Introduction
Hepatocellular carcinoma (HCC) is one of the most
lethal cancers worldwide and is the main cause of death among
cirrhotic patients (1–3). Patients diagnosed with HCC have a
poor prognosis because of the aggressive nature of the disease
(4,5), and surgical resection or local
ablation therapy is effective only at an early stage (6). Furthermore, ~70% of these patients
develop recurrent tumors within five years after curative surgery
(7). Recurrence of HCC, including
multicentric hepatocarcinogenesis and intrahepatic metastasis, is a
key prognostic factor, but it is difficult to distinguish patients
at high risk for recurrence and subsequent adverse prognosis using
only clinical staging systems comprising tumor characteristics and
liver function (1,8). Therefore, a novel approach for
predicting progression and recurrence of HCC is urgently
required.
Liver damage and the increased incidence of HCC
(chronic viral hepatitis B and C, alcohol consumption and
aflatoxin) have multiple causes (5,9,10).
Furthermore, as with other malignancies, the initiation of HCC is a
multistep process, and because it is characterized by high
molecular variability, clinical management requires a more complex
approach (11).
Although recent research along with the development
of new genomic technologies establishes that the development and
progression of HCC are caused by the accumulation of genetic and
epigenetic alterations (12–14),
the detailed underlying mechanisms have not been determined.
Therefore, identifying molecular markers for HCC, particularly
those that may predict recurrence, is important, because
stratification of patients at risk for recurrence facilitates
individualized management, including intensive surveillance and
aggressive adjuvant therapy for high-risk patients.
Prenyl diphosphate synthase subunit 2 (PDSS2) was
identified in 2005 (15), it
encodes the second subunit of prenyl diphosphate synthase, which is
an essential enzyme involved in the biosynthesis of coenzyme Q10
(CoQ10), and PDSS2 determines the side-chain length of mammalian
ubiquinones (16). CoQ10 is
synthesized from mevalonic acid in the liver and plays a vital role
in the mitochondrial respiratory chain, pyrimidine nucleoside
biosynthesis and the modulation of cell apoptosis (17). Aberrant expression of PDSS2
in the liver may cause DNA damage and disrupt the cell cycle
through inhibition of CoQ10 synthesis, leading to initiation and
progression of HCC (18,19). Furthermore, chronic inflammation
caused by hepatitis virus infection might affect PDSS2
expression. Although evidence indicates that PDSS2 suppresses the
development of malignant melanoma and lung cancer (16,20),
the clinical significance and regulatory mechanisms of PDSS2
expression in HCC remain undefined.
Therefore, we attempted to answer these questions in
the present study by analyzing PDSS2 expression in HCC to
identify novel, clinically significant biomarkers for progression
and recurrence of HCC. To the best of our knowledge, this is the
first report to determine PDSS2 expression levels in
HCC.
Materials and methods
Sample collection
Nine HCC cell lines (Hep3B, HepG2, HLE, HLF, HuH1,
HuH2, HuH7, PLC/PRF/5 and SK-Hep1) were obtained from the American
Type Culture Collection (ATCC, Manassas, VA, USA) and were
maintained as previously described (21). Primary HCC and non-cancerous
tissues were collected from 151 patients who underwent liver
resection for HCC at Nagoya University Hospital between January
1998 and July 2012. Clinicopathological data were collected from
medical records. Specimens were classified histologically according
to the 7th Edition of the Union for International Cancer Control
classification (22).
Tissue samples were immediately flash-frozen in
liquid nitrogen and stored at −80°C. RNA was extracted from ~5
mm2 diameter tumor samples without detectable necrotic
areas comprising >80% tumor cells. The corresponding
non-cancerous liver tissue samples that lacked regenerative or
dysplastic nodules were collected from the same patient that were
>2 cm distant from the edge of the tumor. The median duration of
patient follow-up was 37.9 months (range, 0.37–147 months).
Postoperative follow-up included physical examinations, measurement
of serum tumor markers every three months, and enhanced computed
tomography scans every six months. Treatment after recurrence was
generally selected from one of the options as follows: surgery,
radiofrequency ablation, transcatheter arterial chemoembolization,
and chemotherapy, according to tumor status and liver function.
Enrollees granted written informed consent for the use of clinical
samples and data as required by the Institutional Review Board of
Nagoya University, Japan.
Quantitative real-time
reverse-transcription polymerase chain reaction (qRT-PCR).
PDSS2
mRNA levels were determined using qRT-PCR. Total RNA
(10 μg) was isolated from 9 HCC cell lines, 151 primary HCCs and
adjacent non-cancerous tissues, and was used as a template for cDNA
synthesis. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH)
mRNA (TaqMan, GAPDH control reagents; Applied Biosystems, Foster
City, CA, USA) was quantified in each sample for standardization.
Quantitative real-time RT-PCR was performed using the
SYBR® Green PCR Core Reagents kit (Applied Biosystems)
as follows: one cycle at 95°C for 10 min, 40 cycles at 95°C for 5
sec, and 60°C for 60 sec. Real-time detection of the
SYBR® Green fluoresence was conducted using an ABI
StepOnePlus™ Real-Time PCR System (Applied Biosystems). Triplicate
samples of 9 HCC cell lines and 151 clinical samples were analyzed.
Samples without templates served as negative controls. The
expression level of each sample is shown as the value of the
PDSS2 amplicon divided by that of GAPDH (23). The primer sequences are listed in
Table I. PDSS2 mRNA levels
were considered downregulated in tumor tissues when they were
<50% compared with those of the corresponding non-cancerous
tissues.
 | Table IPrimers and annealing
temperatures. |
Table I
Primers and annealing
temperatures.
| Gene | Experiment | Type | Sequence
(5′-3′) | Product size
(bp) | Annealing
temperature (°C) |
|---|
| PDSS2 | qRT-PCR | Forward |
GAATCAGGTAGTGTCAGAGG | 181 | 60 |
| | Reverse |
GAGGCTATTCCAGCTGTCATG | | |
| MSP methylated | Forward |
TCGAAGTTGGATTCGAGGAT | 263 | 62 |
| | Reverse |
AAACGTCGAACGAAAACACC | | |
| MSP
unmethylated | Forward |
GGTTGGTGGTGATAGTGATA | 195 | 56 |
| | Reverse |
CAACAAATAATCCCTCTACAC | | |
| Bisulfite
sequencing | Forward |
TGTTTGGTTGGGTTTTGAGG | 152 | 58 |
| | Reverse | CACCAACCCCTA ACA
ATA AC | | |
| HNF4α | qRT-PCR | Forward |
CGTGGTGGACAAAGACAAGA | 128 | 60 |
| | Reverse |
CATAGCTTGACCTTCGAGTGC | | |
| CDX2 | qRT-PCR | Forward |
GGAACCTGTGCGAGTGGAT | 128 | 60 |
| | Reverse |
GAAACTCCTTCTCCAGCTCC | | |
| GAPDH | qRT-PCR | Forward |
GAAGGTGAAGGTCGGAGTC | 226 | 60 |
| | Probe |
CAAGCTTCCCGTTCTCAGCC | | |
| | Reverse |
GAAGATGGTGATGGGATTTC | | |
Analysis of the promoter region of
PDSS2
The nucleotide sequence of the PDSS2 promoter
region was analyzed to determine the presence or absence of CpG
islands defined as follows: at least a 200-bp region of DNA with a
high HCC content (>50%) and an Observed CpG/Expected CpG ratio
≥0.6 (24,25). CpG Island Searcher software
(http://cpgislands.usc.edu/) was employed
to determine the locations of CpG islands (26).
Methylation-specific PCR (MSP) and
bisulfite sequence analysis
PDSS2 possesses a CpG island near its
promoter region, and we hypothesized that aberrant methylation
regulates PDSS2 transcription in HCC. DNA samples from nine
HCC cell lines treated with bisulfite were subjected to MSP.
Genomic bisulfite-treated DNA from HCC cell lines was sequenced to
ascertain whether the MSP amplification was reliable. The primer
sequences used for MSP and bisulfite sequencing are listed in
Table I. Bisulfite treatment and
the sequencing procedure were performed as reported (27).
5-Aza-2′-deoxycytidine (5-aza-dC)
treatment
To assess the relation of promoter hypermethylation
to PDSS2 transcription, HCC cells (1.5×106) were
treated with 5-aza-dC (Sigma-Aldrich, St. Louis, MO, USA) to
inhibit DNA methylation and cultured for 6 days with medium changes
on days 1, 3 and 5. RNA was extracted, and RT-PCR was performed as
previously described (27).
Expression of genes encoding proteins
potentially associated with PDSS2
The expression levels of hepatocyte nuclear factor
4α (HNF4α) and caudal-type homeobox transcription factor 2 (CDX2),
which may associate with PDSS2 (20,28)
were determined in HCC cell lines using qRT-PCR. The sequences of
primers specific for HNF4α and CDX2 are listed in Table I.
Immunohistochemistry (IHC)
IHC analysis of the localization of PDSS2 was
performed using a mouse monoclonal antibody against PDSS2
(ab119768; Abcam, Cambridge, UK) diluted 1:150 in antibody diluent
(Dako, Glostrup, Denmark) to probe 30 representative sections of
well-preserved HCC tissue previously described (2). Staining patterns were compared
between HCCs and the corresponding normal adjacent tissues. To
avoid subjectivity, the specimens were randomized and coded before
analysis by two independent observers who were unaware of the
status of the samples. Each observer evaluated all specimens at
least twice to minimize intraobserver variation (29).
Statistical analysis
Correlations between the levels of PDSS2 mRNA
with those of HNF4α and CDX2 were a nalyzed using the Spearman rank
correlation test. Relative levels of mRNA expression
(PDSS2/GAPDH) between HCC and non-cancerous tissues were
analyzed using the Mann-Whitney U test. The χ2 test was
used to analyze the significance of the association between the
expression and methylation status of PDSS2 and
clinicopathological parameters. Disease-specific and disease-free
survival rates were calculated using the Kaplan-Meier method, and
the difference in survival curves was analyzed using the log-rank
test. We performed multivariate regression analysis using the Cox
proportional hazards model to detect prognostic factors, and
variables with P<0.05 were entered into the final model. All
statistical analyses were performed using JMP 10 software (SAS
Institute, Inc., Cary, NC, USA). P<0.05 was considered
statistically significant.
Results
Identification of a CpG island in the
PDSS2 promoter region
A CpG island was identified in the PDSS2
promoter region (Fig. 1A), leading
to the hypothesis that hypermethylation of the CpG islands
regulates the expression of PDSS2 in HCC.
PDSS2 mRNA expression and regulatory
mechanisms in HCC cell lines
Significant decrease in PDSS2 mRNA levels was
detected in 6 (67%) of 9 HCC cell lines compared with the mean
expression level in 151 non-cancerous liver tissues (Fig. 1B). Hypermethylation of the
PDSS2 promoter was detected in Hep3B, HuH2, HuH7 and SK-Hep1
cells (Fig. 1B). To determine
whether hypermethylation of the PDSS2 promoter inhibited
expression, PDSS2 mRNA expression levels were compared
before and after treatment with the methylation inhibitor 5-aza-dC.
PDSS2 mRNA levels were restored in cells with downregulated
PDSS2 expression accompanying hypermethylation after
5-aza-dC treatment (Fig. 1B),
indicating that promoter hypermethylation inhibited PDSS2
transcription in HCC.
Expression of genes encoding proteins
potentially associated with PDSS2 in HCC cell lines
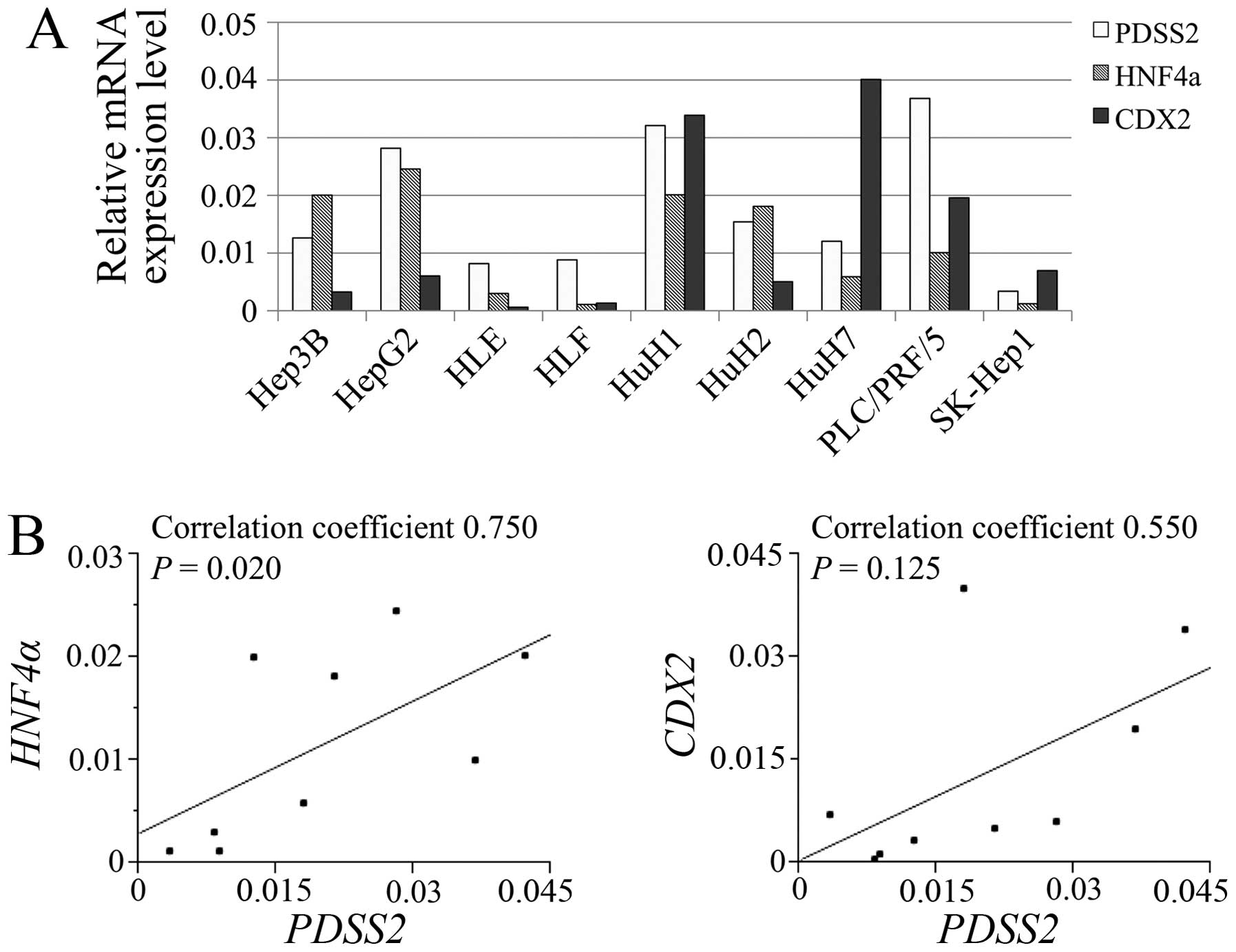
The relative mRNA expression levels of PDSS2,
HNF4α and CDX2 in HCC cell lines are shown in
Fig. 2A. PDSS2 expression
levels significantly correlated with those of HNF4α
(Fig. 2B).
Patient characteristics
The mean age of the 151 patients was 64.6±9.7 years
(range, 34–84 years), the male to female ratio was 5:1, and there
were 37 and 84 patients with hepatitis B and C virus infections,
respectively. Of the patients without HCC, 10, 87 and 54 presented
with normal liver function, chronic hepatitis or cirrhosis,
respectively. We classified 140 and 11 patients as Child-Pugh class
A or B, respectively.
Expression levels of PDSS2 mRNA and
protein in resected tissues
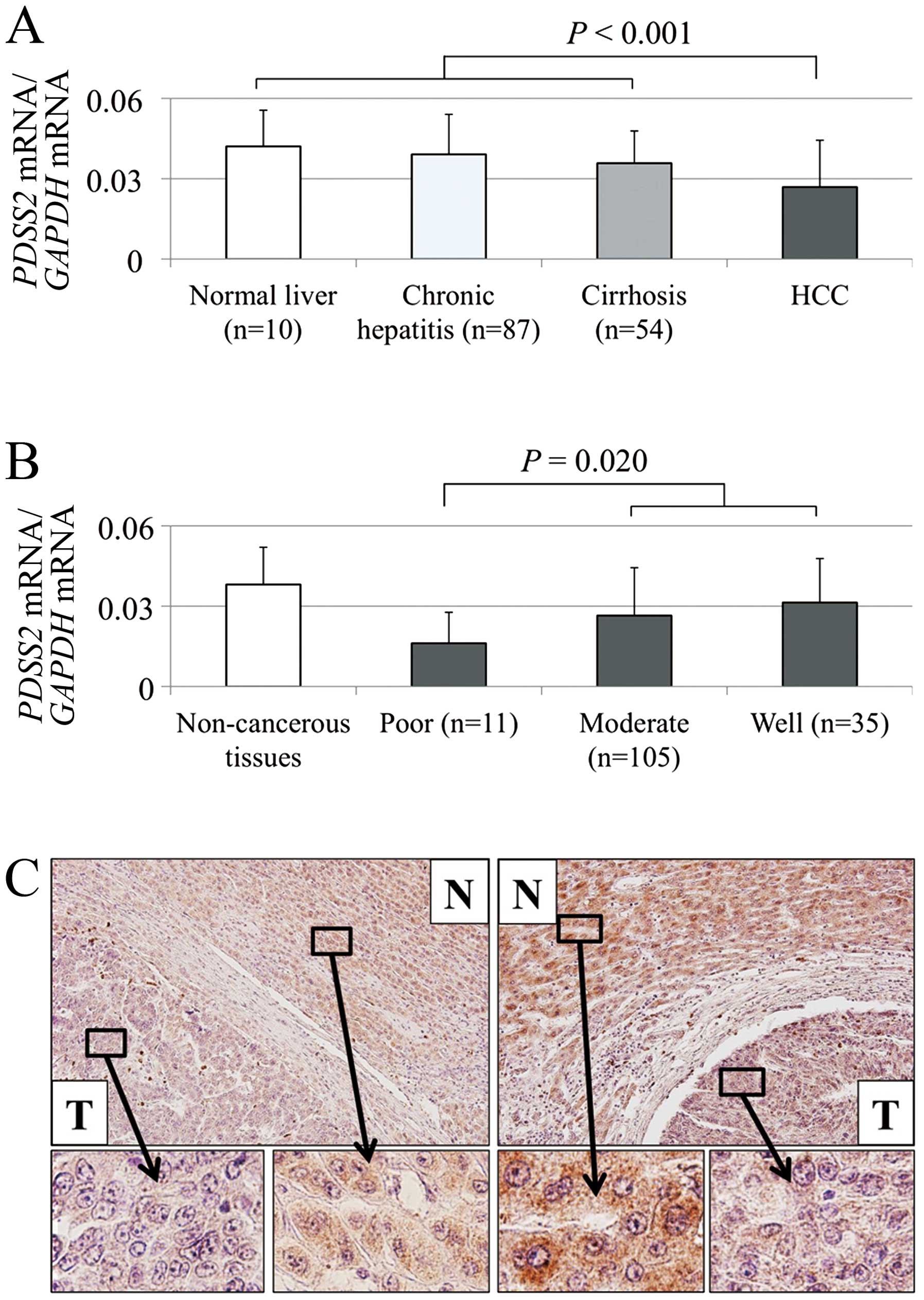
The mean level of PDSS2 mRNA compared with
that of non-cancerous liver diminished gradually in the order of
normal liver, chronic hepatitis and cirrhosis, indicating that
chronic inflammation and fibrosis of non-cancerous liver decreased
PDSS2 expression (Fig. 3A).
In contrast, the type of hepatitis virus infection (hepatitis virus
B, C or none) did not influence PDSS2 expression in
non-cancerous liver tissues. PDSS2 mRNA expression was
decreased in HCC tissues of 122 (81%) of 151 patients compared with
that of the corresponding non-cancerous tissues. The mean
expression level of PDSS2 mRNA was significantly lower in
HCC tissues compared with that of the corresponding non-cancerous
tissues (P<0.001; Fig. 3A).
Moreover, poorly differentiated tumor cells expressed relatively
lower levels of PDSS2 mRNA (Fig. 3B).
The expression of PDSS2 was analyzed using IHC.
Representative sections with reduced PDSS2 staining in HCC tissues
are shown in Fig. 3C. The overall
staining intensities of 30 samples were consistent with mRNA levels
detected using qRT-PCR.
Prognostic implications of PDSS2 mRNA
expression levels
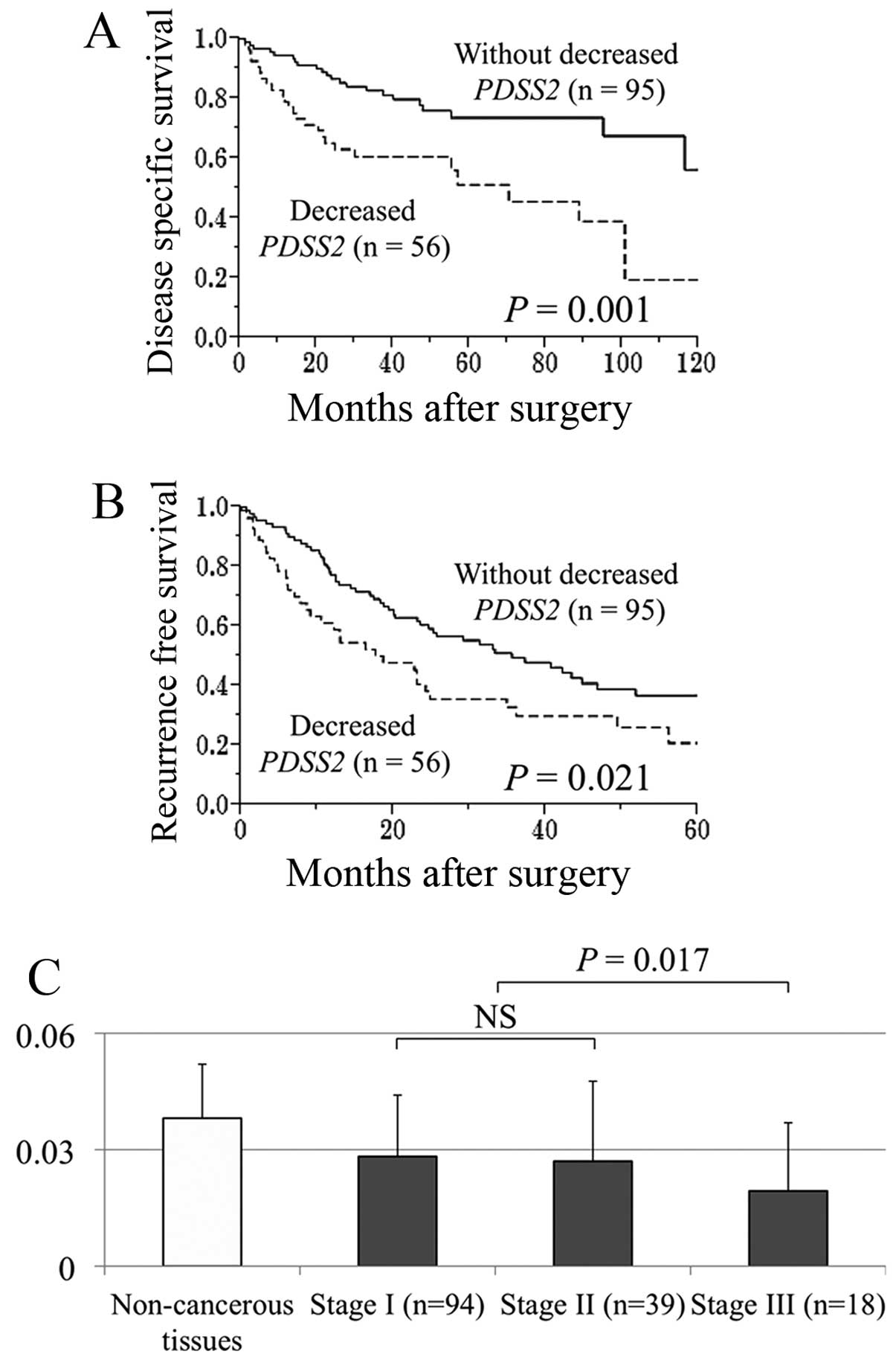
Fifty-six (37%) of 151 patients were categorized
with decreased PDSS2 mRNA levels in HCC tissues compared
with noncancerous tissues. The disease-specific survival rate of
patients with HCC with decreased PDSS2 mRNA was
significantly lower compared with those without this factor (5-year
survival rates, 51 and 74%, respectively, P=0.001; Fig. 4A). Decreased PDSS2 mRNA
expression in patients with HCCs was significantly associated with
uninvolved liver status, preoperative serum α-fetoprotein >20
ng/ml, tumor size ≥3.0 cm, tumor differentiation (moderate to
poor), serosal infiltration, septum formation and advanced UICC
stage (Table II). Multivariate
analysis identified decreased PDSS2 mRNA expression as an
independent prognostic factor (hazard ratio 2.45, 95% confidence
interval 1.27–4.80, P=0.008; Table
III). Patients with HCC with decreased PDSS2 mRNA levels
experienced significantly earlier recurrences compared with those
without (2-year recurrence-free survival rates, 38 and 59%,
respectively, P=0.021; Fig. 4B). A
stepwise decrease of PDSS mRNA expression in patients with HCC
correlated with UICC stage (Fig.
4C).
 | Table IIAssociation between expression levels
of PDSS2 mRNA and clinicopathological parameters in 151
patients with hepatocellular carcinoma (HCC). |
Table II
Association between expression levels
of PDSS2 mRNA and clinicopathological parameters in 151
patients with hepatocellular carcinoma (HCC).
| Clinicopathological
parameters | Decreased
PDSS2 in HCCs (n=56) | Others (n=95) | P-value |
|---|
| Age (years) |
| <65 | 26 | 41 | 0.696 |
| ≥65 | 30 | 54 | |
| Gender |
| Male | 50 | 76 | 0.128 |
| Female | 6 | 19 | |
| Background
liver |
| Normal liver | 1 | 9 | |
| Chronic
hepatitis | 38 | 49 | 0.046a |
| Cirrhosis | 17 | 37 | |
| Pugh-Child’s
classification |
| A | 52 | 88 | 0.959 |
| B | 4 | 7 | |
| Hepatitis
virus |
| Absent | 13 | 17 | |
| HBV | 15 | 22 | 0.551 |
| HCV | 28 | 56 | |
| AFP (ng/ml) |
| ≤20 | 18 | 63 | <0.001a |
| >20 | 38 | 32 | |
| PIVKA II
(mAU/ml) |
| ≤40 | 16 | 42 | 0.054 |
| >40 | 40 | 53 | |
| Tumor
multiplicity |
| Solitary | 43 | 74 | 0.875 |
| Multiple | 13 | 21 | |
| Tumor size
(cm) |
| <3.0 | 12 | 35 | 0.045a |
| ≥3.0 | 44 | 60 | |
|
Differentiation |
| Well | 7 | 28 | 0.014a |
| Moderate to
poor | 49 | 67 | |
| Growth type |
| Expansive
growth | 43 | 84 | 0.063 |
| Invasive
growth | 13 | 11 | |
| Serosal
infiltration |
| Absent | 35 | 79 | 0.005a |
| Present | 21 | 16 | |
| Formation of
capsule |
| Absent | 17 | 30 | 0.875 |
| Present | 39 | 65 | |
| Infiltration to
capsule |
| Absent | 24 | 44 | 0.680 |
| Present | 32 | 51 | |
| Septum
formation |
| Absent | 13 | 40 | 0.017a |
| Present | 43 | 55 | |
| Vascular
invasion |
| Absent | 33 | 81 | <0.001a |
| Present | 23 | 14 | |
| UICC pathological
stage |
| I | 27 | 67 | 0.014a |
| II | 18 | 21 | |
| III | 11 | 7 | |
 | Table IIIPrognostic factors of 151 patients
with hepatocellular carcinoma (HCC). |
Table III
Prognostic factors of 151 patients
with hepatocellular carcinoma (HCC).
| | Univariate
analysis | Multivariable
analysis |
|---|
| |
|
|
|---|
| Variables | n | Hazard ratio | 95% CI | P-value | Hazard ratio | 95% CI | P-value |
|---|
| Age (≥65
years) | 84 | 1.92 | 1.07–3.57 | 0.030a | 1.70 | 0.92–3.25 | 0.090 |
| Gender (male) | 126 | 1.27 | 0.60–3.13 | 0.553 | | | |
| Background liver
(cirrhosis) | 54 | 1.58 | 0.88–2.81 | 0.123 | | | |
| Pugh-Child’s
classification (B) | 11 | 0.93 | 0.28–2.32 | 0.889 | | | |
| AFP (>20
ng/ml) | 70 | 1.90 | 1.07–3.42 | 0.029a | 1.09 | 0.57–2.10 | 0.785 |
| PIVKA II (>40
mAU/ml) | 93 | 2.10 | 1.14–4.07 | 0.016a | 1.20 | 0.62–2.49 | 0.595 |
| Tumor multiplicity
(multiple) | 34 | 2.09 | 1.11–3.76 | 0.023a | 1.80 | 0.92–3.41 | 0.085 |
| Tumor size (≥3.0
cm) | 104 | 2.20 | 1.13–4.71 | 0.020a | 1.40 | 0.63–3.43 | 0.418 |
| Tumor
differentiation (well) | 35 | 0.55 | 0.25–1.10 | 0.095 | | | |
| Growth type
(invasive growth) | 24 | 1.44 | 0.69–2.76 | 0.318 | | | |
| Serosal
infiltration | 37 | 2.51 | 1.32–4.61 | 0.006a | 1.14 | 0.56–2.25 | 0.712 |
| Formation of
capsule | 104 | 1.05 | 0.57–2.02 | 0.884 | | | |
| Infiltration to
capsule | 83 | 1.20 | 0.67–2.18 | 0.537 | | | |
| Septum
formation | 98 | 0.87 | 0.49–1.60 | 0.651 | | | |
| Vascular
invasion | 37 | 3.40 | 1.87–6.07 | <0.001a | 1.85 | 0.94–3.66 | 0.076 |
| Margin status
(positive) | 28 | 2.64 | 1.42–4.73 | 0.003a | 2.60 | 1.36–4.84 | 0.005a |
| Decreased
PDSS2 in HCC | 56 | 2.52 | 1.41–4.52 | 0.002a | 2.45 | 1.27–4.80 | 0.008a |
Discussion
In the present study, our data support the role of
PDSS2 as a suppressor of HCC. PDSS2 mRNA was differentially
expressed by HCC cell lines and was decreased in 81% of the HCC
tissues. Hypermethylation of the PDSS2 promoter was detected
in four HCC cell types that expressed low levels of PDSS2
mRNA. Moreover, PDSS2 mRNA synthesis was reactivated after
demethylation, indicating that promoter hypermethylation regulated
PDSS2 transcription. To the best of our knowledge, the
present study is the first to show a correlation between the
expression and methylation status of PDSS2. However,
decreased transcription of PDSS2 was detected in some HCC
cells without hypermethylation of the PDSS2 promoter.
Because PDSS2 is located within chromosome 6q16.3-21, a site
of frequent loss of heterozygosity (LOH) in HCC (30,31),
we consider LOH as a possible alternative factor leading to
dysregulation.
Recent in silico pathway analysis suggests
that PDSS2 interacts with HNF4α, a nuclear transcription factor
that acts as a tumor suppressor and regulates the expression of
many genes involved in cell growth and proliferation (20,32).
CDX2 is a tumor suppressor of various malignancies and interacts
with HNF4α (28,33). Accordingly, the correlations of
mRNA expression between PDSS2, HNF4α and CDX2
were evaluated, and we found that the expression of PDSS2
correlated positively with that of HNF4α. These findings support
the function of PDSS2 as a suppressor of HCC, and further
pathway analysis will be required to support this conclusion.
PDSSs are heterotetrameric enzymes comprising
subunits encoded by PDSS1 (10p12.1) and PDSS2
(15,16). In the absence of prenyl diphosphate
synthase activity, CoQ10 is not synthesized (15). Therefore, decreased expression of
PDSS2 in the liver tissues may lead to reduced synthesis of
CoQ10 by hepatocytes, resulting in the inhibition of tumor
suppressors (17,18).
Similar to patients with malignant melanoma and lung
cancer, most patients with HCC harbored decreased levels of
PDSS2 mRNA in HCC tissues, and their mean level of
PDSS2 expression was significantly decreased in HCC tissues
compared with non-cancerous liver tissues (16,20).
Notably, we show here a stepwise decrease of PDSS2
expression accompanied with chronic inflammation and fibrosis of
uninvolved liver. Moreover, our data link the level of PDSS2
expression to tumor differentiation. Taken together, these results
suggest that PDSS2 plays a role in suppressing multistep
hepatocarcinogenesis.
Because the IHC and qRT-PCR data were consistent, we
used the latter to assess the prognostic significance of
PDSS2 mRNA levels in a quantitative manner (24,29).
We found that decreased PDSS2 mRNA expression in HCCs was
significantly associated with more aggressive tumor features,
including elevated preoperative serum α-fetoprotein, larger tumor
size and serosal infiltration, and therefore, was identified as an
independent prognostic factor. Moreover, patients with decreased
PDSS2 mRNA levels in their tumor tissues experienced
significantly earlier recurrence after curative hepatectomy.
Furthermore, the degree of the decrease in PDSS2 expression
correlated with UICC stage, indicating that the level of
PDSS2 expression reflects the severity of the malignant
phenotype of HCC.
Our evidence that PDSS2 act as a tumor suppressor is
as follows: i) decreased expression of PDSS2 was frequently
detected and the mean level of PDSS2 expression was
significantly lower in HCC tissues, and ii) decreased expression of
PDSS2 was associated with shorter recurrence free survival
and subsequent poor prognosis. PDSS2 expression levels in
biopsy or resected tissues may be useful for the prediction of
recurrence and poor prognosis, which will facilitate efforts to
devise an efficacious therapeutic strategy.
Our data support the conclusion that PDSS2 acts as a
tumor suppressor and that its expression is regulated by promoter
hypermethylation in HCC. Moreover, decreased expression of
PDSS2 mRNA may represent a novel biomarker for predicting
the progression and recurrence of HCC.
References
|
1
|
Minguez B and Lachenmayer A: Diagnostic
and prognostic molecular markers in hepatocellular carcinoma. Dis
Markers. 31:181–190. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kanda M, Nomoto S, Oya H, et al:
Downregulation of DENND2D by promoter hypermethylation is
associated with early recurrence of hepatocellular carcinoma. Int J
Oncol. 44:44–52. 2014.PubMed/NCBI
|
|
3
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
4
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: epidemiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shiraha H, Yamamoto K and Namba M: Human
hepatocyte carcinogenesis (review). Int J Oncol. 42:1133–1138.
2013.PubMed/NCBI
|
|
6
|
El-Serag HB: Hepatocellular carcinoma:
recent trends in the United States. Gastroenterology. 127:S27–S34.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kanda M, Nomoto S, Nishikawa Y, et al:
Correlations of the expression of vascular endothelial growth
factor B and its isoforms in hepatocellular carcinoma with
clinico-pathological parameters. J Surg Oncol. 98:190–196. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Llovet JM, Burroughs A and Bruix J:
Hepatocellular carcinoma. Lancet. 362:1907–1917. 2003. View Article : Google Scholar
|
|
9
|
Kanda M, Nomoto S, Okamura Y, et al:
Promoter hypermethylation of fibulin 1 gene is associated with
tumor progression in hepatocellular carcinoma. Mol Carcinog.
50:571–579. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
El-Serag HB and Mason AC: Rising incidence
of hepatocellular carcinoma in the United States. N Engl J Med.
340:745–750. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Villanueva A, Newell P, Chiang DY,
Friedman SL and Llovet JM: Genomics and signaling pathways in
hepatocellular carcinoma. Semin Liver Dis. 27:55–76. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Miki D, Ochi H, Hayes CN, Aikata H and
Chayama K: Hepatocellular carcinoma: towards personalized medicine.
Cancer Sci. 103:846–850. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kanda M, Nomoto S, Okamura Y, et al:
Detection of metallothionein 1G as a methylated tumor suppressor
gene in human hepatocellular carcinoma using a novel method of
double combination array analysis. Int J Oncol. 35:477–483.
2009.
|
|
14
|
Herath NI, Leggett BA and MacDonald GA:
Review of genetic and epigenetic alterations in
hepatocarcinogenesis. J Gastroenterol Hepatol. 21:15–21. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Saiki R, Nagata A, Kainou T, Matsuda H and
Kawamukai M: Characterization of solanesyl and decaprenyl
diphosphate synthases in mice and humans. FEBS J. 272:5606–5622.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fung JM, Smith R, Brown MA, et al:
Identification and characterization of a novel melanoma tumor
suppressor gene on human chromosome 6q21. Clin Cancer Res.
15:797–803. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Turunen M, Olsson J and Dallner G:
Metabolism and function of coenzyme Q. Biochim Biophys Acta.
1660:171–199. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Quinzii CM, DiMauro S and Hirano M: Human
coenzyme Q10 deficiency. Neurochem Res. 32:723–727. 2007.
View Article : Google Scholar
|
|
19
|
DiMauro S, Quinzii CM and Hirano M:
Mutations in coenzyme Q10 biosynthetic genes. J Clin Invest.
117:587–589. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen P, Yu J, Knecht J and Chen Q:
Decrease of PDSS2 expression, a novel tumor suppressor, in
non-small cell lung cancer. Cancer Epidemiol. 37:166–171. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Takami H, Kanda M, Oya H, et al:
Evaluation of MAGE-D4 expression in hepatocellular carcinoma in
Japanese patients. J Surg Oncol. 108:557–562. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sobin LH, Gospodarowicz MK and Wittekind
Ch: International Union Against Cancer: TNM Classification of
Malignant Tumors. 7th edition. Wiley-Blackwell; New York: 2009
|
|
23
|
Oya H, Kanda M, Takami H, et al:
Overexpression of melanomaassociated antigen D4 is an independent
prognostic factor in squamous cell carcinoma of the esophagus. Dis
Esophagus. Oct 2–2013.(Epub ahead of print). View Article : Google Scholar
|
|
24
|
Kanda M, Shimizu D, Nomoto S, et al:
Prognostic impact of expression and methylation status of DENN/MADD
domaincontaining protein 2D in gastric cancer. Gastric Cancer. Apr
3–2014.(Epub ahead of print).
|
|
25
|
Shimizu D, Kanda M, Nomoto S, et al:
Identification of intragenic methylation in the TU SC1 gene as a
novel prognostic marker of hepatocellular carcinoma. Oncol Rep.
31:1305–1313. 2014.PubMed/NCBI
|
|
26
|
Takai D and Jones PA: The CpG island
searcher: a new WWW resource. In Silico Biol. 3:235–240.
2003.PubMed/NCBI
|
|
27
|
Hibino S, Kanda M, Oya H, et al: Reduced
expression of DENND2D through promoter hypermethylation is an
adverse prognostic factor in squamous cell carcinoma of the
esophagus. Oncol Rep. 31:693–700. 2014.PubMed/NCBI
|
|
28
|
Saandi T, Baraille F, Derbal-Wolfrom L, et
al: Regulation of the tumor suppressor homeogene Cdx2 by HNF4alpha
in intestinal cancer. Oncogene. 32:3782–3788. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kanda M, Shimizu D, Nomoto S, et al:
Clinical significance of expression and epigenetic profiling of TU
SC1 in gastric cancer. J Surg Oncol. 110:136–144. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nagai H, Pineau P, Tiollais P, Buendia MA
and Dejean A: Comprehensive allelotyping of human hepatocellular
carcinoma. Oncogene. 14:2927–2933. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li SP, Wang HY, Li JQ, et al: Genome-wide
analyses on loss of heterozygosity in hepatocellular carcinoma in
Southern China. J Hepatol. 34:840–849. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Walesky C, Edwards G, Borude P, et al:
Hepatocyte nuclear factor 4 alpha deletion promotes
diethylnitrosamine-induced hepatocellular carcinoma in rodents.
Hepatology. 57:2480–2490. 2013. View Article : Google Scholar
|
|
33
|
Ehehalt F, Rummele P, Kersting S, et al:
Hepatocyte nuclear factor (HNF) 4alpha expression distinguishes
ampullary cancer subtypes and prognosis after resection. Ann Surg.
254:302–310. 2011. View Article : Google Scholar
|