Introduction
C-mannosylation is a unique type of
glycosylation in which α-D-mannose is directly attached to the
indole C2 carbon atom of a tryptophan residue via a C-C
linkage (1,2). It is observed within the sequence
motif Trp-Xaa-Xaa-Trp/Cys (Xaa represents any amino acids) in
proteins, and the N-terminus tryptophan may be
C-mannosylated (3,4). Some proteins have been reported to be
C-mannosylated, for example, in ribonuclease 2 (5), F-spondin (6), thrombospondin (7), a disintegrin and metalloproteinase
with thrombospondin motif (ADAMTS)-like 1/punctin-1 (8) and interleukin-21 receptor (9); however, the biological functions of
C-mannosylation remain largely unknown. Recently, the
responsible C-mannosyltransferase for thrombospondin type-1
repeats (TSR-1), dpy19, was identified in C. elegans
(10). C-mannosylation is
expected to affect protein polarity because the polar mannose is
attached to the non-polar tryptophan. Moreover, the attachment of
mannose to tryptophan is expected to induce a conformational change
of the target proteins. Therefore, C-mannosylation might
affect protein functions, such as protein stability, secretion,
intracellular localization and even enzymatic activity. In fact, it
was reported that C-mannosylation interferes with the
secretion of ADAMTS-like 1/punctin-1 (8). Moreover, it is also known that
C-mannosylated peptides, derived from TSR-1, enhance
lipopolysaccharide-induced signaling, such as tumor necrosis factor
alpha and c-jun N-terminal kinase in RAW264.7 cells (11,12).
However, little is known about other functions of
C-mannosylation, so further detailed investigainvestigations
are required to clarify the roles of its modification.
Hyaluronic acid (HA) is a component of extracellular
matrices (ECMs). Hyaluronidases (HYALs) hydrolyze the β1–4 linkage
between N-acetylglucosamine and glucuronic acid of HA
polymers. ECM degradation enzymes, such as matrix
metalloproteinases and heparanase, have been known to promote
cancer metastasis and invasion (13–15).
In the same way, upregulation of HYALs, especially HYAL1, has been
reported to correlate with tumor cell proliferation, migration,
invasion and angiogenesis in various cancers, including breast,
prostate and ovarian cancers (16–18).
Moreover, HYAL1 is known to correlate with juvenile idiopathic
arthritis (JIA). Defects of HYAL1 are not only the cause of
mucopolysaccharidosis but are present in JIA (19).
In this investigation, we examined the presence of
C-mannosylation in human HYAL1 and its role for HYAL1
functions. As a result, we show possible roles for secretion and
enzymatic activity by C-mannosylation of HYAL1.
Materials and methods
Cell culture
Human fibrosarcoma HT1080 cells, purchased from
Japanese Cancer Research Resources Bank (JCRB), were cultured in
Dulbecco’s modified Eagle’s medium (DMEM; Nissui, Tokyo, Japan),
supplemented with 10% (v/v) fetal bovine serum, 200 U/ml penicillin
G, 200 mg/l kanamycin, 600 mg/l L-glutamine, and 2.25 g/l
NaHCO3 at 37°C in a humidified incubator with 5%
CO2.
Establishment of the HYAL1-overexpressing
cell line
The human HYAL1-myc-his6 gene was
amplified from a human prostate cancer LNCaP cell cDNA library and
subcloned into the pCI-neo vector (Promega, Madison, WI). The
permanent cell line expressing HYAL1-myc-his6 was
established by transfecting the vector into HT1080 cells, followed
by 400 μg/ml G418 (Roche Applied Sciences, Indianapolis, IN)
selection. The clonal cells that expressed high levels of
myc-his6-tagged HYAL1 were designated HT1080-HYAL1-MH
cells. The cells that were transfected with pCI-neo vector were
designated HT1080-neo.
Western blot analysis
To perform western blot analysis, we used a slightly
modified version of a previously described methods (20–23).
Cells were lysed in a lysis buffer [50 mM Tris-HCl, pH 7.5, 150 mM
NaCl, 0.1% sodium dodecyl sulfate (SDS), 1% Triton X-100, 1% sodium
deoxycholate and 1 mM phenylmethylsulfonyl fluoride] at 4°C with
sonication. The lysates were centrifuged at 14,000 rpm for 10 min,
and the amount of protein was measured by staining with Coomassie
Brilliant Blue (CBB) G-250 (Bio-Rad Laboratories, Hercules, CA).
Loading buffer (350 mM Tris-HCl, pH 6.8, 30% glycerol, 0.012%
bromophenol blue, 6% SDS and 30% 2-mercaptoethanol) was added to
each lysate, which was subsequently boiled for 3 min and
electrophoresed on SDS-polyacrylamide gels. Proteins were
transferred to PVDF membranes and immunoblotted with anti-c-myc
(Santa Cruz Biotechnology, Inc., Santa Cruz, CA) or anti-α-tubulin
(Sigma, St. Louis, MO) antibodies. Detection was performed with
enhanced chemiluminescence reagent (Millipore Corporation,
Billerica, MA).
Purification of recombinant protein from
conditioned medium and whole-cell lysate
To purify recombinant HYAL1 from the conditioned
medium, HT1080-HYAL1-MH cells were cultured in serum-free DMEM for
24 h, and the conditioned media was concentrated on an
ultrafiltration membrane and incubated with Ni-NTA agarose (Qiagen,
Hilden, Germany) for 2 h at 4°C. The Ni-NTA agarose was washed five
times with phosphate-buffered saline (PBS) and eluted with 500 mM
imidazole.
To purify recombinant HYAL1 from the whole-cell
lysate, HT1080-HYAL1-MH cells were lysed with binding buffer (50 mM
Tris-HCl, pH 7.5, 0.5 M NaCl, 8 M Urea, 20 mM imidazole), and cell
lysate was incubated with Ni-NTA agarose for 2 h at 4°C. The Ni-NTA
agarose was washed four times with PBS and eluted with 500 mM
imidazole.
The obtained samples were electrophoresed on
SDS-polyacrylamide gels and stained with CBB R-250. The purified
proteins were used for mass spectrometry (24,25).
Mass spectrometry
Purified recombinant HYAL1 was subjected to
SDS-polyacrylamide gels. After CBB staining, the bands were excised
and treated with 0.05 μg of sequencing-grade modified
trypsin (Promega) at 37°C for 12 h in 0.1 M Tris-HCl, pH 8.0. The
digests were desalted using Zip TipC18μ (Millipore Corporation) and
applied to matrix-assisted laser desorption ionization time of
flight mass spectrometry (MALDI-TOF MS) on a Ultraflex TOF/TOF MS
(Bruker Daltonics, Bremen, Germany) in reflector mode using
α-cyano-4-hydroxycinnamic acid as the matrix. The selected peaks
were subjected to MS/MS analysis in LIFT mode.
Measurement of hyaluronidase
activity
To measure HYAL1 activity by in-gel digestion assay,
we carried out the experiment as previously reported (26,27).
Equal numbers (2.0×106 cells) of HT1080-neo and
HT1080-HYAL1-MH cells were cultured in serum-free DMEM for 24 h and
concentrated by using Ni-NTA agarose. Ni-NTA-bound proteins were
eluted, and the samples were electrophoresed on an
SDS-polyacrylamide gel containing rooster comb HA (0.2 mg/ml;
Sigma) at 4°C. The gel was washed twice with SDS extraction buffer
(50 mM Tris-HCl, pH 7.5, 0.1 M NaCl, 2.5% Triton X-100) for 1 h and
incubated in assay buffer (50 mM sodium formate, pH 4.0, 150 mM
NaCl) at 37°C for 24 h. After incubation, the gel was stained with
0.5% alcian blue (Sigma) containing 20% ethanol and 10% acetic acid
solution for 2 h and destained with 25% methanol and 7.5% acetic
acid solution. HYAL1 activity can be observed as a transparent band
in the blue pigment background.
Docking model of HYAL1 and HA by computer
simulation
We examined the conformational changes of HYAL1 when
it was C-mannosylated and unmannosylated by using the
Molecular Operating Environment (MOE; Chemical Computing Group
Inc., Montreal, Canada) per previously reported methods (28,29).
The Protein Data Bank sequences Apis melliflora HYAL-HA
complex (PDB ID: 1FCV) and human HYAL1 (PDB ID: 2PE4) were loaded
into MOE. Hydrogen atoms were added, and the protonation states
were assigned using the Protonate 3D tool of MOE. Using
AMBER12:EHT, one of the calculation methods of force field, energy
was minimized. The 2PE4 sequence was aligned with 1FCV, removing
the Apis melliflora HYAL and HA structure. Energy
minimization was applied to human HYAL1 and HA. A 3D structure of
the refined model was used for the Site Finder module of MOE, which
can identify possible ligand-binding sites. Protein docking was
performed by alpha sphere and excluded volume-based ligand-receptor
docking (ASE-Dock), which is based on ligand-receptor interaction
energies, and the resulting score was calculated as
UDOCK. UDOCK is expressed by the sum of
Urefine, the entire free energy of ligand-receptor
interaction, and Ustrain, the free energy of ligand
stability. In the ASE-Dock module, ligand atoms have alpha spheres
within 1 Å. Utilizing this property, models are created, and ligand
atoms from many conformations that are generated by superposition
with these points can be evaluated and scored by maximum overlap
with alpha spheres and minimum overlap with the receptor atoms.
Protein docking between human HYAL1 and dummy ligand atoms, which
were created at chosen LBSs as centroids, was performed. The
resulting UDOCK was calculated. An α-D-mannose was
properly added to Trp130 of human HYAL1 to be
C-mannosylated within MOE, and then the energy of
C-mannosylated human HYAL1 was minimized. Protein docking
between C-mannosylated human HYAL1 and dummy ligand atoms
was performed. The resulting UDOCK was calculated.
Results
Secreted HYAL1 has enzymatic activity,
but is not C-mannosylated
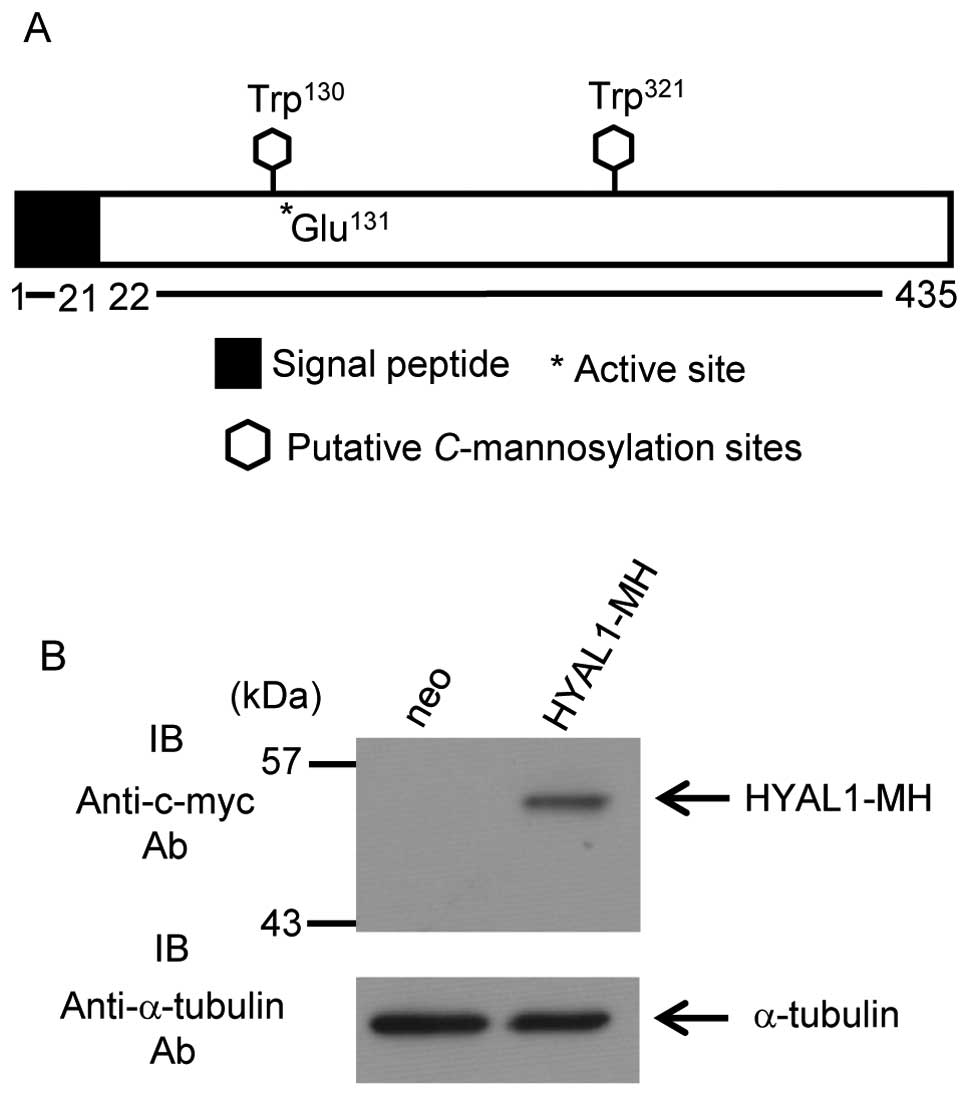
Human HYAL1 contains two predicted
C-mannosylation consensus sequences, Trp130 and
Trp321. The active site of HYAL1 (Glu131) is
located next to Trp130 (Fig. 1A). To examine whether HYAL1 is
C-mannosylated or not, we established HT1080 cells that
stably expressed HYAL1 (Fig.
1B).
HYAL1 has a signal peptide at the N-terminus domain
(Fig. 1A) and is predicted to
secrete. We examined whether HYAL1 secretes or not and confirmed
HYAL1 secretion (Fig. 2A).
Furthermore, secreted HYAL1 possessed enzymatic activity (Fig. 2A; see alcian blue staining). We
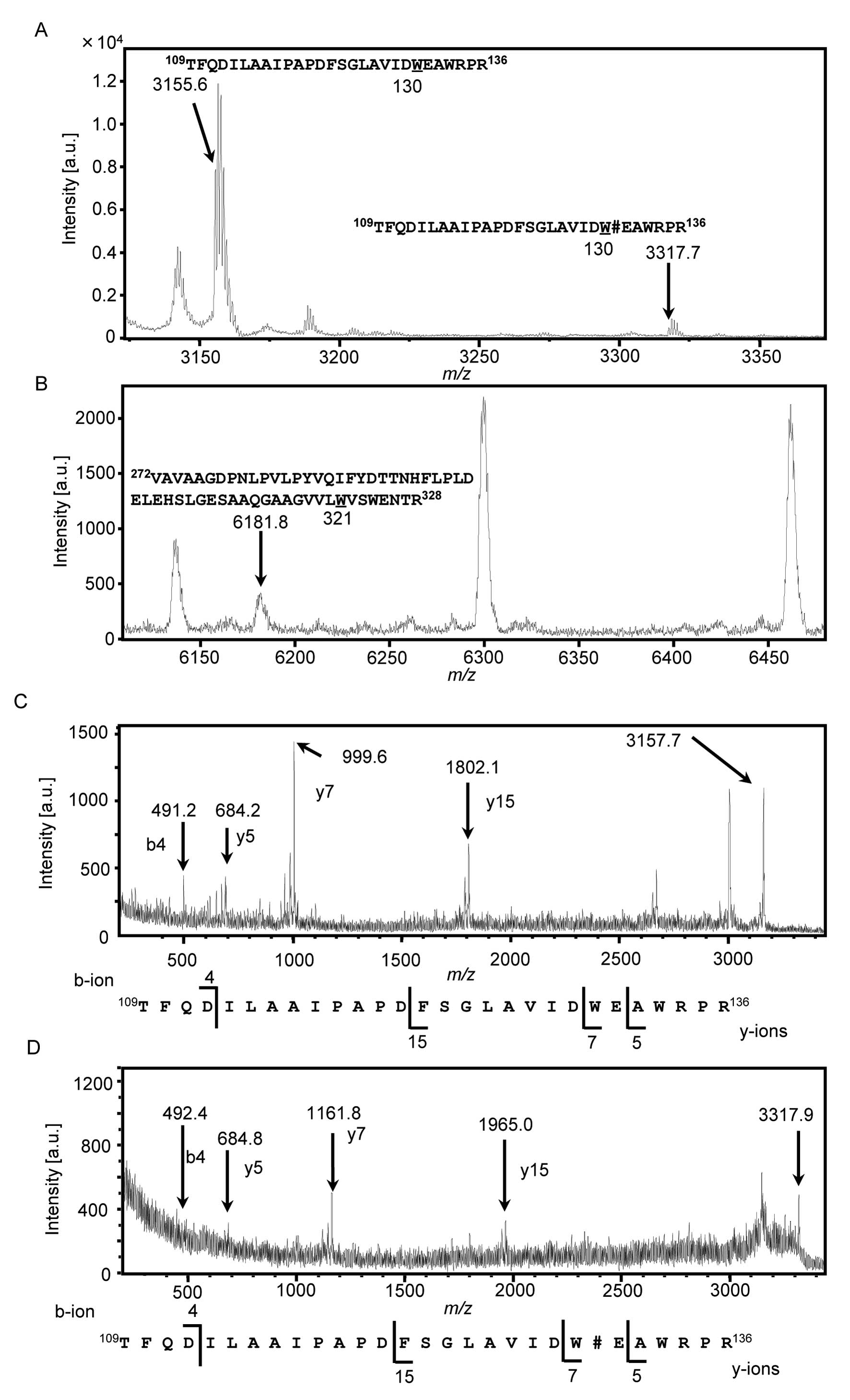
undertook MALDI-TOF MS analysis to determine whether secreted HYAL1
is actually C-mannosylated (30). To obtain recombinant HYAL1 protein,
we purified HYAL1 from conditioned medium of HT1080-HYAL1-MH cells
by using Ni-NTA agarose (data not shown). Purified HYAL1 was
treated with trypsin, and the resulting mixture of the peptides was
analyzed by MALDI-TOF MS (Fig. 2B and
C). C-mannosylation, the attachment of one mannose to a
tryptophan residue, should provoke an increase of m/z 162.
The peptides containing Trp130 were observed at
m/z 3,156.0, but no peaks around at m/z 3,318, which
is the mass resulting from C-mannosylation of the peptide,
were detected (Fig. 2B). In the
same way, we performed MS analysis of the peptides containing
Trp321, and the peak was observed at m/z 6,181.3,
which indicated that it was an unmannosylated peptide (Fig. 2C). However, the peak that was
located at about m/z 6,343, which is the mass resulting from
C-mannosylation of the peptide, was not detected (Fig. 2C). These results revealed that
secreted HYAL1 was not C-mannosylated, although secreted
HYAL1 possessed enzymatic activity.
Determination of the mannose attachment
residue within HYAL1 by using MALDI-TOF MS
Since HYAL1 is reported to lie mainly in lysosomes
(31), we tried to examine whether
intracellular HYAL1 is C-mannosylated. We purified HYAL1
proteins from whole-cell lysates of HT1080-HYAL1-MH cells (data not
shown), and the obtained recombinant HYAL1 was treated with
trypsin. The resulting mixture of peptides that were digested by
trypsin was analyzed by MALDI-TOF MS (Fig. 3A and B). The peptides containing
Trp130 were observed at m/z 3,155.6, which were
unmannosylated peptides (Fig. 3A).
Moreover, a peak at m/z 3,317.7, which was an increase of
m/z 162, was also observed (Fig. 3A). It suggested that the peptides
containing Trp130 were C-mannosylated. We
performed MS analysis on the peptides containing Trp321,
and a peak was observed at only m/z 6,181.8, which was
predicted to be an unmannosylated peptide (Fig. 3B). These results indicated that
intracellular HYAL1 was C-mannosylated at only
Trp130. We further analyzed the peptides containing
Trp130 by MALDI-TOF MS/MS to confirm the mannose
attachment residue (Fig. 3C and
D). Unmannosylated peptides (Fig.
3C) and mannosylated peptides (Fig. 3D) were analyzed, and b4 and y5 ions
were observed at the same position. However, y7 and y15 ions were
observed at the m/z 162-increased positions in mannosylated
peptides (Fig. 3D) compared with
unmannosylated peptides (Fig. 3C).
These results confirmed that C-mannosylation of HYAL1
occurred at Trp130 only in the cell lysate, not the
secreted HYAL1, suggesting that C-mannosylation might
suppress HYAL1 secretion.
HYAL1 conformation is highly affected by
C-mannosylation
Since secreted HYAL1 showed enzymatic activity
(Fig. 2A), C-mannosylation
may not be essential for HYAL1 activity. However, since
C-mannosylated Trp130 lies next to
Glu131, the active site of HYAL1, we assumed that
C-mannosylation might regulate HYAL1 enzymatic activity. In
order to examine the effect of C-mannosylation on HYAL1
enzymatic activity, we employed MOE, a computer simulation software
that could calculate structural changes or free energies of
interacting proteins. The crystal structure of human
hyaluronidase-HA has not been reported, so we used the Protein Data
Bank sequences for Apis melliflora hyaluronidase-HA complex
(PDB: 1FCV) and human HYAL1 (PDB: 2PE4) in reference to a previous
report (27) and performed protein
alignment by the MOE tool to construct the human HYAL1-HA complex.
The position of the active pocket of the constructed model accorded
with 1FCV. We performed ASE-Dock between human HYAL1 and dummy
atoms in the possible active pocket. As expected, HA was stably
located at the active pocket, and the active site Glu131
nearly faced the HA cleavage site (Fig. 4A and B; docking energy, −38.6
kcal/mol). We next calculated the effect of C-mannosylation
of HYAL1 on conformation or free energy. The docking simulation
revealed that C-mannosylation changed the conformation,
especially the nearby active pocket of HYAL1 (Fig. 4A and C). Moreover, the active site
Glu131 faced in the opposite direction toward its
substrate, HA (Fig. 4A and C;
docking energy, −2.45 kcal/mol). These results suggest that
C-mannosylation may cause the instability of HYAL1
conformation, resulting in suppression of HYAL1 enzymatic
activity.
Discussion
In this report, we demonstrated that intracellular
HYAL1 was C-mannosylated at Trp130 (Fig. 3), which has possible roles for
secretion and enzymatic activity (Figs. 2 and 4). Some C-mannosylated proteins
are found among TSR-1 superfamily proteins, such as thrombospondin,
mindin, F-spondin and ADAMTS-like 1/punctin-1 (8); however, HYAL1 does not belong to the
TSR-1 superfamily. This is the first report revealing the presence
of C-mannosylation among the HYAL family. Although some
reports demonstrated that C-mannosylation regulates
secretion, other functions of C-mannosylation were scarcely
reported; however, we suggest that C-mannosylation of HYAL1
might attenuate its enzymatic activity. Attachment of α-D-mannose
to the tryptophan residue by C-mannosylation changes protein
polarity, because the polar D-mannose is attached to the non-polar
tryptophan. Moreover, conformational change is also expected. These
changes are predicted to affect some sorts of functions on
C-mannosylated proteins, not only secretion, but also
protein stability and/or enzymatic activity. Therefore, we
anticipated that HYAL1 functions, such as secretion and enzymatic
activity, were regulated by C-mannosylation. Surprisingly,
however, secreted HYAL1 was not C-mannosylated at all
(Fig. 2), although intracellular
HYAL1 was partially modified (Fig.
3). These results suggest that C-mannosylation of HYAL1
negatively regulates the secretion of HYAL1 and that there is an
unidentified enzyme that detaches mannose from
C-mannosylated HYAL1.
MOE has been used widely in research for many
purposes, such as homology modeling or docking simulation (32,33).
In this research, MOE demonstrated that C-mannosylation
highly changed HYAL1 conformation and induced protein instability,
so that we presumed that C-mannosylation had roles in
enzymatic activity. By C-mannosylation of HYAL1, the active
site Glu131 faced in the opposite direction toward HA,
and Glu131 did not face the cleavage position of HA.
Moreover, C-mannosylated HYAL1 could no longer recognize HA
as a ligand, according to Site Finder, which can search for
possible active pockets of proteins (data not shown). Therefore,
C-mannosylation disables HYAL1 to degrade HA. Although
mannose is very small, the attachment of mannose to a
Trp130 residue next to the active site Glu131
caused profound conformational changes. Furthermore, a previous
report demonstrated that HYAL1 enzymatic activity was influenced by
site-directed mutagenesis for various sites, such as the active
site, disulfide bond sites or some N-glycosylation sites
(27). This result means that
proper conformation is required for its enzymatic activity and
supports our prediction that C-mannosylation, which causes
conformational change, is also important for its functions.
According to MOE, secretion and enzymatic activity
of HYAL1 is predicted to be inhibited by C-mannosylation,
therefore, we have established C-mannosylation-defective
W130A mutant HYAL1 expressing cell line to evaluate the effects of
C-mannosylation for HYAL1 functions. It is predicted that
secretion and enzymatic activity of W130A mutant HYAL1 will be
increased compared with wild-type HYAL1 although the ratio of
C-mannosylated HYAL1 is small. However, secretion of W130A
mutant HYAL1 was decreased compared with wild-type HYAL1 (data not
shown). Moreover, we evaluated the enzymatic activity of purified
secreted wild-type and W130A mutant HYAL1 purified from conditioned
media, which were both unmannosylated. As a result, enzymatic
activity of W130A mutant HYAL1 was also decreased compared with
wild-type HYAL1 (data not shown). These results were inconsistent
with our prediction, and we had concluded that these effects were
not because of C-mannosylation but substitution from Trp to
Ala. Therefore, in this report, we did not evaluate the effect of
C-mannosylation by using C-mannosylation-defective
mutant HYAL1.
Collectively, we demonstrated that HYAL1 was
C-mannosylated at Trp130, and suggest the
possible roles of C-mannosylation for secretion and
enzymatic activity of HYAL1. Since HYAL1 is known to correlate with
tumor malignancy, C-mannosylation of HYAL1 can be target for
cancer therapeutics.
Acknowledgements
This study was supported in part by
grants from the programs Grants-in-Aid for Scientific Research (B)
(nos. 23310163 and 24310167) and for JSPS Fellows (254256). Y.N. is
a Research Fellow of the Japan Society for the Promotion of
Science.
References
|
1.
|
Furmanek A and Hofsteenge J: Protein
C-mannosylation: facts and questions. Acta Biochim Pol. 47:781–789.
2000.PubMed/NCBI
|
|
2.
|
Doucey MA, Hess D, Cacan R and Hofsteenge
J: Protein C-mannosylation is enzyme-catalysed and uses
dolichyl-phosphate-mannose as a precursor. Mol Biol Cell.
9:291–300. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Krieg J, Hartmann S, Vicentini A, Gläsner
W, Hess D and Hofsteenge J: Recognition signal for C-mannosylation
of Trp-7 in RNase 2 consists of sequence Trp-x-x-Trp. Mol Biol
Cell. 9:301–309. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Julenius K: NetCGlyc 1.0: prediction of
mammalian C-mannosylation sites. Glycobiology. 17:868–876. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Hofsteenge J, Müller DR, de Beer T,
Löffler A, Richter WJ and Vliegenthart JF: New type of linkage
between a carbohydrate and a protein: C-glycosylation of a specific
tryptophan residue in human RNase Us. Biochemistry. 33:13524–13530.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Gonzalez de Peredo A, Klein D, Macek B,
Hess D, Peter-Katalinic J and Hofsteenge J: C-mannosylation and
o-fucosylation of thrombospondin type 1 repeats. Mol Cell
Proteomics. 1:11–18. 2002.PubMed/NCBI
|
|
7.
|
Hofsteenge J, Huwiler KG, Macek B, Hess D,
Lawler J, Mosher DF and Peter-Katalinic J: C-mannosylation and
O-fucosylation of the thrombospondin type 1 module. J Biol Chem.
276:6485–6498. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Wang LW, Leonhard-Melief C, Haltiwanger RS
and Apte SS: Post-translational modification of thrombospondin
type-1 repeats in ADAMTS-like 1/punctin-1 by C-mannosylation of
tryptophan. J Biol Chem. 284:30004–30015. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Hamming OJ, Kang L, Svensson A, Karlsen
JL, Rahbek-Nielsen H, Paludan SR, Hjorth SA, Bondensgaard K and
Hartmann R: Crystal structure of interleukin-21 receptor (IL-21R)
bound to IL-21 reveals that sugar chain interacting with WSXWS
motif is integral part of IL-21R. J Biol Chem. 287:9454–9460. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Buettner FF, Ashikov A, Tiemann B, Lehle L
and Bakker H: C. elegans DPY-19 is a C-mannosyltransferase
glycosylating thrombospondin repeats. Mol Cell. 50:295–302. 2013.
View Article : Google Scholar
|
|
11.
|
Ihara Y, Manabe S, Ikezaki M, Inai Y,
Matsui I-SL, Ohta Y, Muroi E and Ito Y: C-mannosylated peptides
derived from the thrombospondin type 1 repeat interact with Hsc70
to modulate its signaling in RAW264.7 cells. Glycobiology.
20:1298–1310. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Muroi E, Manabe S, Ikezaki M, Urata Y,
Sato S, Kondo T, Ito Y and Ihara Y: C-mannosylated peptides derived
from the thrombospondin type 1 repeat enhance
lipopolysaccharide-induced signaling in macrophage-like RAW264.7
cells. Glycobiology. 17:1015–1028. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Overall CM and Lopez-Otin C: Strategies
for MMP inhibition in cancer: innovations for the post-trial era.
Nat Rev Cancer. 2:657–672. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Simizu S, Ishida K, Wierzba MK and Osada
H: Secretion of heparanase protein is regulated by glycosylation in
human tumor cell lines. J Biol Chem. 279:2697–2703. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Simizu S and Niwa Y: Practical molecular
targets for suppression of metastasis. For Immunopathol Dis Therap.
4:43–51. 2013. View Article : Google Scholar
|
|
16.
|
Chao KL, Muthukumar L and Herzberg O:
Structure of human hyaluronidase-1, a hyaluronan hydrolyzing enzyme
involved in tumor growth and angiogenesis. Biochemistry.
46:6911–6920. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Tan JX, Wang XY, Su XL, Li HY, Shi Y, Wang
L and Ren GS: Upregulation of HYAL1 expression in breast cancer
promoted tumor cell proliferation, migration, invasion and
angiogenesis. PLoS One. 6:e228362011. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Tan JX, Wang XY, Li HY, Su XL, Wang L, Ran
L, Zheng K and Ren GS: HYAL1 overexpression is correlated with the
malignant behavior of human breast cancer. Int J Cancer.
128:1303–1315. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Imundo L, Leduc CA, Guha S, Brown M,
Perino G, Gushulak L, Triggs-Raine B and Chung WK: A complete
deficiency of hyaluronoglucosaminidase 1 (HYAL1) presenting as
familial juvenile idiopathic arthritis. J Inherit Metab Dis.
34:1013–1022. 2011. View Article : Google Scholar
|
|
20.
|
Kuroda M, Funasaki S, Saitoh T, Sasazawa
Y, Nishiyama S, Umezawa K and Simizu S: Determination of
topological structure of ARL6ip1 in cells: identification of the
essential binding region of ARL6ip1 for conophylline. FEBS Lett.
587:3656–3660. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Yasukagawa T, Niwa Y, Simizu S and Umezawa
K: Suppression of cellular invasion by glybenclamide through
inhibited secretion of platelet-derived growth factor in ovarian
clear cell carcinoma ES-2 cells. FEBS Lett. 586:1504–1509. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Miyazaki I, Simizu S, Okumura H, Takagi S
and Osada H: A small-molecule inhibitor shows that pirin regulates
migration of melanoma cells. Nat Chem Biol. 6:667–673. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Simizu S, Umezawa K, Takada M, Arber N and
Imoto M: Induction of hydrogen peroxide production and Bax
expression by caspase-3(-like) proteases in tyrosine kinase
inhibitor-induced apoptosis in human small cell lung carcinoma
cells. Exp Cell Res. 238:197–203. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Simizu S, Suzuki T, Muroi M, Lai NS,
Takagi S, Dohmae N and Osada H: Involvement of disulfide bond
formation in the activation of heparanase. Cancer Res.
67:7841–7849. 2007. View Article : Google Scholar
|
|
25.
|
Niwa Y, Suzuki T, Dohmae N, Umezawa K and
Simizu S: Determination of cathepsin V activity and intracellular
trafficking by N-glycosylation. FEBS Lett. 586:3601–3607. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Guntenhöner MW, Pogrel MA and Stern R: A
substrate-gel assay for hyaluronidase activity. Matrix. 12:388–396.
1992.
|
|
27.
|
Zhang L, Bharadwaj AG, Casper A, Barkley
J, Barycki JJ and Simpson MA: Hyaluronidase activity of human Hyal1
requires active site acidic and tyrosine residues. J Biol Chem.
284:9433–9442. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Yamaguchi H, Kidachi Y, Kamiie K, Noshita
T and Uetsu H: Structural insight into the ligand-receptor
interaction between glycyrrhetinic acid (GA) and the high-mobility
group protei B1 (HMGB1)-DNA complex. Bioinformation. 23:1147–1153.
2012. View Article : Google Scholar
|
|
29.
|
Susana DL, Lídia MG, Teresa AFC, Henrique
FC, Rui M and Rita CG: Structure based virtual screening for
discovery of novel human neutrophil elastase inhibitors. Med Chem
Comm. 3:1299–1304. 2012. View Article : Google Scholar
|
|
30.
|
Wilkins MR, Gasteiger E, Gooley AA,
Herbert BR, Molloy MP, Binz PA, Ou K, Sanchez JC, Bairoch A,
Williams KL and Hochstrasser DF: High-throughput mass spectrometric
discovery of protein post-translational modifications. J Mol Biol.
289:645–657. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Frost GI, Csóka AB, Wong T, Stern R and
Csóka TB: Purification, cloning, and expression of human plasma
hyaluronidase. Biochem Biophys Res Commun. 236:10–15. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Azuma M, Kabe Y, Kuramori C, Kodo M,
Yamaguchi Y and Handa H: Adenine nucleotide translocator transports
haem precursors into mitochondria. PLoS One. 3:e30702008.
View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Fukiage C, Nakajima E, Ma H, Azuma M and
Shearer TR: Characterization and regulation of lens-specific
calpain Lp82. J Biol Chem. 277:20678–20685. 2002. View Article : Google Scholar : PubMed/NCBI
|