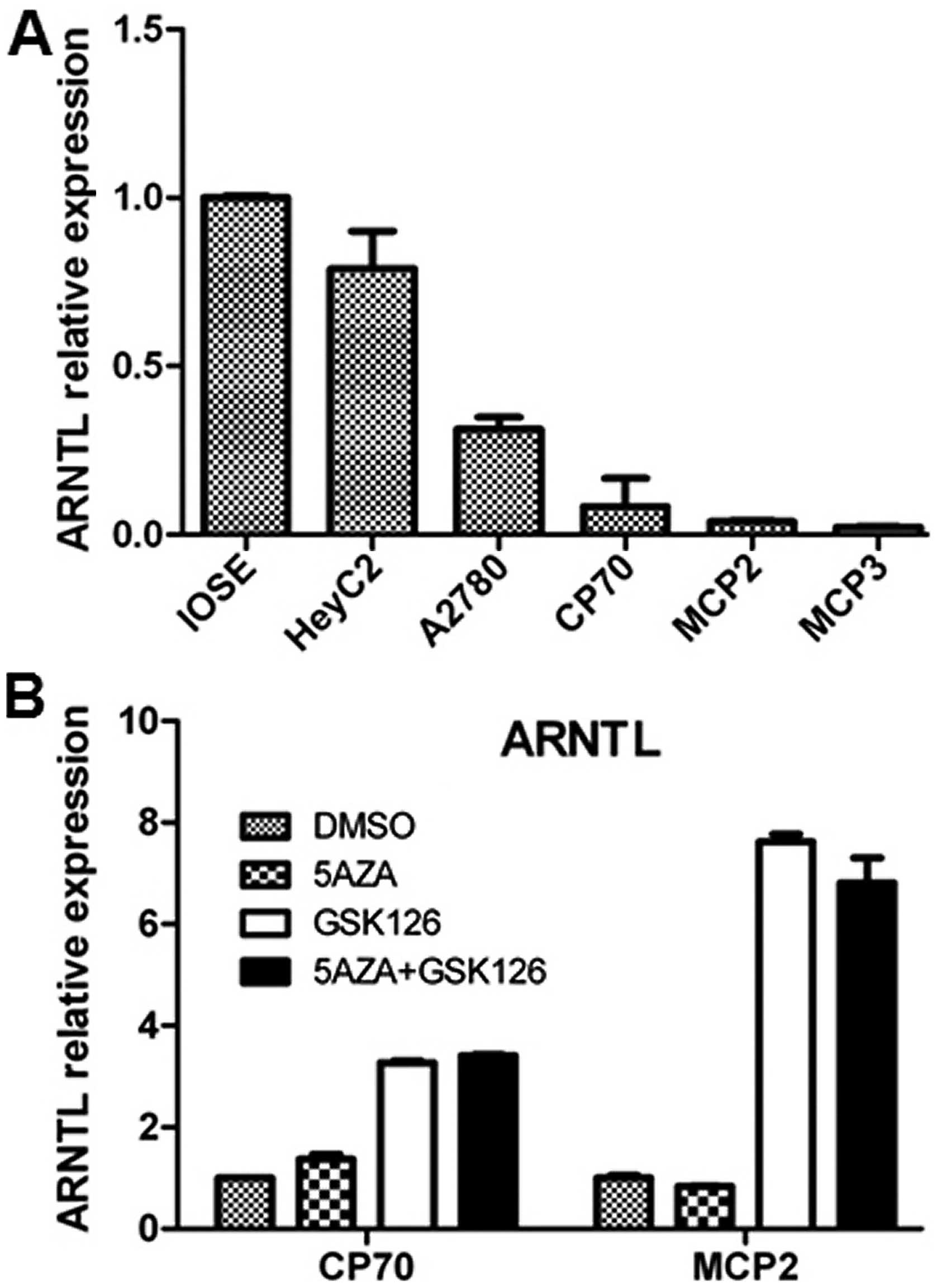
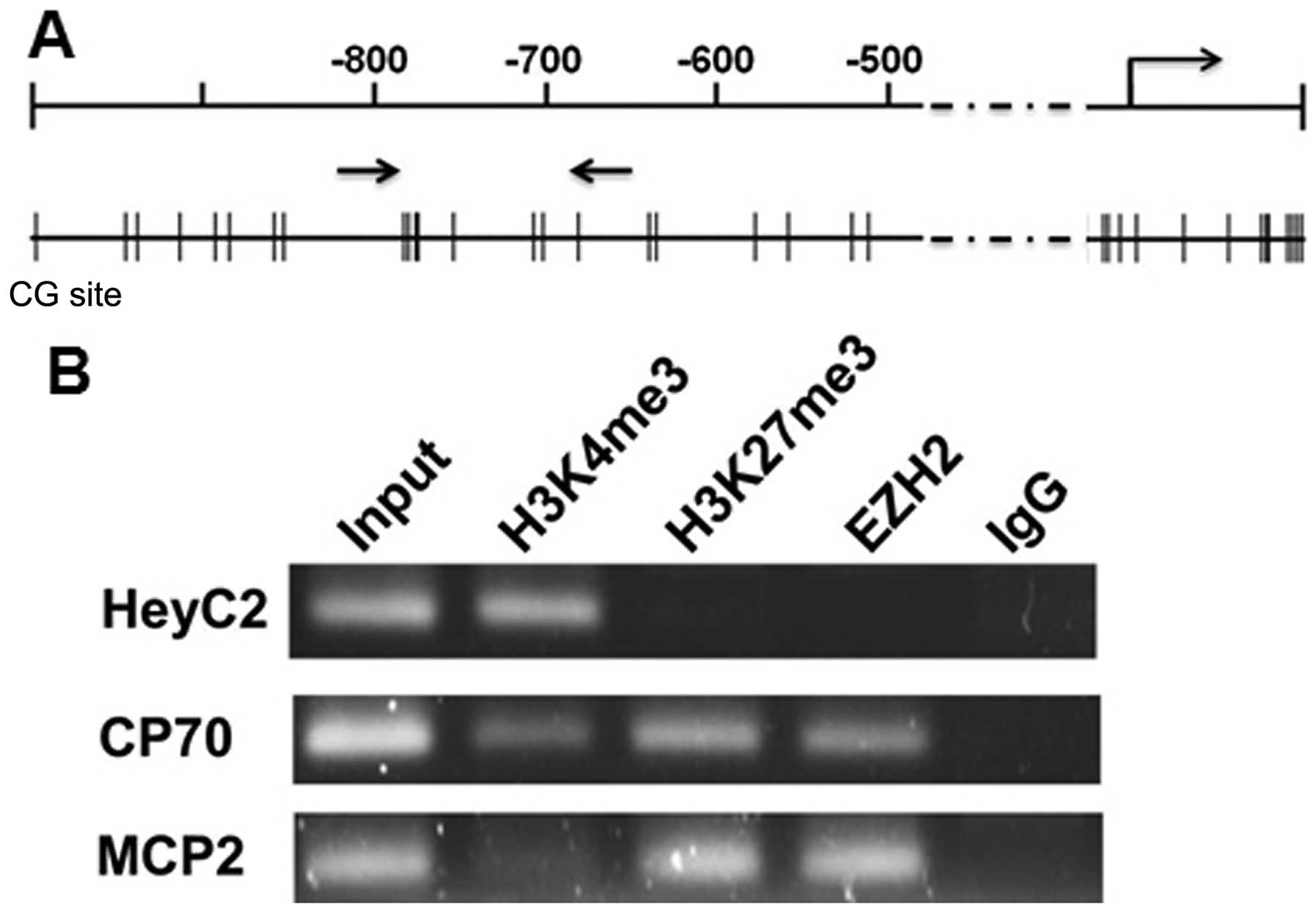
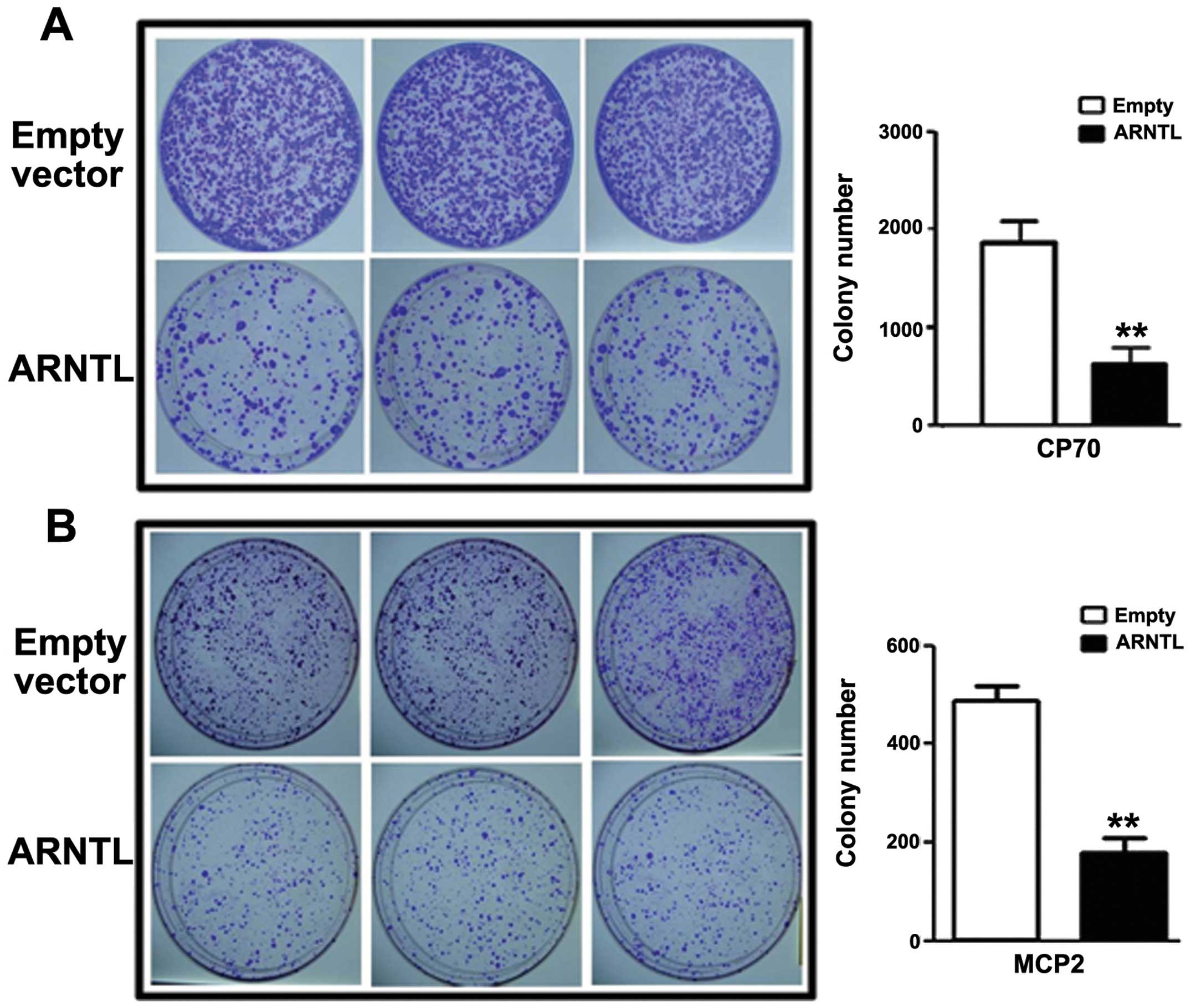
|
1
|
Sellix MT and Menaker M: Circadian clocks
in the ovary. Trends Endocrinol Metab. 21:628–636. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Harrington ME: Exercise strengthens
circadian clocks. J Physiol. 590:59292012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Green CB and Menaker M: Circadian rhythms.
Clocks on the brain. Science. 301:319–320. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Falcon J: Cellular circadian clocks in the
pineal. Prog Neurobiol. 58:121–162. 1999. View Article : Google Scholar
|
|
5
|
Stevens RG and Rea MS: Light in the built
environment: potential role of circadian disruption in endocrine
disruption and breast cancer. Cancer Causes Control. 12:279–287.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hansen J: Risk of breast cancer after
night- and shift work: current evidence and ongoing studies in
Denmark. Cancer Causes Control. 17:531–537. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Schernhammer ES, Laden F, Speizer FE, et
al: Night-shift work and risk of colorectal cancer in the nurses’
health study. J Natl Cancer Inst. 95:825–828. 2003.
|
|
8
|
Canaple L, Kakizawa T and Laudet V: The
days and nights of cancer cells. Cancer Res. 63:7545–7552.
2003.PubMed/NCBI
|
|
9
|
Levin RD, Daehler MA, Grutsch JF, et al:
Circadian function in patients with advanced non-small-cell lung
cancer. Br J Cancer. 93:1202–1208. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gery S and Koeffler HP: Circadian rhythms
and cancer. Cell Cycle. 9:1097–1103. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Leonardi GC, Rapisarda V, Marconi A, et
al: Correlation of the risk of breast cancer and disruption of the
circadian rhythm (Review). Oncol Rep. 28:418–428. 2012.PubMed/NCBI
|
|
12
|
Yu EA and Weaver DR: Disrupting the
circadian clock: gene-specific effects on aging, cancer, and other
phenotypes. Aging (Albany, NY). 3:479–493. 2011.PubMed/NCBI
|
|
13
|
Rana S and Mahmood S: Circadian rhythm and
its role in malignancy. J Circadian Rhythms. 8:32010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Paschos GK and FitzGerald GA: Circadian
clocks and vascular function. Circ Res. 106:833–841. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Takahashi JS, Hong HK, Ko CH and McDearmon
EL: The genetics of mammalian circadian order and disorder:
implications for physiology and disease. Nat Rev Genet. 9:764–775.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hunt T and Sassone-Corsi P: Riding tandem:
circadian clocks and the cell cycle. Cell. 129:461–464. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ripperger JA and Schibler U: Rhythmic
CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp
transcription and chromatin transitions. Nat Genet. 38:369–374.
2006. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hsu CM, Lin SF, Lu CT, Lin PM and Yang MY:
Altered expression of circadian clock genes in head and neck
squamous cell carcinoma. Tumour Biol. 33:149–155. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tokunaga H, Takebayashi Y, Utsunomiya H,
et al: Clinicopathological significance of circadian rhythm-related
gene expression levels in patients with epithelial ovarian cancer.
Acta Obstet Gynecol Scand. 87:1060–1070. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang MY, Chang JG, Lin PM, et al:
Downregulation of circadian clock genes in chronic myeloid
leukemia: alternative methylation pattern of hPER3. Cancer Sci.
97:1298–1307. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Baylin SB and Jones PA: A decade of
exploring the cancer epigenome - biological and translational
implications. Nat Rev Cancer. 11:726–734. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Seisenberger S, Peat JR and Reik W:
Conceptual links between DNA methylation reprogramming in the early
embryo and primordial germ cells. Curr Opin Cell Biol. 25:281–288.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Caslini C, Capo-chichi CD, Roland IH,
Nicolas E, Yeung AT and Xu XX: Histone modifications silence the
GATA transcription factor genes in ovarian cancer. Oncogene.
25:5446–5461. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yu Y, Fujii S, Yuan J, et al: Epigenetic
regulation of ARHI in breast and ovarian cancer cells. Ann NY Acad
Sci. 983:268–277. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chan MW, Wei SH, Wen P, et al:
Hypermethylation of 18S and 28S ribosomal DNAs predicts
progression-free survival in patients with ovarian cancer. Clin
Cancer Res. 11:7376–7383. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chan MW, Huang YW, Hartman-Frey C, et al:
Aberrant transforming growth factor beta1 signaling and SMAD4
nuclear translocation confer epigenetic repression of ADAM19 in
ovarian cancer. Neoplasia. 10:908–919. 2008.PubMed/NCBI
|
|
27
|
Chou JL, Su HY, Chen LY, et al: Promoter
hypermethylation of FBXO32, a novel TGF-beta/SMAD4 target gene and
tumor suppressor, is associated with poor prognosis in human
ovarian cancer. Lab Invest. 90:414–425. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yeh KT, Chen TH, Yang HW, et al: Aberrant
TGFbeta/SMAD4 signaling contributes to epigenetic silencing of a
putative tumor suppressor, RunX1T1 in ovarian cancer. Epigenetics.
6:727–739. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Su HY, Lai HC, Lin YW, Chou YC, Liu CY and
Yu MH: An epigenetic marker panel for screening and prognostic
prediction of ovarian cancer. Int J Cancer. 124:387–393. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gillan L, Matei D, Fishman DA, Gerbin CS,
Karlan BY and Chang DD: Periostin secreted by epithelial ovarian
carcinoma is a ligand for αVβ3 and αVβ5 integrins and promotes cell
motility. Cancer Res. 62:5358–5364. 2002.PubMed/NCBI
|
|
31
|
McCabe MT, Ott HM, Ganji G, et al: EZH2
inhibition as a therapeutic strategy for lymphoma with
EZH2-activating mutations. Nature. 492:108–112. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fu LN, Pelicano H, Liu JS, Huang P and Lee
CC: The circadian gene period2 plays an important role in tumor
suppression and DNA-damage response in vivo. Cell. 111:1055. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fu L and Lee CC: The circadian clock:
pacemaker and tumour suppressor. Nat Rev Cancer. 3:350–361. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hoffman AE, Zheng T, Ba Y and Zhu Y: The
circadian gene NPAS2, a putative tumor suppressor, is
involved in DNA damage response. Mol Cancer Res. 6:1461–1468.
2008.
|
|
35
|
Gery S, Komatsu N, Kawamata N, et al:
Epigenetic silencing of the candidate tumor suppressor gene
Per1 in non-small cell lung cancer. Clin Cancer Res.
13:1399–1404. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Taniguchi H, Fernandez AF, Setien F, et
al: Epigenetic inactivation of the circadian clock gene BMAL1 in
hematologic malignancies. Cancer Res. 69:8447–8454. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Shih MC, Yeh KT, Tang KP, Chen JC and
Chang JG: Promoter methylation in circadian genes of endometrial
cancers detected by methylation-specific PCR. Mol Carcinog.
45:732–740. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Mullenders J, Fabius AW, Madiredjo M,
Bernards R and Beijersbergen RL: A large scale shRNA barcode screen
identifies the circadian clock component ARNTL as putative
regulator of the p53 tumor suppressor pathway. PloS one.
4:e47982009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jung CH, Kim EM, Park JK, et al: Bmal1
suppresses cancer cell invasion by blocking the phosphoinositide
3-kinase-Akt-MMP-2 signaling pathway. Oncol Rep. 29:2109–2113.
2013.PubMed/NCBI
|
|
40
|
Varambally S, Dhanasekaran SM, Zhou M, et
al: The polycomb group protein EZH2 is involved in progression of
prostate cancer. Nature. 419:624–629. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kleer CG, Cao Q, Varambally S, et al: EZH2
is a marker of aggressive breast cancer and promotes neoplastic
transformation of breast epithelial cells. Proc Natl Acad Sci USA.
100:11606–11611. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Vire E, Brenner C, Deplus R, et al: The
Polycomb group protein EZH2 directly controls DNA methylation.
Nature. 439:871–874. 2006. View Article : Google Scholar : PubMed/NCBI
|