Introduction
Nucleophosmin (NPM) is a ubiquitous nucleolar
phosphoprotein with multifunction. NPM has been shown to associate
with the assembly and transport of ribosome, the duplication of
centrosome and DNA damage response (1,2). NPM
regulates the nucleolus activities and cell proliferation,
indicating its crucial roles in cell growth. However, the roles of
NPM in cell differentiation and the mechanisms of its
transportation and relocation are still largely unknown. Recent
data suggest NPM is tightly correlated with the development of a
tumor (1), its expression level is
enhanced in various cancers, such as bladder cancer and prostate
cancer (3–5). As a potential proto-oncogene, NPM can
affect cell growth through multiple pathways (1). Nevertheless, how NPM affects the
reversal of malignant type of tumor cells is still unknown.
In our previous study, we found NPM was a nuclear
matrix protein and its expression was downregulated in the induced
differentiation of human osteosarcoma, human neuroblastoma tumors
and human liver cancer (6–8), indicating NPM functioned in
regulating the differentiation of tumor cells (9). Liver cancer is the most common
malignant gastrointestinal cancer and the screening for functional
proteins related to liver tumor cell differentiation will
contribute to the elucidation of the mechanism of liver cancer
development and its early diagnosis. Hereby, based on the induced
effect of hexamethylene bisacetamide (HMBA) on the differentiation
of human liver cancer cells, we extended our study to investigate
the location and expression of NPM in the nuclear matrix of human
liver cancer cells as well as its relationship with several
oncogenes and tumor suppressor genes. Our aim was to provide more
evidence to reveal the roles of NPM on regulating liver cancer cell
proliferation and differentiation.
Materials and methods
Ethics statement
The use of human liver cancer tissues in this study
was approved by the ethics committees at the Medical College of
Xiamen University. Written consent was obtained from each patient
with liver cancer.
Cell line, tissues and other
reagents
Human liver cancer SMMC-7721 cells were purchased
from China Center for Type Culture Collection. Human liver cancer
tissues and matched non-cancerous tissues were obtained from fresh
surgical material of patients with liver cancer from the First
Affiliated Hospital of Xiamen University (all liver tissues were
histologically confirmed). All antibodies used were purchased from
Santa Cruz Biotechnology (Santa Cruz, CA, USA) or Cell Signaling
Technology, Inc. (CST); the immunohistological kit was purchased
from Zhongshan Goldenbridge Biotechnology, Co., Ltd. The PRMI-1640
and bovine serum for cell culture were products of Hyclone (Thermo
Scientific). HMBA was purchased from Sigma. RNAiso Plus kit,
PrimeScript™ RT reagent kit and SYBR® Premix Ex Taq™
were products of Takara.
Cell culture
SMMC-7721 cells were routinely cultured in RPMI-1640
supplemented with 10% heat-inactivated fetal bovine serum (Hyclone)
at 37°C in a humidified atmosphere containing 5% CO2.
Twelve hours after seeding, SMMC-7721 cells were maintained in the
culture medium containing 5 mmol/l HMBA for 7 days to induce
differentiation.
Preparation of whole cell lysates and
nuclear matrix lysates
The whole cell protein samples were prepared as
follows: cells were washed in ice-cold phosphate buffered saline
(PBS) and then lysed in lysis buffer [7 mol/l urea, 2 mol/l
thiourea, 4%
3-[(3-cholanidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS),
1.5% Triton X-100, 1% pharmalyte (pH 3–10, Bio-Rad), 65 mmol/l
DL-Dithiothreitol (DTT), 40 mmol/l Tris, complete protease
inhibitor cocktail tablets (Roche)]. The suspension was then
sonicated for 15 min at 0°C and centrifuged at 12,000 × g for 20
min.
The protein samples of liver tissues were prepared
as follows: 0.5 g of tissue from both cancer and matched normal
tissues was frozen in liquid nitrogen and ground with mortar and
pestle to yield tissue powder and then suspended in ice-cold RIPA
lysis buffer. The suspension was then sonicated for 15 min at 0°C
and centrifuged at 12,000 × g for 20 min.
Nuclear matrix proteins were prepared using a
modified method described by Michishita et al (10). After washing in ice-cold PBS twice,
SMMC-7721 cells were suspended in cytoskeleton (CSK) buffer [100
mmol/l KCl, 3 mmol/l MgCl2, 5 mmol/l ethylene glycol
tetraacetic acid (EGTA), 10 mmol/l
piperazine-N,N’-bis(2-ethanesulfonic acid) (PIPES), 300 mmol/l
sucrose, 0.5% Triton X-100, and 2 mmol/l PMSF, pH 6.8] for 10 min
at 0°C. After being centrifuged at 600 × g for 5 min, the pellet
was resuspended in digestion buffer (identical to CSK buffer except
for 50 mmol/l NaCl instead of KCl) containing 400 μg/ml
DNaseI for 30 min at room temperature and centrifuged at 600 × g.
Cold ammonium sulfate at a final concentration of 0.25 mol/l was
used to precipitate proteins. After centrifugation, the pellet was
dissolved in lysis buffer [7 mol/l urea, 2 mol/l thiourea, 4%
CHAPS, 1.5% Triton X-100, 1% Pharmalyte (pH 3–10 Bio-Rad), 65
mmol/l DTT, 40 mmol/l Tris, complete protease inhibitor cocktail
tablets (Roche)] and then sonicated at 0°C for 20 min. Finally, the
suspension was centrifuged at 10,000 × g and 4°C for 30 min and the
supernatants were used as nuclear matrix extracts. Protein
concentrations were determined by Bradford assay.
2-DE, MALDI-TOF-MS analysis and protein
identification
2-D PAGE was performed as follows. Protein lysates
were diluted in sample buffer with 2% Immobiline™ DryStrip gel
(IPG) buffer, pH 3–10, nonlinear (GE Healthcare). The samples were
applied to IPG Drystrips (17 cm, pH 3–10, GE Healthcare). After
isoelectric focusing, the strips were equilibrated and the second
dimensional SDS-polyacrylamide gel electrophoresis was carried out.
The triplicate sets of silver-stained gels were scanned using a
Umax Power Look III photometer and analyzed with the PD Quest 8.0
software (Bio-Rad). The 2-DE gel images were normalized and
compared by matching method. Differentially expressed spots were
analyzed.
The spots were cut and digested using modified
sequencing grade trypsin (Promega). The digested peptides were
eluted and dried in freeze drying equipment. After that, the
samples were dissolved with 2 μl of matrix solution
containing 10 mg/ml alpha cyano-4-hydroxy cinnamic acid (CHCA,
Sigma) and were submitted to Bruker III MALDI-TOF mass
spectrometer. The spectra were internally calibrated using the
trypsin autolysis products by Flex Analysis software and searched
against Swiss-Prot and NCBI database using the Mascot tool from
Matrix Science. All the searches were analyzed with a 50-ppm mass
tolerance.
Western blotting
For western blot experiments, 20 μg cell
lysates were loaded and separated on polyacrylamide gels and then
transferred to polyvinylidene fluoride (PVDF) membranes (Millipore)
according to standard protocol. The membranes were blocked for 1 h
at room temperature in 5% albumin from bovine serum (BSA). The
target proteins were probed with primary antibodies and horseradish
peroxidase (HRP)-labeled secondary antibodies (Santa Cruz
Biotechnology). β-actin was used as an indicator for equality of
lane loading. Antibody positive bands were visualized using ECL
western blot detection reagents (Pierce). Quantification of the
immunoreactive bands was performed with Quantity One software.
Real-time quantitative RT-PCR
Total RNA was isolated from the cells and tissues
using RNAiso Plus kit (Takara). A two-step reverse
transcription-PCR procedure was performed using the PrimeScript RT
reagent kit (Takara) following the manual instructions. The
resultant cDNA was then quantified by Rotor Gene 6000 with SYBR
Premix Ex Taq (Takara). The expression level of NPM was normalized
to β-actin mRNA. The PCR primers used were: i) NPM-F (5′-GGAGGTG
GTAGCAAGGTTCC) and NPM-R (5′-TTCACTGGCGCT TTTTCTTCA); ii) β-actin-F
(5′-CATGTACGTTGCTAT CCAGGC) and β-actin-R (5′-CTCCTTAATGTCACGCAC
GAT). All PCR reactions were performed in triplicates to ensure
reproducibility.
Immunohistochemistry analysis
Immunohistochemical analyses were performed by the
following methods. Sections of 5 μm were taken from tissue
block and affixed to polylysine coated slides and air-dried
overnight at 37°C. After dewaxing and antigen retrieval, endogenous
peroxidase was quenched with 3% hydrogen peroxide for 10 min. After
washing in distilled water three times, the cover slips were
blocked for 30 min with 3% BSA at room temperature and then
incubated in a mixture of primary antibodies (mouse monoclonal
anti-NPM, 1:200 dilution, Santa Cruz Biotechnology) diluted in 2%
BSA. After overnight incubation, the cover slips were washed
thoroughly with PBST (PBS, 0.05% Tween-20). The next steps were
performed according to the manual of the 2-step plus®
poly-HRP anti-mouse/rabbit IgG detection system (ZSGB-Bio Co.,
Ltd.). The antigen-antibody complex was visualized with
diaminobenzidine (DAB) substrate. Images of the stained sections
(three sections per sample) were captured and the quantitative
analysis was performed with Image-Pro Plus 6.0 software.
For the samples of immunofluorescence analysis, the
CY3-conjugated goat anti-mouse IgG was used as secondary antibody
in 2% BSA for 1 h at room temperature in the dark. After three
washes in PBS, the sections were sealed with nail polish to prevent
movement while observed under a laser confocal scanning
microscope.
Laser-scanning confocal microscopy for
double-immunofluorescence analysis
Cells grown on cover slips were fixed in 4%
parafomaldehyde for 10 min at room temperature and then washed in
PBS. For permeabilization the cells were immersed in PBS containing
0.5% Triton X-100 for 10 min and then washed in PBS three times for
5 min. After blocking in 1% BSA in PBST (PBS containing 0.05%
Tween-20), cells were incubated in a mixture of two primary
antibodies (mouse anti-human NPM/rabbit anti-human c-Fos, mouse
anti-human NPM/rabbit anti-human c-Myc, mouse anti-human NPM/rabbit
anti-human P53, mouse anti-human NPM/rabbit anti-human Rb) in 1%
BSA in PBST in a humidified chamber for 1 h at room temperature.
After washed three times in PBS for 5 min cells were incubated with
a mixture of two secondary antibodies (FITC-conjugated goat
anti-mouse IgG/CY3-conjugated goat anti-rabbit IgG) in 1% BSA for 1
h at room temperature in the dark. After three washes in PBS, the
cells were incubated in DAPI for 5 min in the dark and rinsed in
PBS, then mounted with a drop of mounting medium and sealed with
nail polish to prevent movement under the microscope. Image
acquisition was performed with laser confocal scanning microscopy
(TCS-SP2 MP, Leica). For the sections of cancer tissues and
adjacent normal tissues, after dewaxing and antigen retrieval, the
following steps were in accord with the standard method of cell
samples.
Construction of GST-NPM expression vector
and GST pull-down assay
Full-length NPM was isolated from SMMC-7721 cells by
RT-PCR using NPM-specific primers. The primers used were: NPM-F
(5′-CGCGGATCCATGGAAGATTCG ATGGACAT) and NPM-R (5′-CCGCTCGAGAAAGAGAC
TTCCTCCACTGCC). The full-length NPM was then sequenced and cloned
into the expression vector pGEX-4T-2 with glutathione S-transferase
(GST) tag at the N terminus. The E. coli-expressed
GST-fusion (bait) protein, GST-NPM, is immobilized onto the
MagneGST Particles (Promega) to affinity purify any proteins (prey)
from SMMC-7721 cell lysates according to the technical manual of
Promega MagneGST™ pull-down system. The bait-prey protein complexes
contained in eluted samples were further analyzed by SDS-PAGE and
western blotting.
Co-immunoprecipitation (Co-IP) assay
A typical experiment used 1000 μl of whole
cell lysate (WCL) of liver tumor cells (containing 1000 μg
of soluble protein), and was incubated with 1 μg of purified
NPM IgG for 4 h or overnight at 4°C. Control immunoprecipitation
experiments included 1 μg of purified pre-immune IgG or
irrelevant IgG. WCL (50 μl) was used as positive control. A
80-μl aliquot of a 50% (v/v) slurry of protein A-coupled
Sepharose beads was then added and incubated for 2 h at 4°C. The
beads was pelleted in a micro-centrifuge (3,000 × g for 30 sec) and
washed three times in wash buffer (identical to lysis buffer). Then
the beads were boiled in reducing SDS-PAGE sample loading buffer
and analyzed by SDS-PAGE and western blotting.
Results
Identification of NPM in nuclear matrix
lysates of liver cells by MALDI-TOF-MS
The nuclear matrix lysates of SMMC-7721 cells before
and after HMBA treatment were subjected to 2-DE PAGE followed by
silver-staining of the gel. All procedures (extraction of nuclear
matrix proteins, 2-DE PAGE and silver-staining) were independently
repeated three times to ensure the reproducibility of 2-DE PAGE.
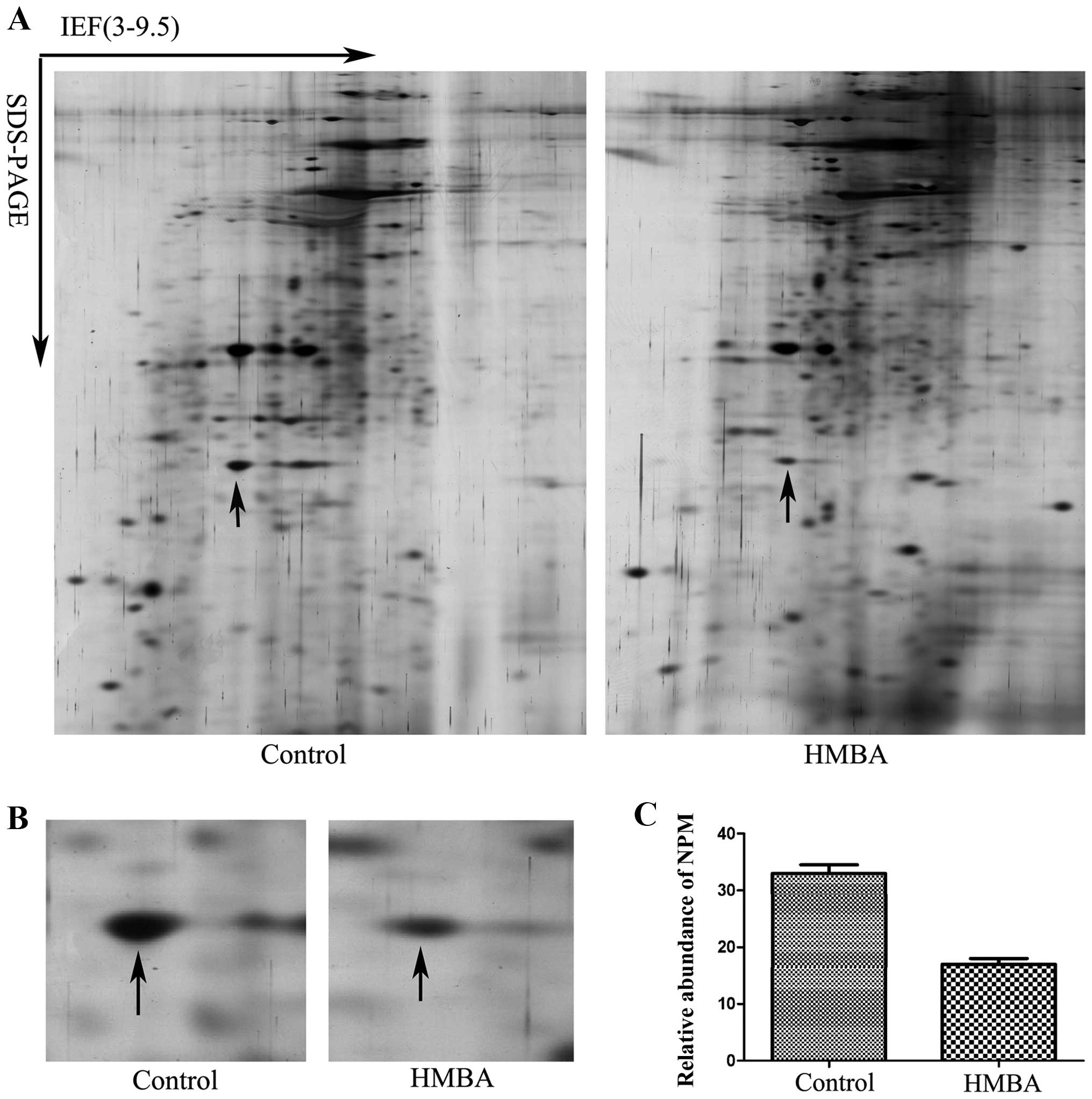
Representative gel images are shown in Fig. 1A. PD Quest 8.0 software (Bio-Rad)
was used for quantification analysis. The differential protein
spots in intensity were excised and digested with typsin and then
were identified by mass spectrometry. After searching against
MASCOT database (www.matrixscience.com) the protein spot labeled with
an arrow in Fig. 1B was identified
as NPM (Table I). Fig. 1B is the enlarged maps of NPM from
2-DE gels and Fig. 1C shows the
relative expression level of NPM in cells before and after HMBA
treatment.
 | Table I.NPM protein was identified by
searching MASCOT database (www.matrixscience.com). |
Table I.
NPM protein was identified by
searching MASCOT database (www.matrixscience.com).
| Protein name | NCBI entry | Mol. Mass calc
(Da) | pI (calc) | Score |
|---|
| Nucleophosmin | gi83641870 | 28497 | 4.56 | 176 |
Upregulation of NPM protein level both in
SMMC-7721 cells and liver cancer tissues
To verify the existence and abberant expression of
NPM in nulcear matrix and liver tissues, western blot analysis was
performed. The whole cell lysates and nuclear matrix lysates of
SMMC-7721 cells and human liver tissues and matched normal tissues
were separated by SDS-PAGE and then transferred to PVDF membrane in
semi-dry conditions according to the standard protocol. The results
showed that when compared with that in the differentiated cells the
protein level of NPM in liver tumor SMMC-7721 cells was improved
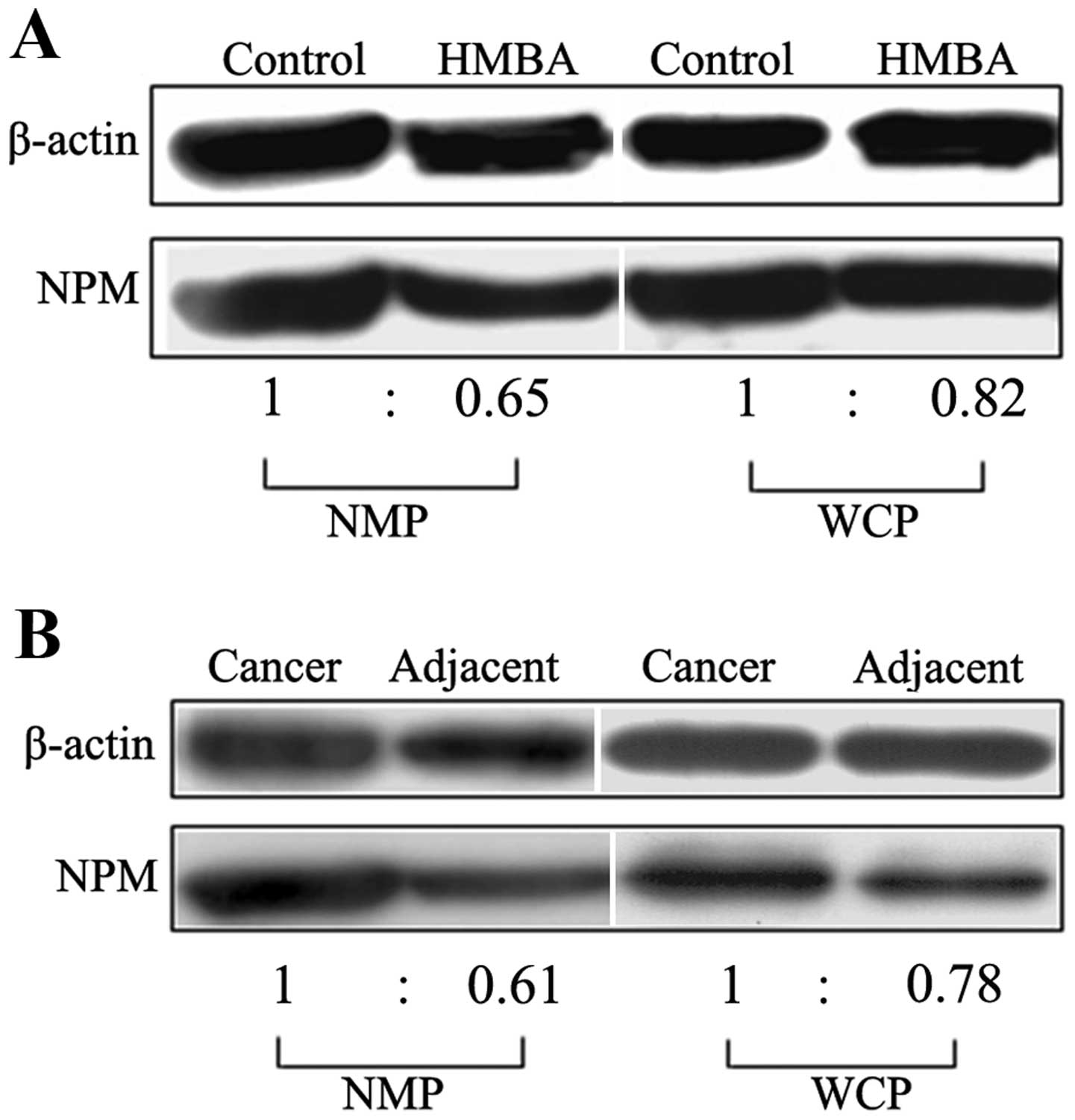
both in nuclear matrix and whole cell proteins (Fig. 2A). For uncovering the possible
changes of NPM in liver cancer development, we further probed the
expression level of NPM in human liver cancer tissues. We found the
expression level of NPM in liver cancer tissues was much higher
than that in adjacent non-cancerous tissues among 30 patients who
were clinically diagnosed with liver cancer (Fig. 2B).
Upregulation of NPM mRNA level both in
SMMC-7721 cells and liver cancer tissues
Western blot data suggested that NPM protein was
overexpressed in SMMC-7721 cells and liver cancer tissues. To
investigate whether the mRNA level of NPM was changed, the mRNA
expression of NPM was examined in SMMC-7721 cells with or without
HMBA-treatment and in liver cancer tissues and matched normal
tissues. Consistent with the protein expression, NPM was highly
expressed in SMMC-7721 cells compared with differentiated cells
(Fig. 3A). Of the paired liver
cancer tissues analyzed, all the cases showed enhanced NPM mRNA
expression compared with the matched normal tissues (Fig. 3B–D).
Evaluation of NPM expression in liver
cancer tissues
To investigate the expression of NPM in liver cancer
tissues, immunohistochemistry and immunofluorescence was performed
to detect NPM expression in paired primary liver cancer and their
adjacent non-cancer tissues. In addition, results showed that the
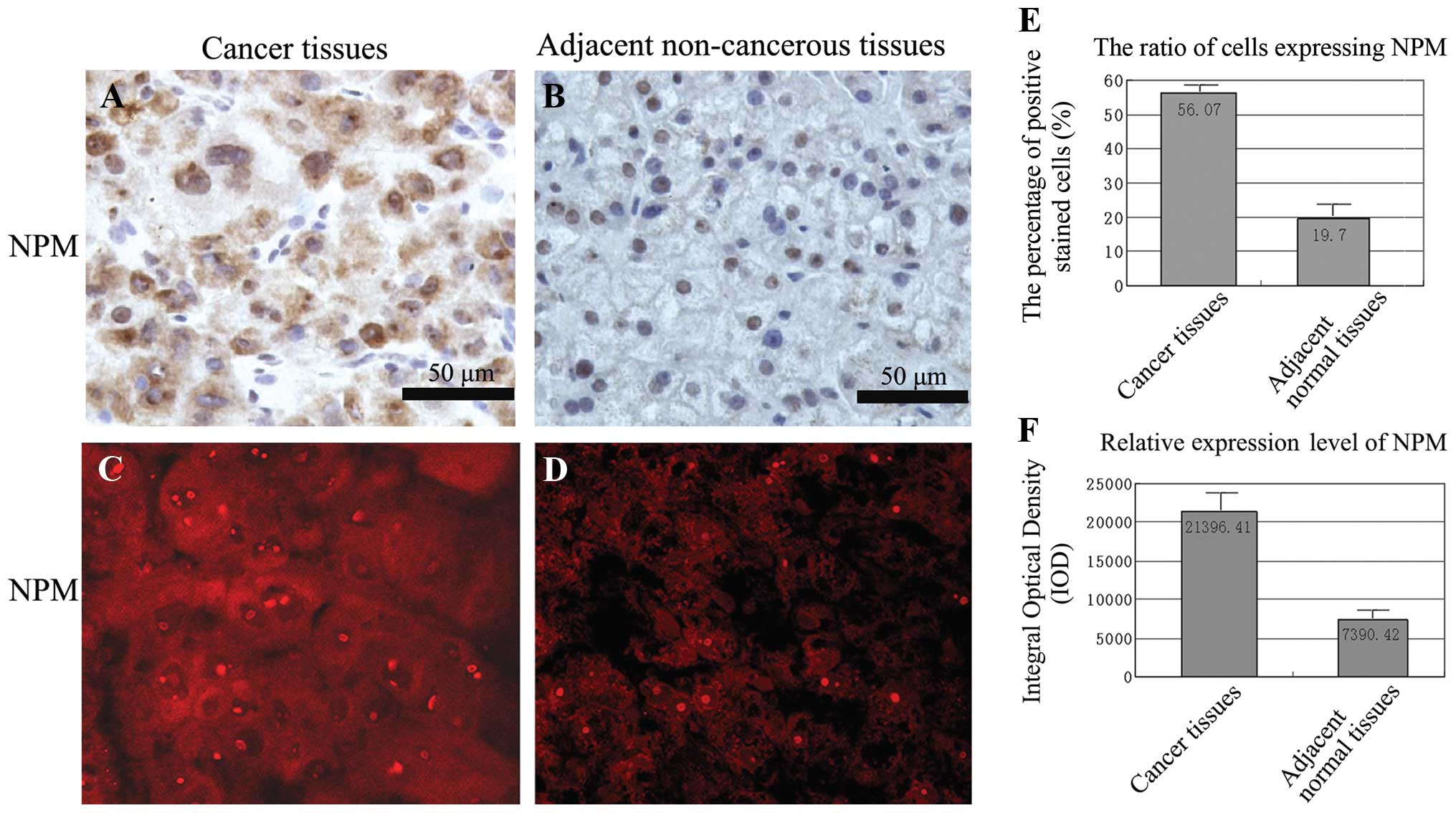
brown-yellow granules (Fig. 4A and
B) and the red fluorescence (Fig.
4C and D) of NPM were mainly expressed in the nucleus and the
cytoplasm of cancer tissues whereas weakly expressed in the nucleus
of adjacent non-cancer tissues. Quantitative analysis of NPM
expression with Image-Pro Plus software showed that the ratio of
positively stained cell altered from 56.07% in cancer tissues to
19.70% in adjacent normal tissues and the relative expression level
of NPM decreased by 65% (Fig. 4E and
F).
Intracellular colocalization of NPM with
c-Fos, c-Myc, P53 and Rb
To corroborate the physical proximity of NPM with
c-Fos, c-Myc, P53 and Rb at the subcellular level, the double
immunofluorescent staining method was employed. The green
fluorescence representing NPM was observed in the nucleus
especially around the periphery of the nucleolus, and some weak
fluorescent signal could be detected in the cytoplasm in control
SMMC-7721 cells; after HMBA treatment the green fluorescence was
mainly assembled in the nucleolus, the intensity of green
fluorescence decreased. The yellow fluorescence indicates the
colocalization of NPM with other proteins labeled by red Cy3.
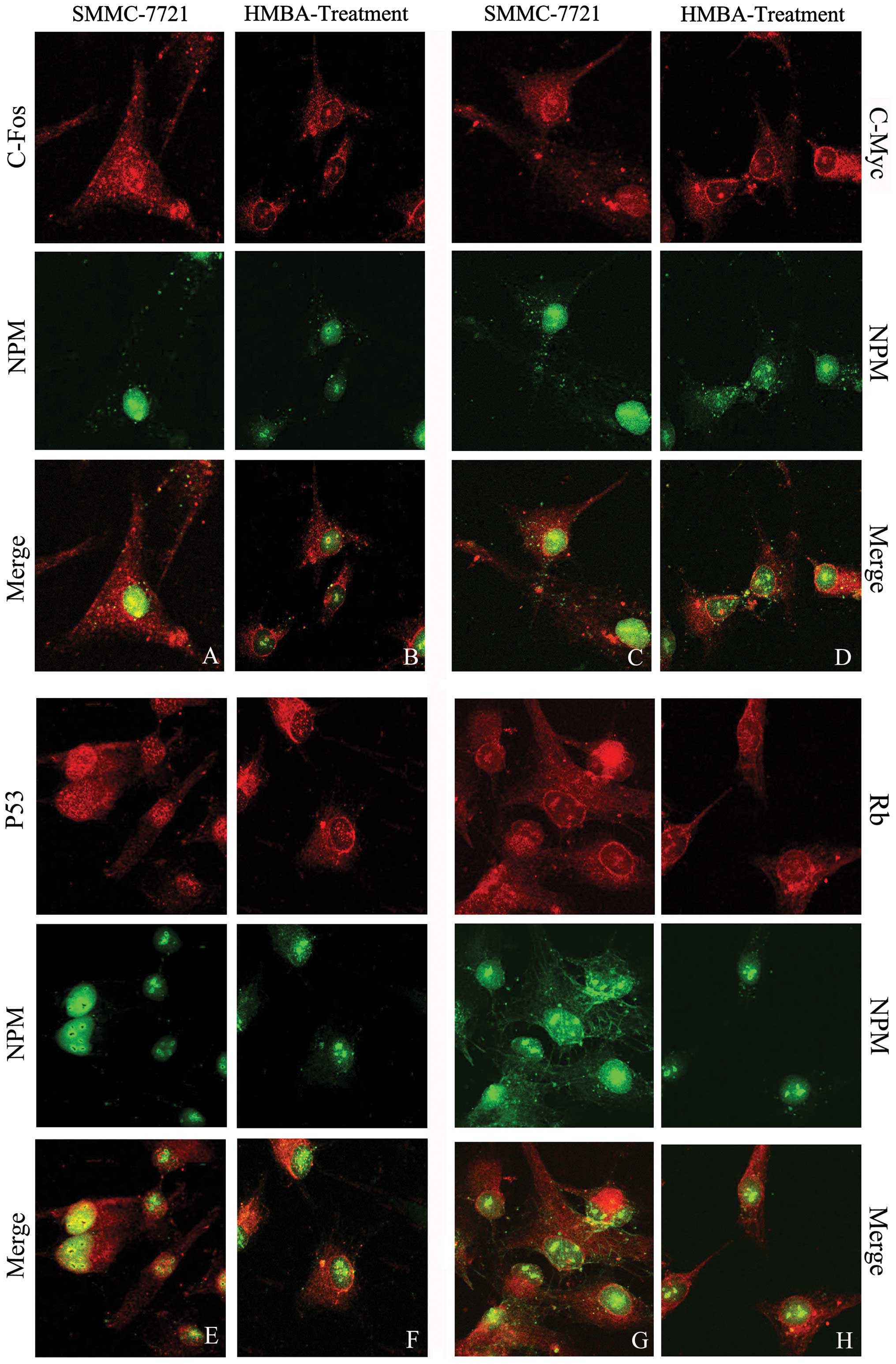
Colocalization of NPM with c-Fos in liver
tumor cells
The red fluorescence representing c-Fos accumulated
both in nucleus and in the cytoplasm. NPM colocalized with c-Fos in
the nucleus, especially around the regions of the nucleolus
(Fig. 5A). After HMBA treatment
the intensity of yellow fluorescence in nucleus was dramatically
weakened and the colocalization is mainly located in the nucleolus
(Fig. 5B).
Colocalization of NPM with c-Myc in liver
tumor cells
The observation of laser confocal microscope showed
the c-Myc red fluorescence was distributed through the whole cell.
The merged picture shows NPM obviously colocalized with c-Myc in
the nucleolar periphery (Fig. 5C);
HMBA treatment resulted in the decreased fluorescent intensity of
c-Myc in the nucleus, and the fluorescent intensity in the
cytoplasm was much stronger than that in the nucleus (Fig. 5D). The colocalization of NPM with
c-Myc was observed in the cytoplasm and nucleolus in the
differentiated cells, which demonstrated a tendency for the
colocalization of NPM and c-Myc to transfer from the nucleolus to
the cytoplasm.
Colocalization of NPM with P53 in liver
tumor cells
In SMMC-7721 cells the red fluorescence representing
P53 was distributed both in the nucleus and cytoplasm, but the
fluorescent intensity in the nucleus was much stronger than that in
the cytoplasm. The merged image reveals that the strong
colocalization of NPM with P53 widely distributed in the nucleus
especially in the nucleolar periphery (Fig. 5E). In HMBA-treated cells, the
merged results show the yellow fluorescence as weak in the nuclear
region, indicating the colocalization between P53 and NPM was
decreased and transferred outside the nucleus (Fig. 5F).
Colocalization of NPM with Rb in liver
tumor cells
The red fluorescence representing Rb accumulated
both in nucleus and cytoplasm (Fig.
5G). After HMBA treatment the Rb red fluorescence dispersed in
the cytoplasm, especially around the periphery of the nucleus. The
yellow fluorescence showed the colocalization of NPM and Rb mainly
distributed in the nucleolus (Fig.
5H).
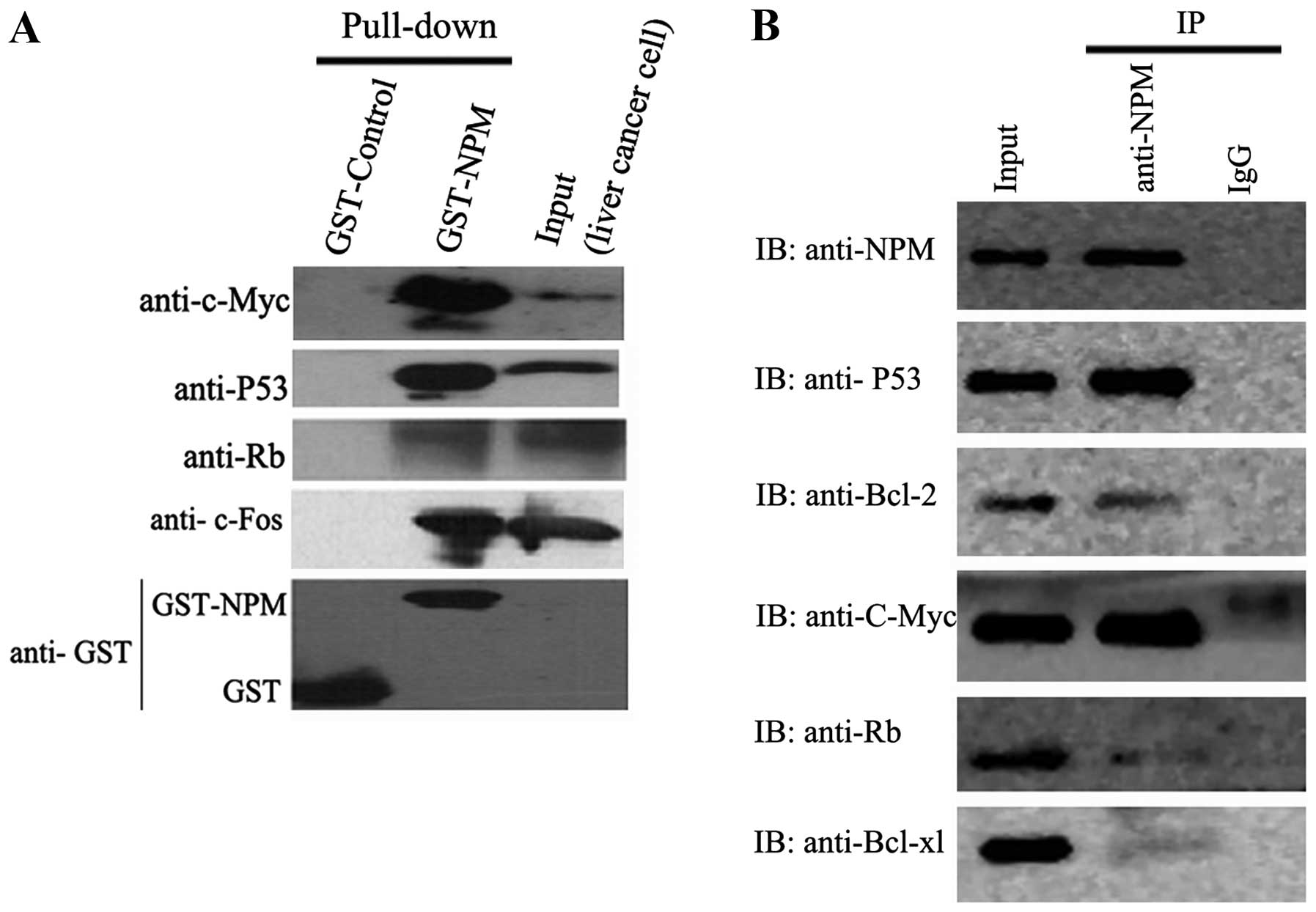
Interaction of NPM with c-Myc, c-Fos,
P53, Bcl-2,Bcl-xl and Rb in liver cancer cells
To verify the potential interactions indicated by
colocalization between NPM and other proteins, GST-NPM fusion
protein was constructed and used in a pull-down assay. For western
blot analysis, GST protein expressed from pGEX-4T-2 vector was used
as negetive control and the cell lysates of SMMC-7721 cells was
used as positive control. Pull-down results showed that NPM
interacts with c-Myc, c-Fos, P53 and Rb (Fig. 6A). Co-IP assay further verified the
interactions between NPM and these proteins in vivo.
Moreover, co-IP assay found the interactions between NPM and Bcl-2
or Bcl-xl (Fig. 6B).
Discussion
Expression changes of NPM in nuclear
matrix during the differentiation of liver tumor cells
As a ubiquitous nucleolar phosphoprotein, NPM is
involved in a series of biological processes, such as the assembly
and transport of ribosome, the duplication of centrosome and the
DNA damage response. The location and expression changes of NPM are
closely related to cell proliferation and carcinogenesis. In the
present study proteomic analysis revealed that NPM was included in
the protein components of liver cancer cell nuclear matrix,
moreover, its expression level was downregulated after HMBA-induced
differentiation, which was further confirmed by western blot assay.
Hsu et al (11) reported
the down regulation of NPM in RA-induced HL-60 cells, and in our
previous study we also found decreased level of NPM during the
differentiation of tumor cells (6), which was consistent with the results
of the present study. Overexpression of NPM resulted in high
cellular growth rate (12). Data
in this study further confirmed that NPM was a nuclear matrix
protein and its expression level was decreased in differentiated
tumor cells, proving that NPM played crucial roles on regulating
the differentiation of liver cancer cells.
Expression alterations of NPM both in
liver cancer cells and liver cancer tissues and matched non-cancer
tissues
The aberrant expression of NPM in human cancer
tissues has been widely reported. Accumulated data showed the
overexpression of NPM in tumor cells (13–15).
In this study we found NPM was overexpressed in human liver cancer
cells both at protein and at mRNA levels; the immunocytochemical
results showed NPM was mainly expressed in the nucleolus and
nucleoplasm, and it was concentrated into the nucleolus after HMBA
treatment. For further revealing the expression and location of NPM
in human liver cancer tissues, we performed immunohistochemistry on
the paraffin sections of cancer tissues. The results, for the first
time, displayed that NPM expression was strongly positive in the
cytoplasm of liver cancer tissues while it was weakly positive in
the nucleus of matched non-cancer tissues, indicating differential
location and expression level of NPM during human liver
carcinogenesis, which corroborated the data from liver cell lines.
Development of bladder cancer and liver cancer associated with the
enhanced expression of NPM (4,16),
and higher NPM protein level resulted in more advanced tumor
stages, grades, poor prognosis, and likelihood of recurrence
(4), moreover, the mRNA levels of
NPM was higher in cancerous tissues when compared with normal
tissues (17), and our results
further confirmed the enhanced mRNA level of NPM in liver cancer
tissues.
In recent years, studies have been carried out on
the functional roles of NPM in the differentiation of tumor cells,
however, the detailed mechanisms are still unknown. Our present
study showed the differential location and expression level of NPM
either in liver cell lines or in human liver cancer tissues,
indicating its crucial roles in liver carcinogenesis.
Interaction and altered colocalization of
NPM with oncogenes and tumor suppressor genes
As an important functional regulatory protein, NPM
can regulate the activities of several genes, such as p53 and p14
(18). NPM might regulate the
differentiation of tumor cells through interaction with some
proteins of oncogenes and tumor suppressor genes.
In the present study, laser confocal scanning
microscope revealed NPM colocalized with c-Fos, c-Myc, P53 and Rb
at the nucleolar periphery of liver tumor cells; moreover, the
colocalized regions were translocated due to the HMBA-induced
differentiation. The colocalized relationship indicates the
potential interaction of NPM with c-Fos, c-Myc, P53 and Rb.
The oncogene c-myc, a member of myc gene family, is
closely related to cell proliferation. The protein of c-myc can
directly interact with NPM (19–22).
Our results corroborated the deductions and found that NPM
colocalized with c-Myc in the periphery of nucleolus. In the
differentiated liver cells, NPM coexpressed with c-Myc in the
nucleoli and the cytoplasm, indicating the interaction between NPM
and c-Myc was a dynamic process. How NPM and c-Myc shuttled from
the nucleoli to the cytoplasm needs more consideration. C-Fos can
improve the transcriptional activities of several genes related to
cell proliferation through binding with c-Jun. We, for the first
time, reported that NPM interacted with c-Fos in the nucleolus and
nucleoplasm of tumor cells, and the colocalized locus changed
during the differentiation of tumor cells. These results indicated
NPM may directly or indirectly interact with c-Fos and further
regulated cell differentiation. We also report here that NPM
interacted with Bcl-2 and Bcl-xl, both of which are
apoptosis-related proteins, indicating the potential role of NPM in
apoptosis regulation. There is no previous report on the
interactions between Bcl-2, Bcl-xl and NPM. It was reported that
NPM was an inhibitor of p53, NPM could suppress the activity of p53
in DNA damage response (23). NPM
directly interacted with p53 and coexpressed at the nucleolus
(24). Our results further
corroborated the interactions between NPM and p53. Lin et al
(25) found NPM also interacted
with the over-phosphorylated Rb protein. We found NPM colocalized
with Rb at the nucleolar periphery. We provided evidence for
further probing the detailed regulatory mechanism of NPM through
interaction with Rb.
Our research proved the expression changes of NPM in
the differentiation of human liver tumor cells, as well as its
colocalization and interaction with c-Fos, c-Myc, P53, Rb, Bcl-2
and Bcl-xl. As a regulatory factor, NPM played pivotal roles in
regulating liver tumor cell differentiation induced with HMBA,
indicating NPM may be the potential target protein of HMBA. The
location and expression changes of NPM significantly affected the
proliferation and differentiation of liver tumor cells. NPM
regulated cell differentiation through interacting with the
proteins from oncogenes and tumor suppressor genes. Exploring the
undiscovered roles of NPM is of great significance for revealing
the regulation of human liver cell carcinogenesis and its
reversal.
Collectively, the present study showed that NPM was
overexpressed both in liver tumor cells and in liver cancer
tissues; its expression level and location were altered during the
HMBA-induced differentiation. NPM colocalized and interacted with
c-Myc, c-Fos, P53 and Rb and the colocalizations were eliminated or
translocated by HMBA treatment, indicating the functional roles of
NPM during the differentiation of liver tumor cells. Further
investigation on the molecular mechanism of NPM regulating cancer
cell differentiation will help to reveal its detailed functional
roles.
Acknowledgements
This study was supported by National
Natural Science Foundation of China (Grant numbers: 81272245,
81272921 and 81201305), Natural Science Foundation of Fujian
Province (Grant numbers: 2011J01256 and 2013J01359) and Joint
Programme by Healthy Care System and Educational Department in
Fujian Province (Grant number: WKJ-FJ-16).
References
|
1.
|
Grisendi S, Mecucci C, Falini B and
Pandolfi PP: Nucleophosmin and cancer. Nat Rev Cancer. 6:493–505.
2006. View
Article : Google Scholar
|
|
2.
|
Lindström MS: Elucidation of motifs in
ribosomal protein S9 that mediate its nucleolar localization and
binding to NPM1/nucleophosmin. PLoS One. 7:e524762012.PubMed/NCBI
|
|
3.
|
Lim MJ and Wang XW: Nucleophosmin and
human cancer. Cancer Detect Prev. 30:481–490. 2006. View Article : Google Scholar
|
|
4.
|
Tsui KH, Juang HH, Lee TH, Chang PL, Chen
CL and Yung BY: Association of nucleophosmin/B23 with bladder
cancer recurrence based on immunohistochemical assessment in
clinical samples. Acta Pharmacol Sin. 29:364–370. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Hsu CY and Yung BY: Over-expression of
nucleophosmin/B23 decreases the susceptibility of human leukemia
HL-60 cells to retinoic acid-induced differentiation and apoptosis.
Int J Cancer. 88:392–400. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Li QF, Shi SL, Liu QR, Tang J, Song J and
Liang Y: Anticancer effects of ginsenoside Rg1, cinnamic acid, and
tanshinone IIA in osteosarcoma MG-63 cells: nuclear matrix
downregulation and cytoplasmic trafficking of nucleophosmin. Int J
Biochem Cell Biol. 40:1918–1929. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Liang Y, Li QF, Zhang XY, Shi SL and Jing
GJ: Differential expression of nuclear matrix proteins during the
differentiation of human neuroblastoma SK-N-SH cells induced by
retinoic acid. J Cell Biochem. 106:849–857. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Tang J, Niu JW, Xu DH, Li ZX, Li QF and
Chen JA: Alteration of nuclear matrix-intermediate filament system
and differential expression of nuclear matrix proteins during human
hepato-carcinoma cell differentiation. World J Gastroenterol.
13:2791–2797. 2007.
|
|
9.
|
Derenzini M, Sirri V, Trere D and Ochs RL:
The quantity of nucleolar proteins nucleolin and protein B23 is
related to cell doubling time in human cancer cells. Lab Invest.
73:497–502. 1995.
|
|
10.
|
Michishita E, Kurahashi T, Suzuki T, et
al: Changes in nuclear matrix proteins during the senescence-like
phenomenon induced by 5-chlorodeoxyuridine in HeLa cells. Exp
Gerontol. 37:885–890. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Hsu CY and Yung BY: Down-regulation of
nucleophosmin/B23 during retinoic acid-induced differentiation of
human promyelocytic leukemia HL-60 cells. Oncogene. 16:915–923.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Dergunova NN, Bulycheva TI, Artemenko EG,
et al: A major nucleolar protein B23 as a marker of proliferation
activity of human peripheral lymphocytes. Immunol Lett. 83:67–72.
2002. View Article : Google Scholar
|
|
13.
|
Subong EN, Shue MJ, Epstein JI, Briggman
JV, Chan PK and Partin AW: Monoclonal antibody to prostate cancer
nuclear matrix protein (PRO:4-216) recognizes nucleophosmin/B23.
Prostate. 39:298–304. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Nozawa Y, Van Belzen N, Van der Made AC,
Dinjens WN and Bosman FT: Expression of nucleophosmin/B23 in normal
and neoplastic colorectal mucosa. J Pathol. 178:48–52. 1996.
View Article : Google Scholar
|
|
15.
|
Zhang Y: The ARF-B23 connection:
implications for growth control and cancer treatment. Cell Cycle.
3:259–262. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Ulanet DB, Torbenson M, Dang CV,
Casciola-Rosen L and Rosen A: Unique conformation of cancer
autoantigen B23 in hepatoma: a mechanism for specificity in the
autoimmune response. Proc Natl Acad Sci USA. 100:12361–12366. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
You BJ, Huang IJ, Liu WH, Hung YB, Chang
JH and Yung BY: Decrease in nucleophosmin/B23 mRNA and telomerase
activity during indomethacin-induced apoptosis of gastric KATO-III
cancer cells. Naunyn Schmiedebergs Arch Pharmacol. 360:683–690.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Gjerset RA: DNA damage, p14ARF,
nucleophosmin (NPM/B23), and cancer. J Mol Histol. 37:239–251.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Li Z, Boone D and Hann SR: Nucleophosmin
interacts directly with c-Myc and controls c-Myc-induced
hyperproliferation and transformation. Proc Natl Acad Sci USA.
105:18794–18799. 2008. View Article : Google Scholar
|
|
20.
|
Yung BY: c-Myc-mediated expression of
nucleophosmin/B23 decreases during retinoic acid-induced
differentiation of human leukemia HL-60 cells. FEBS Lett.
578:211–216. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Arabi A, Rustum C, Hallberg E and Wright
AP: Accumulation of c-Myc and proteasomes at the nucleoli of cells
containing elevated c-Myc protein levels. J Cell Sci.
116:1707–1717. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Grandori C, Gomez-Roman N, Felton-Edkins
ZA, et al: c-Myc binds to human ribosomal DNA and stimulates
transcription of rRNA genes by RNA polymerase I. Nat Cell Biol.
7:311–318. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Maiguel DA, Jones L, Chakravarty D, Yang C
and Carrier F: Nucleophosmin sets a threshold for p53 response to
UV radiation. Mol Cell Biol. 24:3703–3711. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Colombo E, Marine JC, Danovi D, Falini B
and Pelicci PG: Nucleophosmin regulates the stability and
transcriptional activity of p53. Nat Cell Biol. 4:529–533. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Lin CY, Liang YC and Yung BY:
Nucleophosmin/B23 regulates transcriptional activation of E2F1 via
modulating the promoter binding of NF-kappaB, E2F1 and pRB. Cell
Signal. 18:2041–2048. 2006. View Article : Google Scholar : PubMed/NCBI
|