and Shimizu K: Mutations of E2F-4
trinucleotide repeats in colorectal cancer with microsatellite
instability. Biochem Biophys Res Commun. 227:553–557. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Souza RF, Yin J, Smolinski KN, et al:
Frequent mutation of the E2F-4 cell cycle gene in primary human
gastrointestinal tumors. Cancer Res. 57:2350–2353. 1997.PubMed/NCBI
|
|
12.
|
Ikeda M, Orimo H, Moriyama H, et al: Close
correlation between mutations of E2F4 and hMSH3 genes in colorectal
cancers with microsatellite instability. Cancer Res. 58:594–598.
1998.PubMed/NCBI
|
|
13.
|
Moriyama H, Sasamoto H, Kambara T, et al:
E2F-4 mutation in hereditary non-polyposis colorectal cancer. J Exp
Clin Cancer Res. 21:185–189. 2002.PubMed/NCBI
|
|
14.
|
Sardet C, Vidal M, Cobrinik D, Geng Y,
Onufryk C, Chen A and Weinberg RA: E2F-4 and E2F-5, two members of
the E2F family, are expressed in the early phases of the cell
cycle. Proc Natl Acad Sci USA. 92:2403–2407. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Wu CL, Zukerberg LR, Ngwu C, Harlow E and
Lees JA: In vivo association of E2F and DP family proteins. Mol
Cell Biol. 15:2536–2546. 1995.PubMed/NCBI
|
|
16.
|
Perreault N and Beaulieu JF: Use of the
dissociating enzyme thermolysin to generate viable human normal
intestinal epithelial cell cultures. Exp Cell Res. 224:354–364.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Dou QP, Zhao S, Levin AH, Wang J, Helin K
and Pardee AB: G1/S-regulated E2F-containing protein complexes bind
to the mouse thymidine kinase gene promoter. J Biol Chem.
269:1306–1313. 1994.PubMed/NCBI
|
|
18.
|
Jansen-Durr P, Meichle A, Steiner P, et
al: Differential modulation of cyclin gene expression by MYC. Proc
Natl Acad Sci USA. 90:3685–3689. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Oswald F, Lovec H, Moroy T and Lipp M:
E2F-dependent regulation of human MYC: Trans-activation by cyclins
D1 and A overrides tumour suppressor protein functions. Oncogene.
9:2029–2036. 1994.PubMed/NCBI
|
|
20.
|
Hateboer G, Kerkhoven RM, Shvarts A,
Bernards R and Beijersbergen RL: Degradation of E2F by the
ubiquitin-protea-some pathway: Regulation by retinoblastoma family
proteins and adenovirus transforming proteins. Genes Dev.
10:2960–2970. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Hofmann F, Martelli F, Livingston DM and
Wang Z: The retinoblastoma gene product protects E2F-1 from
degradation by the ubiquitin-proteasome pathway. Genes Dev.
10:2949–2959. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Martelli F and Livingston DM: Regulation
of endogenous E2F1 stability by the retinoblastoma family proteins.
Proc Natl Acad Sci USA. 96:2858–2863. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Heinen CD, Richardson D, White R and
Groden J: Microsatellite instability in colorectal adenocarcinoma
cell lines that have full-length adenomatous polyposis coli
protein. Cancer Res. 55:4797–4799. 1995.PubMed/NCBI
|
|
24.
|
Boyer JC, Umar A, Risinger JI, et al:
Microsatellite instability, mismatch repair deficiency, and genetic
defects in human cancer cell lines. Cancer Res. 55:6063–6070.
1995.PubMed/NCBI
|
|
25.
|
Takashima H, Matsumoto Y, Matsubara N, et
al: Effect of naturally occurring E2F-4 alterations on
transcriptional activation and proliferation in transfected cells.
Lab Invest. 81:1565–1573. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Martelli F, Hamilton T, Silver DP, et al:
p19ARF targets certain E2F species for degradation. Proc Natl Acad
Sci USA. 98:4455–4460. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Vlach J, Hennecke S and Amati B:
Phosphorylation-dependent degradation of the cyclin-dependent
kinase inhibitor p27. EMBO J. 16:5334–5344. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Diehl JA, Cheng M, Roussel MF and Sherr
CJ: Glycogen synthase kinase-3beta regulates cyclin D1 proteolysis
and subcellular localization. Genes Dev. 12:3499–3511. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Vandel L and Kouzarides T: Residues
phosphorylated by TFIIH are required for E2F-1 degradation during
S-phase. EMBO J. 18:4280–4291. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Dunn EF, Iida M, Myers RA, et al:
Dasatinib sensitizes KRAS mutant colorectal tumors to cetuximab.
Oncogene. 30:561–574. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Chen C and Wells AD: Comparative analysis
of E2F family member oncogenic activity. PLoS One. 2:e9122007.
View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Xu G, Livingston DM and Krek W: Multiple
members of the E2F transcription factor family are the products of
oncogenes. Proc Natl Acad Sci USA. 92:1357–1361. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Rakha EA, Pinder SE, Paish EC, Robertson
JF and Ellis IO: Expression of E2F-4 in invasive breast carcinomas
is associated with poor prognosis. J Pathol. 203:754–761. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Mori S, Chang JT, Andrechek ER, et al:
Anchorage-independent cell growth signature identifies tumors with
metastatic potential. Oncogene. 28:2796–2805. 2009. View Article : Google Scholar : PubMed/NCBI
|