Introduction
Lung cancer (LuCa) is the leading cause of
cancer-related deaths among men and women worldwide and is
responsible for more deaths than breast, colon, and prostate cancer
(PCa) combined (1). LuCas are
broadly classified into small cell lung carcinomas (SCLCs) and
non-small cell lung carcinomas (NSCLCs). About 85% of all LuCas are
identified as NSCLCs; 75% of these are metastatic or advanced at
diagnosis (2). Although patients
presenting with stage I–II diseases are usually treated with
surgery, half of these cases subsequently develop metastatic
disease that proves to be fatal. Despite many efforts, little has
been achieved for the treatment of this deadly disease. Advances in
understanding the factors involved in LuCa progression and
development of prognostic and predictive markers have the potential
to improve therapeutic outcomes.
NSCLC growth and metastases to secondary sites are
highly regulated events, which involve cellular transformation,
establishment of a pro-angiogenic environment, and migration and
invasion of tumor cells. This latter process is analogous to
leukocyte trafficking. To this end, chemokines and their receptors
play a major role. Chemokines are small, 8–10 kDa proteins involved
in directional migration of cells towards a chemokine gradient that
is detected by G-protein-coupled chemokine receptors. These
chemotactic cytokines are further classified into CC, CXC, C and
CX3C family members, based on their cysteine residues and disulfide
bonds. These chemokines are essential for homeostasis and function
of the immune and stem cell systems. In recent years, a new role of
chemokines has emerged, which involves neoplastic transformation of
cells, tumor cell growth and survival, and organ-specific
metastasis during carcinogenesis (3,4). Of
the CXC chemokines, CXCR4 is involved in NSCLC progression. NSCLC
tumors and cell lines express CXCR4, and, in mouse models,
anti-CXCR4 antibody reduces tumor metastases. Further, organs to
which NSCLCs preferentially metastasize constitutively express
CXCL12, a natural ligand for CXCR4 (5). To our knowledge, this is the first
study to demonstrate the association of CXCR5 and CXCL13 with
NSCLC. In contrast to CXCR4, which is expressed by normal and
malignant hematopoietic and non-hematopoietic cells (6), CXCR5 is expressed primarily by
mature, recirculating B cells and by small subsets of
CD4+ and CD8+ T cells. The migration of these
leukocytes into and within lymph nodes is controlled by
CXCR5-CXCL13 interactions (6,7).
Recently, it has been recognized that the CXCR5-CXCL13 axis is
associated with various hematologic (7–10)
and solid tumor malignancies (11–16).
Indeed, CXCR5 and CXCL13 are expressed in prostate, breast,
neuronal, and oral carcinomas (11,13–15).
Previously, we elucidated the molecular mechanisms and functional
significance of CXCR5 and CXCL13, whereby this axis promotes PCa
cell migration, invasion, and differential matrix metalloproteinase
(MMP) expression (17). We also
showed that CXCL13-mediated invasion of PCa cells requires Akt and
ERK1/2 activation, suggesting a new role for DOCK2, a protein
involved in intracellular signaling, in proliferation of
hormone-refractory CXCR5-positive PCa cells (18).
Based on these findings, we investigated the
expression of CXCR5 and CXCL13 in patient samples of NSCLCs,
evaluating the expression of CXCR5 in normal, squamous cell
carcinoma (SCC), and adenocarcinoma (AC) tissues by
immunohistochemical staining. To determine the association of
CXCL13 with NSCLC progression, serum CXCL13 levels were analyzed
for both subtypes of NSCLCs. Furthermore, the expression patterns
of CXCR5 in human LuCa cell lines were determined, and the findings
were correlated with clinicopathological features to evaluate the
role of CXCR5 in NSCLC progression. To understand the biological
significance of CXCR5 over-expression in NSCLCs, the migration
potential of LuCa cells via CXCL13 was analyzed. These results
demonstrate the association with and point to a role of CXCR5 and
CXCL13 in NSCLCs.
Materials and methods
Tissue specimens
Tissue microarray (TMA) slides containing malignant
(n=78), non-neoplastic (n=8), and other (n=12) lung tissues (n=98)
were purchased from AccuMax Array (ISU Abxis Co., Ltd., Seoul,
Korea). These spots were generated from lung biopsies of 45 cases
diagnosed with NSCLCs with histological subtypes of AC (n=27), SCC
(n=12), and others (n=6); and eight non-neoplastic cases. To
construct TMAs, two cores (1 mm in diameter) per patient were
arrayed on a receiver paraffin block, and, concerning the
histopathology, a qualified pathologist validated each core of the
TMAs twice for class and grade of the tumor. LuCa TMAs consisted of
tumors from 45 patients and represented all histopathological
subtypes reported for LuCa. The total of 98 spots represented eight
non-neoplastic, 54 ACs, 24 SCCs, and 12 others.
Immunohistochemistry
TMA slides containing biopsies obtained from
malignant and non-neoplastic cases were stained for CXCR5. Briefly,
TMAs were de-paraffinized in xylene and rehydrated through a graded
series of ethanol (100, 95 and 70%) for 5 min in each series and
washed in distilled water. After de-paraffinization, antigen
retrieval was accomplished by incubating TMAs with 0.01 M EDTA (pH
8.0) in a pressure cooker for 5 min. Slides were then cooled in
running water and transferred to Tris-buffer (pH 7.6). The
endogenous peroxidase activity was blocked by incubating the slides
with 3% H2O2 in phosphate-buffered saline
(PBS) for 5 min. The slides were then rinsed three times with
de-ionized water followed by Tris-buffer (pH 7.6). Following
washing, Fc blocking was accomplished by incubating the slides with
Fc Block (Innovex Biosciences, Inc., Richmond, CA, USA) for 30 min
at room temperature (RT) in a humidity chamber. To reduce
non-specific binding, the sections were washed with Tris-buffer and
incubated with 3% normal goat serum for 1 h at RT. The slides were
then washed with Tris-buffer, and sections were incubated with 5
μg/ml of HRP-conjugated mouse anti-CXCR5 antibody (R&D Systems,
Minneapolis, MN, USA) for 90 min at RT in a humidity chamber.
Negative control slides were incubated with 5 μg/ml of mouse
isotype control antibody (R&D Systems). Following incubation,
sections were washed with Tris-buffer and developed with a
3,3′-diaminobenzidine (DAB) (Vector Laboratories, Inc., Burlingame,
CA, USA) as a chromogen for 25 min at RT. The sections were also
incubated with alkaline phosphatase (AP)-conjugated goat anti-mouse
antibody (Invitrogen Life Technologies, Grand Island, NY, USA) for
20 min at RT and developed with AP-New Magenta (BioFX Laboratories,
Inc., Owings Mills, MD, USA) substrate for 25 min at RT.
Counterstaining was with hematoxylin (Sigma, St. Louis, MO, USA).
Subsequently, sections were washed with water, dehydrated in 70,
95%, and absolute alcohol for 5 min each, passed through xylene
three times for 1 min each, and mounted with Permount (Sigma). The
immunopositivity of the sections was analyzed using an Aperio
ScanScope scanning system (Aperio Technologies, Vista, CA,
USA).
Quantitation of immunohistochemical
staining
To analyze the immunohistochemical staining, virtual
slides were created from the stained samples after scanning each
specimen with an Aperio ScanScope scanning system (Aperio
Technologies). The ScanScope generated true-color digital images of
each stained sample, which were viewed using Aperio ImageScope
v.6.25 software. The algorithm for determining the intensity of
membrane-specific staining was used to calculate, for each sample,
the staining intensity and percent of target labeled by digitally
analyzing the color intensity. A color markup image for each slide
was obtained based on membrane staining intensity. The output was
viewed as determinations of staining intensity ranging from 0–3 to
correlate with conventional manual scoring methods (where 0,
negative and 3, strong staining), and statistical analyses were
performed using the means of these values.
Quantitative enzyme-linked immunosorbent
assay (ELISA) for serum CXCL13
Sera from patients with SCCs (n=17), ACs (n=14), and
healthy controls (n=9) were obtained from the James Graham Brown
Cancer Center, University of Louisville, KY, USA. Healthy donors
had no active lung disease or symptoms at the time of blood
collection. All subjects gave written informed consent and were
approved by the University of Alabama at Birmingham Institutional
Review Board (IRB). Subsequently, the University of Louisville IRB
approved the use of these diagnostic specimens in accordance with
the Department of Health and Human Service Policy for the
Protection of Human Research Subjects 45 CFR 46.101(b) 2 and use of
archived de-identified materials. Serum CXCL13 levels were measured
by human CXCL13 Quantikine ELISA (R&D Systems), following the
manufacturer’s instructions. Briefly, 100 μl of assay diluent
(provided with the kit), followed by 50 μl of standard, control, or
samples (sera from patients and healthy controls) were added in
different wells of a 96-well plate and incubated for 2 h at RT.
Following washing four times with Quantikine Wash Buffer 1
(provided with the kit), 200 μl of conjugate (antibody) was added
to each well, and the plate was incubated for 2 h at RT. The plate
was washed further, and 200 μl of substrate solution (provided with
the kit) was added to each well. The plate was incubated for 30 min
in the dark at RT. Following incubation, 50 μl of stop solution was
added to each well, and the plate was read in an ELISA reader at
450 nm. The ELISA assays were capable of detecting >1 pg/ml of
CXCL13.
Cell cultures
Human NSCLC NCI-H1915 (CRL-5904) and small cell lung
carcinoma SW-1271 (CRL-2177) cell lines were obtained from American
Type Cell Culture (ATCC) (Teddington, UK). NCI-H1915 cells were
cultured in RPMI-1640 media (Mediatech Cellgro, Herndon, VA, USA)
supplemented with 10% heat-inactivated fetal bovine serum (FBS)
(Sigma), 100 μg/ml streptomycin, and 100 U/ml penicillin (Sigma) at
37°C with 5% CO2. SW-1271 cells were cultured in
Leibovitz’s L-15 Medium (Mediatech Cellgro) supplemented with 10%
FBS, 100 μg/ml streptomycin, and 100 U/ml penicillin at 37°C with
100% air. Cells were split twice a week to ensure optimal growth
conditions.
Functional analysis of CXCR5
expression
For both LuCa cell lines, CXCR5 surface expression
was analyzed by flow cytometry. Briefly, LuCa cells were washed
three times in PBS [supplemented with 1% bovine serum albumin
(BSA)], and treated with 1 μg of Fc Block (BD Biosciences, San
Jose, CA, USA) per 105 cells for 15 min at RT.
Fc-blocked cells were incubated with 1 μg of fluorescein-conjugated
mouse anti-human CXCR5 or fluorescein-conjugated mouse IgG2a
isotype control antibodies (R&D Systems) per 105
cells for 1 h at 4°C. Following staining, the unbound antibodies
were removed by washing the cells three times with
fluorescence-activated cell-sorting (FACS) buffer (1% BSA in PBS).
The labeled cells were then fixed in 500 μl of 2% paraformaldehyde
solution and analyzed by flow cytometry using a FACScan flow
cytometer (BD Biosciences). The flow cytometry data were analyzed
with FlowJo software. The stained cells were also analyzed by an
Amnis ImageStream system (Amnis Corp., Seattle, WA, USA).
Migration assay
Cell migration was assessed with a BD BioCoat™
migration chamber system (BD Biosciences). Briefly, Matrigel
inserts were hydrated for 2 h with 500 μl of serum-free Dulbecco’s
modified Eagle’s medium at 37°C with 5% CO2. CXCL13
(PeproTech, Rocky Hill, NJ, USA), at a concentration of 0 or 100
ng/ml, was added to the bottom chamber containing serum-free RPMI
medium. Next, NCI-H1915 and SW-1271 cells were incubated with
isotype control or anti-human CXCR5 antibody at a concentration of
1 μg/ml (both from R&D Systems) for 1 h at 37°C with 5%
CO2 and added to the top chambers in serum-free RPMI
medium at 10,000 cells per well. The cells were allowed to migrate
for 8 h at 37°C under 5% CO2. Non-migrating cells on the
upper surface of the membrane were removed with a cotton swab. The
cells that migrated to the lower surface were fixed with 100%
methanol for 5 min at RT, stained with crystal violet for 2 min,
and rinsed twice with distilled water. The membranes were placed on
glass slides. The migrated cells were counted by microscopy at ×40
magnification. The experiments were performed in triplicate and
repeated three times.
Statistics
The intensity of CXCR5 and CXCL13 expression by lung
TMAs was tested for normality assumptions using the Shapiro-Wilk
test and was transformed to a Log scale. The general linear models
procedure was used to test the association of CXCR5 and CXCL13
expression and disease condition using SAS v.9.1.3 statistical
analysis software. Results were declared significant at α level of
0.001. The experimental data were compared using a two-tailed
Student’s t-test and expressed as mean ± SEM. The results were
analyzed using the Stat View program (Abacus Concepts, Inc.,
Piscataway, NJ, USA) and were labeled statistically significant if
p-values were <0.01. With Cell Quest Software, the
Kolmogorov-Smirnov (K-S) two-sample test was used to calculate the
statistical significance of the CXCR5 histograms.
Results
Immunohistochemical analyses demonstrate
CXCR5 is overexpressed in LuCa tissues relative to non-neoplastic
tissues
LuCa TMAs, consisting of 98 biopsy areas, generated
from biopsies of malignant (SCC and AC) and non-neoplastic cases,
were analyzed for CXCR5 expression by immunohistochemistry. CXCR5
was expressed in tissues collected from SCC and AC cases
(p<0.001) relative to non-neoplastic tissues, in which no signal
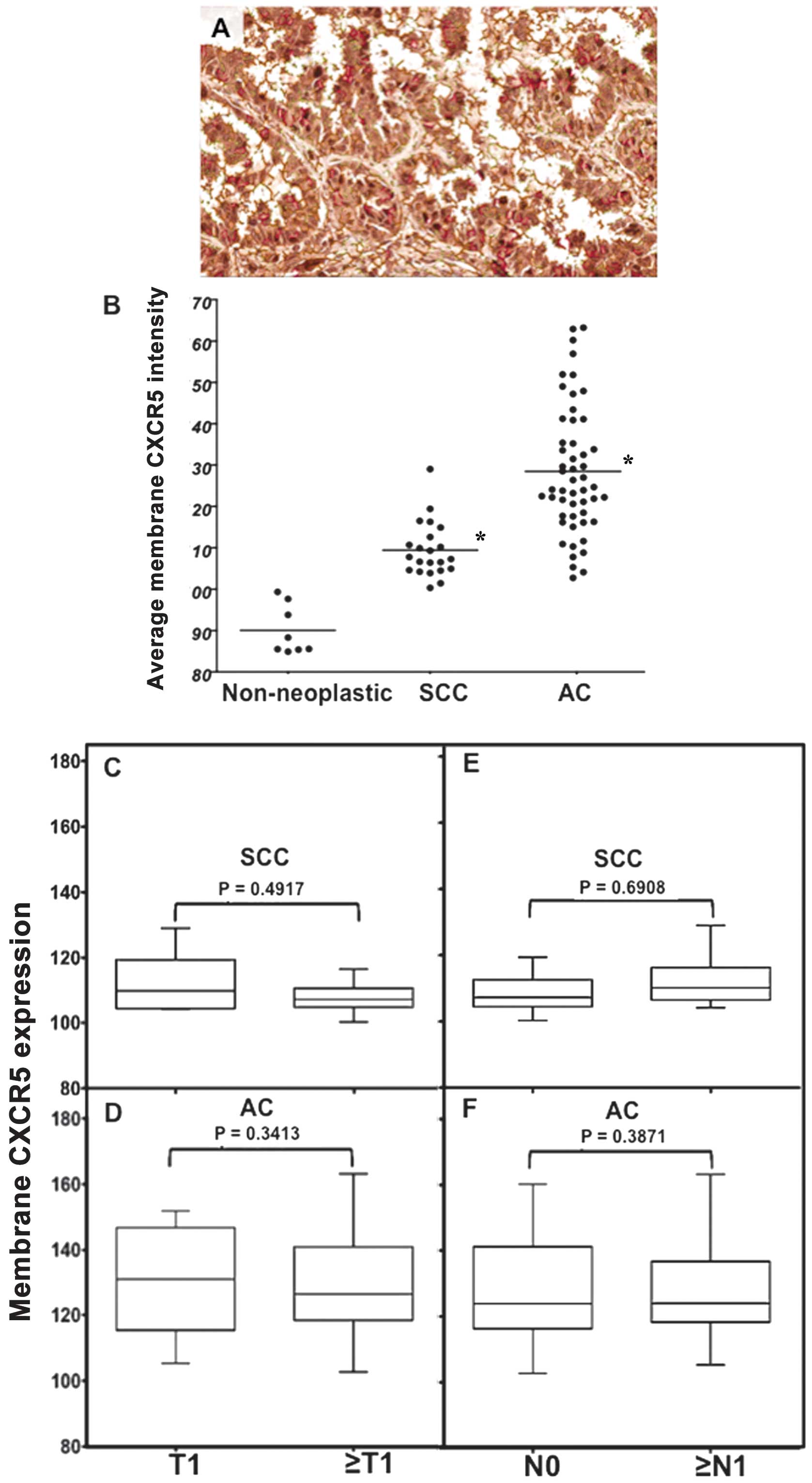
was detected (Fig. 1). Average
positive, nuclear, and membrane CXCR5 intensities were quantified
in non-neoplastic, SCC, and AC cases using image analysis Aperio
ImageScope v.6.25 software (Figs.
2–4). These intensities were
highest in ACs, with median values of 128, 185, and 130,
respectively; followed by SCC with median values of 118, 170, and
110, respectively; and lowest in non-neoplastic tissues with median
values of 92, 142, and 90, respectively. Further, CXCR5 expression
correlated with stage (T) and nodal involvement (N) of tumors in
both SCC and AC tissues. In SCCs, total average CXCR5 expression in
cases with T1 (median value, 116) was essentially equal to cases
with ≥T2 (median value, 115) but lower than ≥T2 in ACs (median
value, 120). However, the average positive pixel count of CXCR5
expression in ACs was higher in T1 (average value, 154) than in ≥T2
(average value, 138) there was higher CXCR5 expression for cases
with ≥N1 (median value, 121) than N0 (median value, 116) in SCCs,
but there was little difference for AC cases (median value 124)
(Fig. 2). Both nuclear and
membrane CXCR5 expression was higher in T1 than in ≥T2 SCCs (median
values, 174 and 110 vs. 162 and 108, respectively). Although
nuclear and membrane CXCR5 expressions in T1 (median values, 186
and 132, respectively) were slightly higher than ≥T2 (median values
185 and 128, respectively) of ACs, similar to SCCs, the maximum
expressions of both nuclear and membrane CXCR5 intensities in ≥T2
ACs (average values 198 and 162, respectively) were higher relative
to the maximum for T1 ACs (average values, 194 and 150,
respectively). Further, both nuclear and membrane CXCR5 intensities
were higher in SCCs with ≥N1 (median values, 172 and 112) relative
to SCCs with N0 (median values, 162 and 108). However, in AC
tissues, both nuclear and membrane CXCR5 intensities were
essentially equal in N0 and ≥N1, with median values of 186 and 122,
respectively (Figs. 3 and 4).
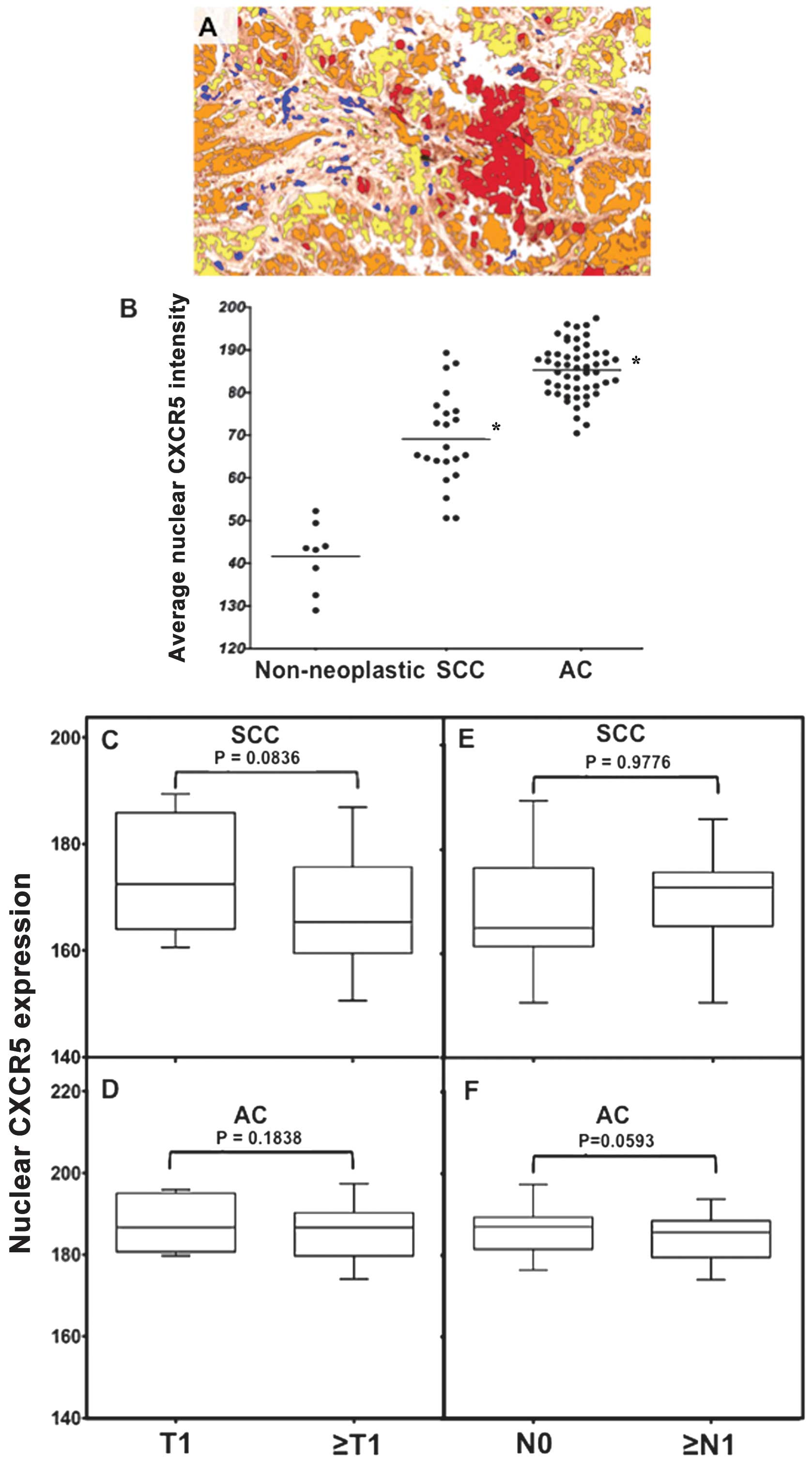 | Figure 3Nuclear CXCR5 expression in lung
cancer (LuCa) tissues. (A) Representative figures of non-neoplastic
(n=8), squamous cell carcinoma (SCC) (n=22), and adenocarcinoma
(AC) (n=52) lung tissues stained with isotype control or anti-CXCR5
antibodies. Brown [3,3′-diaminobenzidine (DAB)] color show CXCR5
staining. An Aperio ScanScope CS system with a 40× objective
captured digital images of each slide. Stained cells with negative
and positive nuclei were counted and categorized according to stain
intensity 0 (Blue), 1+(yellow), 2+(orange) and 3+(Red). (B) The
nuclear intensity of CXCR5, in non-neoplastic (n=8), SCC (n=22),
and AC (n=52) tissues, quantified using a nuclear algorithm of
image analysis Aperio ImageScope v.6.25 software.
*Significant differences (p<0.001) between groups
with LuCa and control. (C–F) Nuclear intensities of CXCR5 in
different tumor stages and in tumors with nodal involvement in SCC
and AC cases, respectively. |
Levels of serum CXCL13 are elevated in
LuCa patients
Serum CXCL13 in SCC and AC patients was quantified
with a commercially available ELISA kit. Similar to
histo-pathological findings of CXCR5, the serum CXCL13 level was
significantly higher for AC followed by SCC patients, relative to
healthy controls. The median values of CXCL13 expression (pg/ml) in
serum of ACs and SCCs were 42 and 32, respectively (Fig. 5). However, the CXCL13 expression in
healthy controls was lower, with a median value of 15.
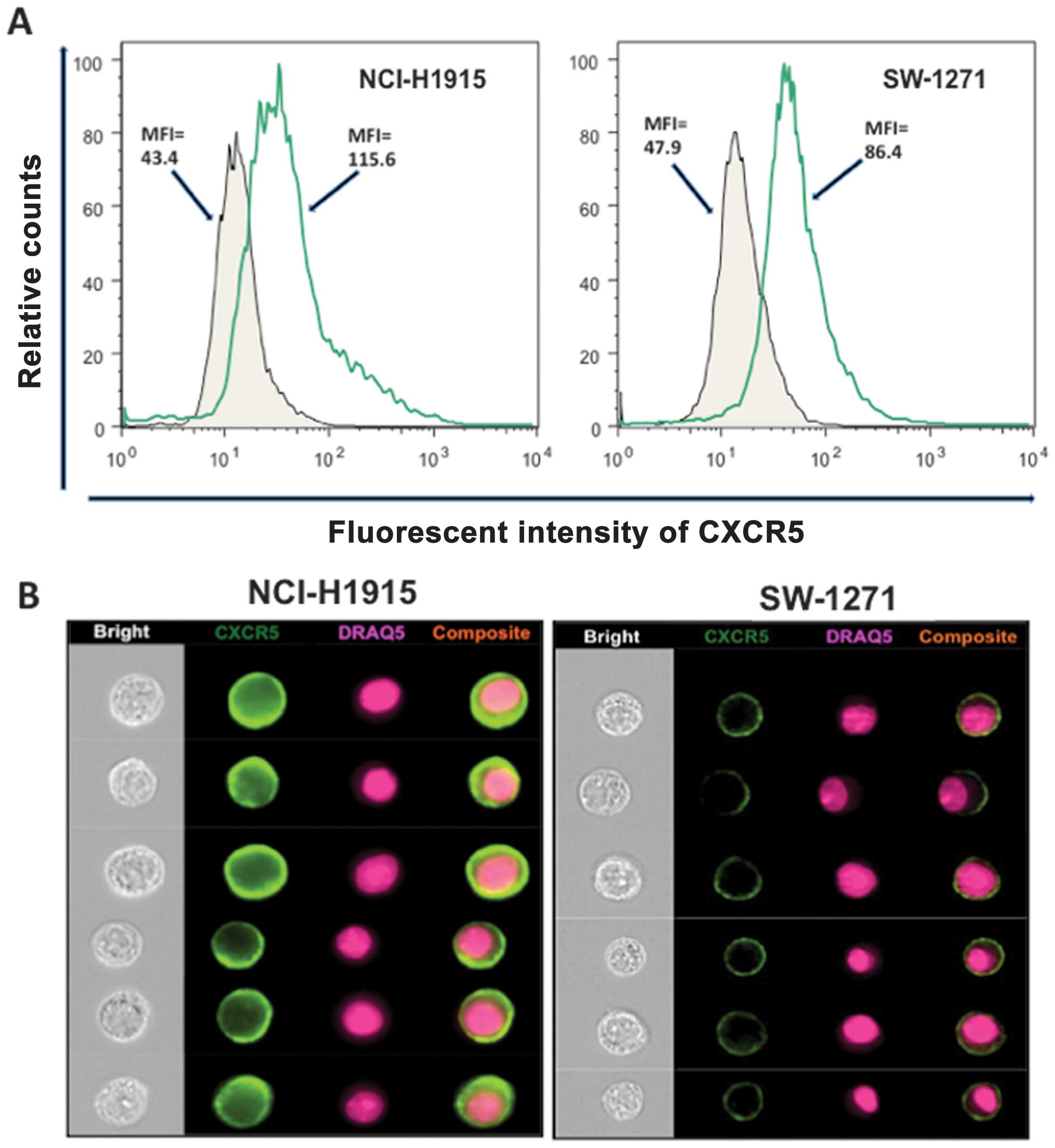
CXCR5 is over-expressed on LuCa cell
lines
CXCR5 expression was evaluated in human NSCLC
(NCI-H1915) and SCLC (SW-1271) cell lines using flow cytometry and
images were captured by a flow-based imaging system (Amnis
ImageStream system). Both cell lines stained positive for CXCR5 and
a nuclear stain, DRAQ5. The intensity of membrane CXCR5 expression,
measured in terms of mean fluorescence intensity (MFI), was higher
in NSCLC (NCI-H1915) cells, relative to SCLC (SW-1271) cells (MFI,
115.6 vs. 86.4) (Fig. 6A). CXCR5
was expressed on membranes of both cell lines, as seen by their
composite images (Fig. 6B).
LuCa cells migrate following the
CXCR5-CXCL13 interaction
The functional significance of CXCR5 expression by
LuCa cell lines was demonstrated by the capacity of NCI-H1915 and
SW-1271 cells to migrate towards CXCL13. Both types of cells
migrated to media with CXCL13, relative to media without CXCL13
(Fig. 7). However, the numbers of
NCI-H1915 cells that migrated in response to CXCL13 was higher than
the numbers of SW-1271 cells. CXCR5-CXCL13 dependent chemotaxis was
abrogated by anti-CXCR5 antibodies. Cells treated with isotype
control antibodies served as controls, in which no inhibition in
the number of migratory cells was observed in response to
CXCL13.
Discussion
Chemokines/chemokine receptors apparently facilitate
tumor dissemination, leading to metastasis, growth, and cell
survival (19–21). Few studies have addressed the
significance of chemokine/chemokine receptor expression in NSCLCs.
Higher expressions of CXCR1, CXCR2, and CXCR4 with their ligands
CXCL5, CXCL8, and CXCL12 appear to be associated with tumor
angiogenesis, metastasis, and poor survival in NSCLCs (5,22–26).
Until now, there were no comprehensive studies regarding the
presence or potential role of CXCR5 in NSCLCs.
CXCR5 is a principal regulator for targeting T
cells, B cells, and dendritic cells into secondary lymphoid organs.
The CXCR5-CXCL13 axis is involved in development and progression of
solid tumors, e.g., breast cancers and neuronal cancers (13,14).
Recently, we demonstrated a differential expression of CXCR5 in PCa
cell lines correlated with PCa progression (17,18).
These findings provided the rationale for the present study, which
characterizes CXCL5 and CXCR13 expression and interactions during
LuCa progression.
The expression of CXCR5 and/or CXCL13 was assessed
in LuCa tissues, serum, and LuCa cell lines. Higher CXCR5
expression in LuCa tissues, relative to non-neoplastic tissues and
elevated serum CXCL13 in LuCa patients relative to healthy controls
correlated with disease progression. SCCs and ACs are two major
histologic types of NSCLCs. Patients with ACs have a poorer
prognosis than those with SCCs (27,28).
However, the differences in biological aggressiveness between SCCs
and ACs of the lung are not well understood. We found higher
nuclear and membrane intensities of CXCR5 in ACs relative to SCCs.
Our findings demonstrate that CXCR5 is differentially expressed in
lung carcinomas depending on stage of the disease. Moreover,
differential nuclear and membrane CXCR5 expression in SCCs and ACs
correlate with their aggressiveness. We further investigated CXCR5
expression in relation to tumor stage (T) and nodal metastasis (N).
Although we did not find a statistically significant correlation
between CXCR5 expression and tumor stage or nodal metastasis, there
was increased CXCR5 expression in SCC cases with nodal metastases.
Hence, it is plausible that the CXCR5-CXCL13 axis is involved in
tumor dissemination to lymph nodes. We did not observe any distinct
change in CXCR5 expression in AC cases with increased tumor stage
and nodal metastasis. Since the present study was limited to a
small number of patients, subsequent studies with more patient
samples from each group (SCC and AC) and subgroups (T1, ≥T2 and N0
and ≥N1) will provide conclusive information correlating CXCR5
expression in NSCLCs with disease progression and survival.
In our previous studies, we showed that serum CXCL13
levels correlated with prostatic disease and mediated PCa cell
migration, integrin clustering, and cell adhesion (12). Here, for NSCLC patients, serum
CXCL13 levels in ACs were higher than in SCCs, which may be
associated with the aggressiveness of the latter disease.
In view of the expression of CXCR5 in LuCa tissues
and to understand the biological significance of CXCR5 expression,
we stained and analyzed CXCR5 expression on two human LuCa cell
lines, NSCLC (NCI-H1915) and SCLC (SW-1271). We also determined the
migration potential of these cells under a chemotactic gradient of
CXCL13. Staining and migration assay results were in accordance
with the hypothesis that the CXCR5-CXCL13 axis is involved in LuCa
cell dissemination and/or metastasis. SW-1271 cells, which had
lower expression of CXCR5 relative to NCI-H1915 cells, were not as
responsive to CXCL13 as were NCI-H1915 cells.
In conclusion, NSCLC tissues expressed CXCR5, which
correlated with stage/grade of the disease. Higher CXCR5 expression
and migration by NSCLC cells suggest a role in migration and
metastasis of primary lung tumors in response to CXCL13. These
findings indicate that differential expression patterns of CXCR5
and CXCL13 in two subtypes (SCC and AC) of NSCLC are associated
with differences in their prognosis and survival. Identification of
a sensitive tumor marker, predictive of diagnosis, prognosis, and
drug sensitivity, would enhance NSCLC treatment and diagnosis. We
propose that CXCR5/CXCL13, either alone or in combination, could be
used as a prognostic biomarker for LuCa; however, this can be
established only by larger studies in which these factors are
evaluated in the same patients.
Acknowledgements
The content of this manuscript benefited from many
fruitful conversations with members of the Morehouse School of
Medicine, Atlanta, GA, USA; the University of Louisville, School of
Medicine; and the University of Alabama at Birmingham. This study
was supported by U54CA118638 and G12MD007602.
References
|
1
|
Esposito L, Conti D, Ailavajhala R, Khalil
N and Giordano A: Lung cancer: are we up to the challenge? Curr
Genomics. 11:513–518. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Molina JR, Yang P, Cassivi SD, Schild SE
and Adjei AA: Non-small cell lung cancer: epidemiology, risk
factors, treatment, and survivorship. Mayo Clin Proc. 83:584–594.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Baird AM, Gray SG and O’Byrne KJ:
Epigenetics underpinning the regulation of the CXC (ELR+)
chemokines in non-small cell lung cancer. PLoS One. 6:e145932011.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lazennec G and Richmond A: Chemokines and
chemokine receptors: new insights into cancer-related inflammation.
Trends Mol Med. 16:133–144. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Su L, Zhang J, Xu H, Wang Y, Chu Y, Liu R
and Xiong S: Differential expression of CXCR4 is associated with
the meta-static potential of human non-small cell lung cancer
cells. Clin Cancer Res. 11:8273–8280. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Burger JA and Kipps TJ: CXCR4: a key
receptor in the crosstalk between tumor cells and their
microenvironment. Blood. 107:1761–1767. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bürkle A, Niedermeier M, Schmitt-Gräff A,
Wierda WG, Keating MJ and Burger JA: Overexpression of the CXCR5
chemokine receptor, and its ligand, CXCL13 in B-cell chronic
lymphocytic leukemia. Blood. 110:3316–3325. 2007.PubMed/NCBI
|
|
8
|
Hussain SK, Zhu W, Chang SC, et al: Serum
levels of the chemokine CXCL13, genetic variation in CXCL13 and its
receptor CXCR5, and HIV-associated non-Hodgkin B-Cell lymphoma
risk. Cancer Epidemiol Biomarkers Prev. 22:295–307. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kurtova AV, Tamayo AT, Ford RJ and Burger
JA: Mantle cell lymphoma cells express high levels of CXCR4, CXCR5,
and VLA-4 (CD49d): importance for interactions with the stromal
microenvironment and specific targeting. Blood. 113:4604–4613.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chan CC, Shen D, Hackett JJ, Buggage RR
and Tuaillon N: Expression of chemokine receptors, CXCR4 and CXCR5,
and chemokines, BLC and SDF-1, in the eyes of patients with primary
intraocular lymphoma. Ophthalmology. 110:421–426. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Singh S, Singh R, Singh UP, et al:
Clinical and biological significance of CXCR5 expressed by prostate
cancer specimens and cell lines. Int J Cancer. 125:2288–2295. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Singh S, Singh R, Sharma PK, et al: Serum
CXCL13 positively correlates with prostatic disease,
prostate-specific antigen and mediates prostate cancer cell
invasion, integrin clustering and cell adhesion. Cancer Lett.
283:29–35. 2009. View Article : Google Scholar
|
|
13
|
Panse J, Friedrichs K, Marx A, et al:
Chemokine CXCL13 is overexpressed in the tumour tissue and in the
peripheral blood of breast cancer patients. Br J Cancer.
99:930–938. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Del Grosso F, Coco S, Scaruffi P, et al:
Role of CXCL13-CXCR5 crosstalk between malignant neuroblastoma
cells and Schwannian stromal cells in neuroblastic tumors. Mol
Cancer Res. 9:815–823. 2011.PubMed/NCBI
|
|
15
|
Yuvaraj S, Griffin AC, Sundaram K,
Kirkwood KL, Norris JS and Reddy SV: A novel function of CXCL13 to
stimulate RANK ligand expression in oral squamous cell carcinoma
cells. Mol Cancer Res. 7:1399–1407. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Airoldi I, Cocco C, Morandi F, Prigione I
and Pistoia V: CXCR5 may be involved in the attraction of human
metastatic neuroblastoma cells to the bone marrow. Cancer Immunol
Immunother. 57:541–548. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
El Haibi CP, Sharma PK, Singh R, Johnson
PR, Suttles J, Singh S and Lillard JW Jr: PI3Kp110-, Src-,
FAK-dependent and DOCK2-independent migration and invasion of
CXCL13-stimulated prostate cancer cells. Mol Cancer.
9:852010.PubMed/NCBI
|
|
18
|
El-Haibi CP, Singh R, Sharma PK, Singh S
and Lillard JW Jr: CXCL13 mediates prostate cancer cell
proliferation through JNK signalling and invasion through ERK
activation. Cell Prolif. 44:311–319. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Iwakiri S, Mino N, Takahashi T, et al:
Higher expression of chemokine receptor CXCR7 is linked to early
and metastatic recurrence in pathological stage I nonsmall cell
lung cancer. Cancer. 115:2580–2593. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rollins BJ: Chemokines. Blood. 90:909–928.
1997.PubMed/NCBI
|
|
21
|
Taub DD: Chemokine-leukocyte interactions.
The voodoo that they do so well. Cytokine Growth Factor Rev.
7:355–376. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Spano JP, Andre F, Morat L, et al:
Chemokine receptor CXCR4 and early-stage non-small cell lung
cancer: pattern of expression and correlation with outcome. Ann
Oncol. 15:613–617. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhu YM, Webster SJ, Flower D and Woll PJ:
Interleukin-8/CXCL8 is a growth factor for human lung cancer cells.
Br J Cancer. 91:1970–1976. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Phillips RJ, Burdick MD, Lutz M, Belperio
JA, Keane MP and Strieter RM: The stromal derived
factor-1/CXCL12-CXC chemokine receptor 4 biological axis in
non-small cell lung cancer metastases. Am J Respir Crit Care Med.
167:1676–1686. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Saintigny P, Massarelli E, Lin S, et al:
CXCR2 expression in tumor cells is a poor prognostic factor and
promotes invasion and metastasis in lung adenocarcinoma. Cancer
Res. 73:571–582. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen JJ, Yao PL, Yuan A, et al:
Up-regulation of tumor interleukin-8 expression by infiltrating
macrophages: its correlation with tumor angiogenesis and patient
survival in non-small cell lung cancer. Clin Cancer Res. 9:729–737.
2003.PubMed/NCBI
|
|
27
|
Ichinose Y, Yano T, Asoh H, Yokoyama H,
Yoshino I and Katsuda Y: Prognostic factors obtained by a
pathologic examination in completely resected non-small-cell lung
cancer. An analysis in each pathologic stage. J Thorac Cardiovasc
Surg. 110:601–605. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Suzuki K, Nagai K, Yoshida J, Nishimura M,
Takahashi K, Yokose T and Nishiwaki Y: Conventional
clinicopathologic prognostic factors in surgically resected
nonsmall cell lung carcinoma. A comparison of prognostic factors
for each pathologic TNM stage based on multivariate analyses.
Cancer. 86:1976–1984. 1999. View Article : Google Scholar
|