1. Introduction
Malignant melanoma (MM) has one of the
fastest-rising incidences of all cancers (1). This, coupled with the poor prognosis
for patients in stages III and IV of the disease, is causing a
growing healthcare burden on society. At the molecular level,
rather than being a single disease, melanoma comprises a
heterogeneous group of disorders that harbour distinct aberrations
in diverse cellular processes such as cell cycle regulation, cell
signalling, cell adhesion, cell differentiation and apoptosis
(2). Such heterogeneity suggests
that multiple mechanisms are involved in disease aetiology and this
is reflected in the contribution of both different mutations and
differential gene and protein expression associated with MM
development and progression (3,4).
Our understanding of genomics is currently
undergoing a paradigm shift. The past decade has seen staggering
advances in next generation sequencing (NGS), precipitating an
explosion of genome-wide transcriptome studies, culminating in 2012
with the completion of the ENCODE project (5). Large scale cDNA sequencing and
interrogation of whole chromosome tiling arrays across a variety of
cell types reveal that >90% of genomic nucleotides are
transcribed (6). Perhaps the most
surprising discovery to emerge from these studies is that only a
tiny percentage of the RNA transcripts synthesised are fated to
code for protein, with the vast majority being comprised of
non-coding RNA. This realisation eschews the central dogma of
molecular biology (7) and has
ushered in a new era of non-coding RNA research that is focused on
unpicking the functional significance of these prevalent
transcripts (8,9).
Non-coding RNAs can be broadly defined according to
their size as either short (<200 nt) or long (>200 nt). Short
non-coding RNAs are well-characterised, particularly those involved
in the production and operation of gene expression machinery such
as ribosomal RNA, transfer RNA, small-nuclear RNA and
small-nucleolar RNA. More recently, short non-coding RNAs that
function to regulate, rather than operate gene expression have been
intensely investigated and can be further divided into three main
subcategories: PIWI-associated RNAs, which regulate transposable
elements, small interfering RNAs and microRNAs (miRNAs), which
mediate post-transcriptional silencing of mRNA and the recently
described transcription initiation RNAs, which appear to instigate
gene expression at the promoter level. Of these, miRNAs are the
best understood and function to regulate both fundamental processes
in biology, such as proliferation, apoptosis and differentiation,
while also contributing significantly to disease aetiology, most
notably cancer (10), but also
cardiovascular disease (11),
stroke (12) and several
neurological disorders (13,14).
The functional role played by miRNAs in the establishment and
metastatic progression of melanoma was recently reviewed (15) and compiling data from several
groups has enabled the coupling of differential miRNA expression in
MM to specific target protein modulation and critically downstream
functional consequence for disease (16–24).
Such studies represent an emerging area of research, indeed, the
prevalence of miRNAs in the human transcriptome continues to expand
as deep-sequencing technologies improve, with current estimates
identifying ~9,000 small non-coding RNAs, ~1,100 of which are
potentially functional miRNA transcripts (5). However, this number is dwarfed by the
pervasiveness of the long non-coding RNA (lncRNA) transcripts, with
GENCODE identifying a staggering 22,444 non-coding transcripts
>200 nt in length and more recent studies estimating this number
to be nearer 32,000 (25).
In contrast to miRNA, our understanding of lncRNA
function is in its infancy, however, huge efforts are being made to
fill this knowledge gap, particularly in relation to their role in
disease. The importance of such study is underlined by what we do
understand about a handful of lncRNAs involved in fundamental
aspects of biology. Most notable of these are the lncRNAs
XIST, which inactivates gene expression from the
X-chromosome during dosage equalisation and HOTAIR, a lncRNA
that acts in trans via the Polycomb repressor complex to
regulate genes at a distance, intimating that lncRNA are capable of
more complex modes of gene regulation (26,27).
In parallel to their role in the regulation of cellular processes
and akin to their smaller cousins, the miRNAs, it is the
involvement of lncRNAs in the development and progression of cancer
that drives much of the current research on these transcripts. This
review will focus on recent advances made in the discovery of
lncRNAs with a functional role in the progression of melanoma
metastasis.
2. Long non-coding RNAs drive metastatic
progression in melanoma
The prognosis for patients with early stage,
localised melanoma is favourable, with 10-year survival rates
approaching 90%, depending on Breslow thickness, ulceration and
mitotic index (28). This
prognosis deteriorates rapidly with the onset of regional
metastasis (10-year survival of around 50%) and for patients with
stage IV melanoma and distant metastasis, 5-year survival rates are
<10% (29). Clearly, metastatic
progression in melanoma severely impacts on patient health and
there is an urgent need to expand our understanding of the
mechanisms underpinning this process. Numerous studies
investigating the functional role played by lncRNA in cancer
suggest that they are functioning to regulate events intimately
associated with the metastatic transition, such as cell migration
and tissue invasion (30,31). Here we will review our current
understanding of lncRNAs known to impact on melanoma and consider
what we understand about the functional mechanisms involved.
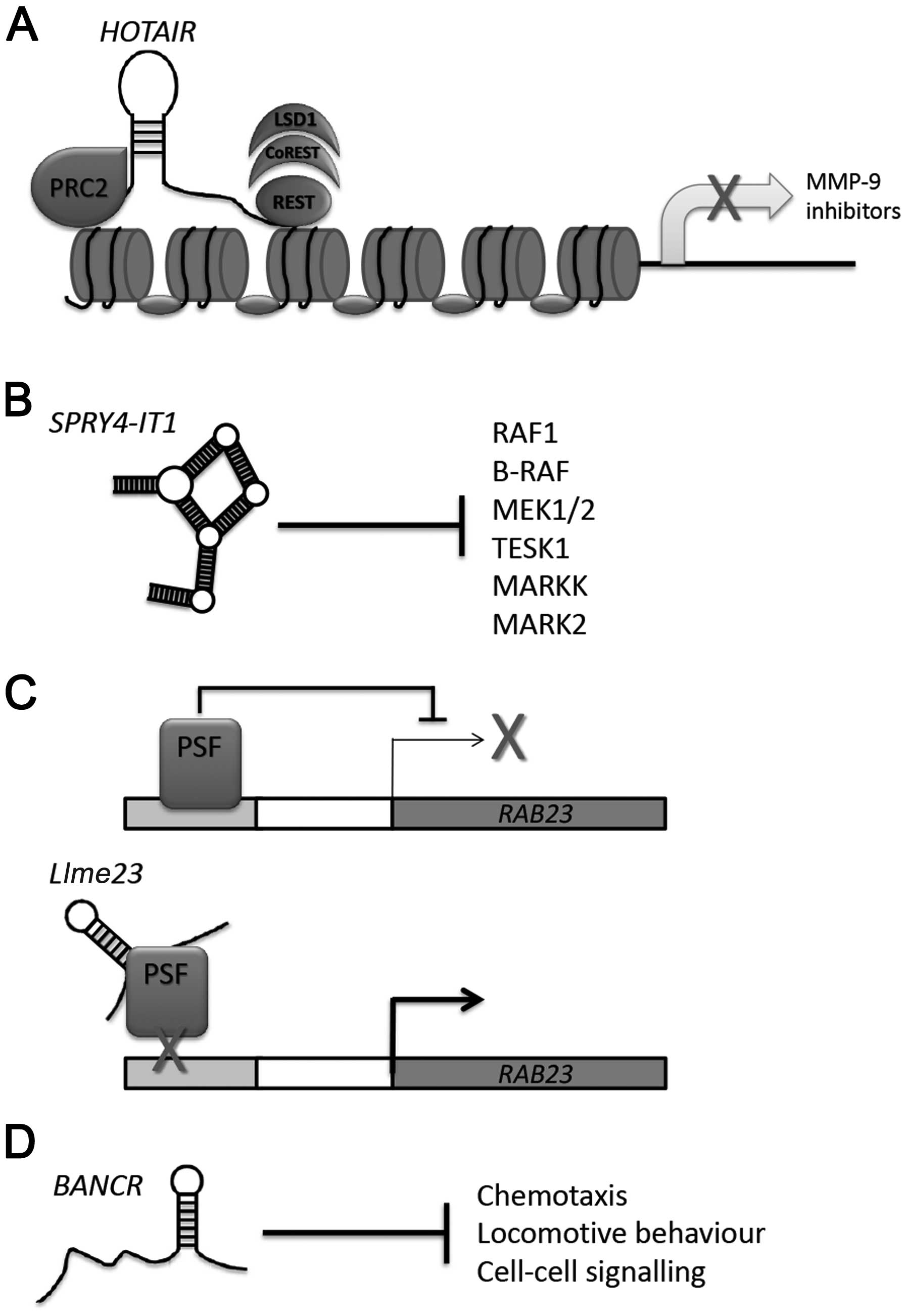
HOTAIR
HOTAIR is a member of an exclusive, but growing,
family of lncRNAs that have comparatively well-defined cellular
functions, in this case the epigenetic regulation of gene
expression. HOTAIR is transcribed from the HOXC cluster and
interacts with the Polycomb repressive complex 2 (PRC2), which it
then trafficks to the HOXD cluster, facilitating transcriptional
silencing of this 40 kb region (27). In addition to regulating the
expression of this specific genomic region, HOTAIR also appears to
regulate the gene expression at hundreds of other genomic locations
via an interaction with the LSD1/CoREST/REST complex, which enables
recruitment of PRC2 and LSD1 to chromatin for coupled histone H3K27
methylation and K4 demethylation (32). With such a pivotal role in
epigenetic modification and transcriptional activity, it is not
surprising that HOTAIR expression is deregulated in numerous
cancers, including breast, lung, colorectal, pancreatic, ovarian
hepatocellular carcinoma and gastrointestinal stromal tumours
(GIST) (33–42), indeed HOTAIR has been shown to
reprogram the cancer epigenome in breast cancer towards a
metastatic state, cementing its position as a molecule of huge
importance in cancer biology (43).
In order to determine if HOTAIR and other lncRNAs
known to be deregulated across a range of cancers are upregulated
in metastatic melanoma, Tang et al carried out quantitative
PCR (qPCR)-based expression profiling on matched primary melanoma
versus lymph node tumour samples from three patients (44). These analyses revealed two things:
i) HOTAIR expression is dramatically upregulated by ~100-fold in
metastatic versus primary melanoma; and ii) several other lncRNAs
(HULC, MALAT-1, MEG3, NEAT1 and UCA1), which are associated with
metastatic progression across a range of cancers were not
differently expressed in metastatic melanoma, suggesting that
lncRNAs contribute to metastasis in a cancer-specific manner
(44). Moreover, it appears that
elevated HOTAIR expression is a determining factor for the
metastatic state, at least in vitro, with siRNA-mediated
depletion of HOTAIR in the metastatic melanoma cell line, A375,
resulting in significant decreases in both cell motility and
invasion. These properties underpin metastasis, which involves the
destruction of the basement membrane and migration of the tumour
cells into the connective tissues before spreading to the lymph
nodes and distal sites. The basement membrane is largely comprised
of type IV collagen, which is degraded by matrix metalloproteinases
(MMPs), specifically MMP-2 and MMP-9 (45). Intriguingly, in situ
zymography assays carried out by Tang et al (44) show that depletion of HOTAIR in A375
cells resulted in reduced activity of MMP-2 and MMP-9, suggesting a
possible mechanism for HOTAIR-mediated metastasis (Fig. 1A), a theory supported by recent
data indicating that MMP gene expression is heavily regulated at
the epigenetic level (46).
Considering HOTAIR’s association with epigenetic
reprogramming and given the potential use of epigenetic drugs that
target MMPs to treat melanoma (47), there is an urgent need to develop a
keener understanding of such mechanisms.
SPRY4-IT1
HOTAIR appears destined to be involved in the
aetiology of the majority of cancers, melanoma included. However,
it was not the first lncRNA to be associated with melanoma, this
accolade goes to the SPRY4-IT1 transcript, a lncRNA that is
derived from an intron of the SPRY4 gene (48). In contrast to HOTAIR, very
little is known about the cellular function of SPRY4-IT1,
which was reported to be differentially expressed in melanoma by
Khaitan et al (48). The
authors utilised a microarray approach to determine differences in
lncRNA expression between the stage III melanoma cell line,
WM1552C, and control melanocytes. Alongside this, patient samples
isolated from stage I–IV disease were also assessed and results
aligned with cell line data to identify lncRNA transcripts that
were differentially expressed in both experimental systems. This
careful approach, validated by NGS, identified four lncRNA
transcripts, three of which were disregarded due to their location
in the 3′ UTR of protein-coding genes and concerns over the
technical difficulties this would present for downstream analysis,
the remaining transcript, which also displayed the greatest
differential expression (~12-fold), was SPRY4-IT1 (48).
In addition to increased expression in melanoma, the
SPRY4-IT1 lncRNA is also of interest due to its predicted
secondary structure. Analysis of SPRY4-IT1 using RNAfold and
RNAstructure (49,50) revealed several putative regulatory
motifs, including three nested helices and two ‘pyknons’ motifs
(48). The presence of pyknons
(non-random, genome-wide motifs) is particularly intriguing as it
has been suggested that these elements play a role in
post-transcriptional gene silencing (51), indicating that SPRY4-IT1 may
directly affect gene expression, something yet to be confirmed
experimentally. Depletion of SPRY4-IT1 in A375 cells using
siRNA and scramble controls resulted in decreased metabolic
viability and increased apoptosis via MTT and Annexin V assay,
respectively. Moreover, SPRY4-IT1-depleted A375 cells
exhibited significant reduction in both cell invasion and cell
motility when compared with scramble siRNA-transfected control.
Importantly, each of these phenotypic changes can be ascribed to
reduced SPRY4-IT1, as knockdown of the lncRNA did not alter
the expression of its ‘host’ gene SPRY4. Furthermore,
overexpression of SPRY4-IT1 in LOX-IMV1, a metastatic
melanoma cell line shown to express only low-levels of
SPRY4-IT1, significantly increased cell motility in a wound
healing assay (48).
A defined cellular function for SPRY4-IT1
remains elusive, however, it has recently been linked with the
aetiology of preeclampsia and oesophageal squamous cell carcinoma
(52,53). One possible clue may reside in the
geographical location of SPRY4-IT1 within the intronic
sequence of the SRYY4 gene, a property that may indicate a
biological function that is linked to that of the ‘host’ gene
(54). SPRY4 is a member of
the Sprouty family of Ras/ERK inhibitors proteins that prevent the
formation of active GTP-RAS and are therefore considered to be
putative tumour suppressors (55,56).
Clearly, SPRY4-IT1 is not operating as a tumour suppressor
in melanoma; however, it may manipulate similar intrinsic cellular
pathways, such as Ras/ERK (Fig.
1B). Future experiments, such as investigating the effect of
Ras/ERK inhibitors on increased cellular motility following
SPRY4-IT1 overexpression will likely prove informative and
are of clinical importance given emerging targeted therapies for
melanoma that focus on MAPK-signalling (57).
Llme23
Thus far, we have considered lncRNAs identified due
to their differential expression in metastatic melanoma. The lncRNA
Llme23 differs in this respect as while it appears to be
exclusively expressed in melanoma, it was first described using
assays designed to identify functional lncRNA partners to the
polypyrimidine tract-binding protein-associated splicing factor
(PSF) (58). The PSF protein is a
somewhat intriguing molecule that was originally thought to operate
as a splicing factor, having been identified in spliceosomal
extracts (59). However,
subsequent analysis has revealed a tumour-suppressor function for
this protein. Study carried out in mouse models demonstrate that
PSF is able to regulate the transcriptional activity in multiple
proto-oncogenes via its DNA-binding domain (DBD), which interacts
with the regulatory regions of these targets, repressing their
expression (60).
Fascinatingly, this tumour-suppressor function is
eradicated by the binding of a mouse retrotransposon lncRNA,
VL30-1, to the RNA-binding domain motifs present in PSF.
Moreover, retroviral transmission of VL30-1 to human
melanoma cells promoted metastatic progression in immunocompromised
mice, suggesting a possible role for PSF in the aetiology of
metastatic melanoma (61,62). While attention-grabbing, the
absence of a human homologue of the mouse VL30-1 lncRNA
raised doubts regarding the existence of a similar intrinsic
mechanism in humans. However, recent study by Wu et al
describes a human lncRNA that interacts directly with PSF in order
to drive melanoma tumour formation (58). Here an RNA-SELEX approach was
utilised to enrich for human RNAs that bind PSF and via subsequent
cDNA library construction and electromobility gel-shift assays, a
1,600 nt lncRNA, termed Llme23, was identified that binds
directly to PSF. These in vitro studies were also confirmed
in vivo by RNA-immunoprecipitations and downstream
functional experiments confirmed that Llme23 binding to PSF
inactivated PSF-mediated repression of RAB23, confirming
that the human Llme23 lncRNA also inhibits PSF’s
tumour-suppressor function (Fig.
1C), as is the case with the murine lncRNA VL30-1 and
PSF in the mouse model system (61).
How then is Llme23 impacting on melanoma aetiology?
Data investigating the effects of Llme23 overexpression and
depletion is more limited than for HOTAIR and
SPRY4-IT1, however, Llme23-depleted YUSAC cells displayed a
significant decrease in their ability to form colonies in soft-agar
and most significantly, these same cells displayed ~75% decrease in
tumour volume at day 38 post-injection into nude mice (58). Taking into consideration the
emerging literature around PSF and cancer (63,64),
it will be extremely interesting to observe the delineation of
Llme23 function in the literature, which to date is
restricted to the article discussed here.
BANCR
As discussed above, derailment of the Ras/ERK MAPK
signalling cascade is extremely common in metastatic melanoma. Our
understanding of the molecular events that underpin such
deregulation took a conceptual leap forward in 2002 with the
discovery of mutations in the v-RAF murine oncogene homologue B
(BRAF) proto-oncogene (65). The
significance of this finding is linked to the prevalence of BRAF
mutations in cutaneous melanoma, with >50% of tumours harbouring
mutations in BRAF and crucially >90% of these cases possessing
the same BRAFV600E substitution (66,67).
This realisation resulted in a focused effort to develop
BRAFV600E-specific inhibitors and has resulted in the
development and release of two drugs, vemurafenib and dabrafenib,
which received approval by the FDA for the treatment of
BRAFV600E mutant melanoma in 2011 and 2013,
respectively. Unfortunately, despite excellent response rates of
~50%, the vast majority of patients treated with these drugs
relapse and progress to chemoresistant disease that is generally
fatal. A number of mechanistic explanations have been identified to
explain this relapse, with the majority linked to the reactivation
of aberrant MARK-signalling, although other pathways (insulin
growth factor receptor and platelet-derived growth factor receptor)
have also been implicated (68).
Clearly, there is an urgent need to better understand how oncogenic
BRAF interacts with the cellular machinery to impact disease.
Flockhart et al set out with just this goal
in mind as outlined in their recent article, which describes the
use of RNA-seq on primary human melanocytes transduced with
lentivirus expressing either BRAFV600E or red
fluorescent protein control in order to specifically identify
differentially expressed lncRNAs (69). This approach represents an
important shift away from other studies, which have generally
focused on lncRNAs that are differentially expressed in cancer,
without first addressing the cause of the observed differential
expression and whether it is driven by oncogenic events or an
artefact of tumour heterogeneity and genomic instability.
In addition to determining global transcriptomic
changes in primary melanocytes expressing BRAFV600E,
RNA-seq data were granted more clinical credence by concurrently
analysing BRAFV600E-positive melanoma tissue samples.
Indeed, results were processed through a rigorous study-flow that
utilised cross-referencing with a publically available melanoma
RNA-seq data set (70) and
interrogation of the ENCODE RNA-seq data in order to validate that
identified transcripts are actively transcribed in melanocytes.
Finally, transcripts of interest were analysed using a previously
described coding potential calculator (CPC) algorithm that
discriminates coding from non-coding transcripts (71). This combinatorial approach
identified a novel lncRNA that is overexpressed in
BRAFV600E-positive melanocytes and melanoma, which the
authors termed BRAF-activated non-coding RNA (BANCR)
(69).
Functional experiments depleting BANCR in
melanoma cells did not result in decreased viability and
proliferation. However, consistent with HOTAIR,
SPRY4-IT1 and Llme23, depletion of BANCR did
significantly reduce melanoma cell motility. Gene expression
profiling carried out on Colo829 BRAFV600E-depleted
melanoma cells using a cDNA microarray revealed that 88 genes were
differentially expressed compared with control and significantly
genes involved in cell motility were overrepresented among this
number (Fig. 1D) (69). In order to gain some functional
insight into how BANCR might be regulating cell migration,
targets that displayed altered gene expression in
BANCR-depleted cells and are associated with cell motility
were investigated further. Interestingly, the chemokine CXCL11, the
expression of which appears to be positively regulated by
BANCR, was able to rescue the reduced cell motility
phenotype observed in BANCR-depleted melanoma cells. These
data suggest a scenario where the BRAFV600E mutation
induces overexpression of BANCR, which in turn then
positively regulates expression of CXCL11 in order to promote cell
migration. This observation may be of clinical importance as
activation of the chemokine receptor, CXCR3, has been linked to
lymph node metastasis in melanoma (72).
3. Conclusions
Over the past decade, the explosion of research on
non-coding transcripts has left us in little doubt that our genomes
exhibit great transcriptional complexity. However, our
understanding of how this complexity links to function, if at all,
remains far less assured. Clearly, lncRNAs are key players in
cancer progression and exhibit huge potential as biomarkers and
novel therapeutic targets for treatment. Perhaps the biggest
challenge facing researchers is determining which of the many
thousands of transcripts are truly functional. Classical single
‘gene’ experimental analysis remains central to this process, but
with improved read-length technologies for NGS on the horizon it is
likely that such studies will benefit greatly from improved
functional annotation. A second challenge relates to our
understanding of how lncRNA structure can be analysed and used to
predict function and accurately identify target genes via
bioinformatics. Certainly there is a long and somewhat uncharted
road ahead. Flockhart et al (69), identified an additional 38
annotated lncRNA transcripts, in addition to BANCR, that are
regulated by BRAFV600E and expressed in melanoma and
similar reservoirs likely remain untapped from other melanoma
RNA-seq data sets. Furthermore, the array of regulatory mechanisms
that impact on non-coding RNA function continues to grow.
Methyl-6-adenosine modification of RNA transcripts was recently
shown to be a reversible event that is thought to regulate mRNA and
lncRNA stability and has ushered yet another area of gene
expression research, RNA epigenetics (73). While the challenges are numerous,
the rewards are significant, characterisation of functional lncRNAs
and their modes of action will provide exciting opportunities to
augment and improve melanoma diagnosis, prognostic monitoring and
targeted therapies.
References
|
1
|
Weyers W: The ‘epidemic’ of melanoma
between under- and overdiagnosis. J Cutan Pathol. 39:9–16.
2012.
|
|
2
|
Lomas J, Martin-Duque P, Pons M and
Quintanilla M: The genetics of malignant melanoma. Front Biosci.
13:5071–5093. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hill VK, Gartner JJ, Samuels Y and
Goldstein AM: The genetics of melanoma: recent advances. Annu Rev
Genomics Hum Genet. 14:257–279. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bougnoux AC and Solassol J: The
contribution of proteomics to the identification of biomarkers for
cutaneous malignant melanoma. Clin Biochem. 46:518–523. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
ENCODE Project Consortium. An integrated
encyclopedia of DNA elements in the human genome. Nature.
489:57–74. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Clark MB, Amaral PP, Schlesinger FJ, et
al: The reality of pervasive transcription. PLoS Biol.
9:e1000625discussion e1001102. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Crick F: Central dogma of molecular
biology. Nature. 227:561–563. 1970. View
Article : Google Scholar
|
|
8
|
Graur D, Zheng Y, Price N, Azevedo RB,
Zufall RA and Elhaik E: On the immortality of television sets:
‘function’ in the human genome according to the evolution-free
gospel of ENCODE. Genome Biol Evol. 5:578–590. 2013.PubMed/NCBI
|
|
9
|
Doolittle WF: Is junk DNA bunk? A critique
of ENCODE. Proc Natl Acad Sci USA. 110:5294–5300. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tessitore A, Cicciarelli G, Del Vecchio F,
et al: microRNAs in the DNA damage/repair network and cancer. Int J
Genomics. 2014:8202482014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Condorelli G, Latronico MV and Cavarretta
E: microRNAs in cardiovascular diseases: current knowledge and the
road ahead. J Am Coll Cardiol. 63:2177–2187. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yin KJ, Hamblin M and Chen YE: Non-coding
RNAs in cerebral endothelial pathophysiology: emerging roles in
stroke. Neurochem Int. April 3–2014.(Epub ahead of print).
|
|
13
|
Lukiw WJ and Alexandrov PN: Regulation of
complement factor H (CFH) by multiple miRNAs in Alzheimer’s disease
(AD) brain. Mol Neurobiol. 46:11–19. 2012.PubMed/NCBI
|
|
14
|
Filatova EV, Alieva AKh, Shadrina MI and
Slominsky PA: MicroRNAs: possible role in pathogenesis of
Parkinson’s disease. Biochemistry (Mosc). 77:813–819.
2012.PubMed/NCBI
|
|
15
|
Völler D, Ott C and Bosserhoff A:
MicroRNAs in malignant melanoma. Clin Biochem. 46:909–917.
2013.
|
|
16
|
Dar AA, Majid S, de Semir D, Nosrati M,
Bezrookove V and Kashani-Sabet M: miRNA-205 suppresses melanoma
cell proliferation and induces senescence via regulation of E2F1
protein. J Biol Chem. 286:16606–16614. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Braig S, Mueller DW, Rothhammer T and
Bosserhoff AK: MicroRNA miR-196a is a central regulator of HOX-B7
and BMP4 expression in malignant melanoma. Cell Mol Life Sci.
67:3535–3548. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mueller DW and Bosserhoff AK: MicroRNA
miR-196a controls melanoma-associated genes by regulating HOX-C8
expression. Int J Cancer. 129:1064–1074. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Levati L, Pagani E, Romani S, et al:
MicroRNA-155 targets the SKI gene in human melanoma cell lines.
Pigment Cell Melanoma Res. 24:538–550. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Deng Y, Deng H, Bi F, et al: MicroRNA-137
targets carboxyl-terminal binding protein 1 in melanoma cell lines.
Int J Biol Sci. 7:133–137. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bemis LT, Chen R, Amato CM, et al:
MicroRNA-137 targets microphthalmia-associated transcription factor
in melanoma cell lines. Cancer Res. 68:1362–1368. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Segura MF, Hanniford D, Menendez S, et al:
Aberrant miR-182 expression promotes melanoma metastasis by
repressing FOXO3 and microphthalmia-associated transcription
factor. Proc Natl Acad Sci USA. 106:1814–1819. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Haflidadóttir BS, Bergsteinsdóttir K,
Praetorius C and Steingrímsson E: miR-148 regulates Mitf in
melanoma cells. PLoS One. 5:e115742010.PubMed/NCBI
|
|
24
|
Karreth FA, Tay Y, Perna D, et al: In vivo
identification of tumor-suppressive PTEN ceRNAs in an oncogenic
BRAF-induced mouse model of melanoma. Cell. 147:382–395. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Volders PJ, Helsens K, Wang X, et al:
LNCipedia: a database for annotated human lncRNA transcript
sequences and structures. Nucleic Acids Res. 41:D246–D251. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Brockdorff N, Ashworth A, Kay GF, et al:
The product of the mouse Xist gene is a 15 kb inactive X-specific
transcript containing no conserved ORF and located in the nucleus.
Cell. 71:515–526. 1992. View Article : Google Scholar
|
|
27
|
Rinn JL, Kertesz M, Wang JK, et al:
Functional demarcation of active and silent chromatin domains in
human HOX loci by noncoding RNAs. Cell. 129:1311–1323. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Balch CM, Gershenwald JE, Soong SJ, et al:
Final version of 2009 AJCC melanoma staging and classification. J
Clin Oncol. 27:6199–6206. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Dickson PV and Gershenwald JE: Staging and
prognosis of cutaneous melanoma. Surg Oncol Clin N Am. 20:1–17.
2011. View Article : Google Scholar
|
|
30
|
Zhang H, Chen Z, Wang X, Huang Z, He Z and
Chen Y: Long non-coding RNA: a new player in cancer. J Hematol
Oncol. 6:372013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cheetham SW, Gruhl F, Mattick JS and
Dinger ME: Long noncoding RNAs and the genetics of cancer. Br J
Cancer. 108:2419–2425. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tsai MC, Manor O, Wan Y, et al: Long
noncoding RNA as modular scaffold of histone modification
complexes. Science. 329:689–693. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wu ZH, Wang XL, Tang HM, et al: Long
non-coding RNA HOTAIR is a powerful predictor of metastasis and
poor prognosis and is associated with epithelial-mesenchymal
transition in colon cancer. Oncol Rep. 32:395–402. 2014.PubMed/NCBI
|
|
34
|
Sørensen KP, Thomassen M, Tan Q, et al:
Long non-coding RNA HOTAIR is an independent prognostic marker of
metastasis in estrogen receptor-positive primary breast cancer.
Breast Cancer Res Treat. 142:529–536. 2013.PubMed/NCBI
|
|
35
|
Qiu JJ, Lin YY, Ye LC, et al:
Overexpression of long non-coding RNA HOTAIR predicts poor patient
prognosis and promotes tumor metastasis in epithelial ovarian
cancer. Gynecol Oncol. 134:121–128. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Niinuma T, Suzuki H, Nojima M, et al:
Upregulation of miR-196a and HOTAIR drive malignant character in
gastrointestinal stromal tumors. Cancer Res. 72:1126–1136. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu XH, Liu ZL, Sun M, Liu J, Wang ZX and
De W: The long non-coding RNA HOTAIR indicates a poor prognosis and
promotes metastasis in non-small cell lung cancer. BMC Cancer.
13:4642013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kogo R, Shimamura T, Mimori K, et al: Long
noncoding RNA HOTAIR regulates polycomb-dependent chromatin
modification and is associated with poor prognosis in colorectal
cancers. Cancer Res. 71:6320–6326. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kim K, Jutooru I, Chadalapaka G, et al:
HOTAIR is a negative prognostic factor and exhibits pro-oncogenic
activity in pancreatic cancer. Oncogene. 32:1616–1625. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Geng YJ, Xie SL, Li Q, Ma J and Wang GY:
Large intervening non-coding RNA HOTAIR is associated with
hepatocellular carcinoma progression. J Int Med Res. 39:2119–2128.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ding C, Cheng S, Yang Z, et al: Long
non-coding RNA HOTAIR promotes cell migration and invasion via
down-regulation of RNA binding motif protein 38 in hepatocellular
carcinoma cells. Int J Mol Sci. 15:4060–4076. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Cui L, Xie XY, Wang H, Chen XL, Liu SL and
Hu LN: Expression of long non-coding RNA HOTAIR mRNA in ovarian
cancer. Sichuan Da Xue Xue Bao Yi Xue Ban. 44:57–59. 2013.(In
Chinese).
|
|
43
|
Gupta RA, Shah N, Wang KC, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Tang L, Zhang W, Su B and Yu B: Long
noncoding RNA HOTAIR is associated with motility, invasion, and
metastatic potential of metastatic melanoma. Biomed Res Int.
2013:2510982013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Watanabe H: Extracellular matrix -
regulation of cancer invasion and metastasis. Gan To Kagaku Ryoho.
37:2058–2061. 2010.(In Japanese).
|
|
46
|
Labrie M and St-Pierre Y: Epigenetic
regulation of mmp-9 gene expression. Cell Mol Life Sci.
70:3109–3124. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Frank A, David V, Aurelie TR, Florent G,
William H and Philippe B: Regulation of MMPs during melanoma
progression: from genetic to epigenetic. Anticancer Agents Med
Chem. 12:773–782. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Khaitan D, Dinger ME, Mazar J, et al:
Themelanoma-upregulated long noncoding RNA SPRY4-IT1 modulates
apoptosis and invasion. Cancer Res. 71:3852–3862. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Reuter JS and Mathews DH: RNAstructure:
software for RNA secondary structure prediction and analysis. BMC
Bioinformatics. 11:1292010. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Hofacker IL: RNA secondary structure
analysis using the Vienna RNA package. Curr Protoc Bioinformatics.
Chapter 12(Unit 12): 22004.
|
|
51
|
Rigoutsos I, Huynh T, Miranda K, Tsirigos
A, McHardy A and Platt D: Short blocks from the noncoding parts of
the human genome have instances within nearly all known genes and
relate to biological processes. Proc Natl Acad Sci USA.
103:6605–6610. 2006. View Article : Google Scholar
|
|
52
|
Zou Y, Jiang Z, Yu X, et al: Upregulation
of long noncoding RNA SPRY4-IT1 modulates proliferation, migration,
apoptosis, and network formation in trophoblast cells HTR-8SV/neo.
PLoS One. 8:e795982013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Xie HW, Wu QQ, Zhu B, et al: Long
noncoding RNA SPRY4-IT1 is upregulated in esophageal squamous cell
carcinoma and associated with poor prognosis. Tumour Biol. May
9–2014.(Epub ahead of print).
|
|
54
|
Guil S and Esteller M: Cis-acting
noncoding RNAs: friends and foes. Nat Struct Mol Biol.
19:1068–1075. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Tennis MA, Van Scoyk MM, Freeman SV,
Vandervest KM, Nemenoff RA and Winn RA: Sprouty-4 inhibits
transformed cell growth, migration and invasion, and
epithelial-mesenchymal transition, and is regulated by Wnt7A
through PPARgamma in non-small cell lung cancer. Mol Cancer Res.
8:833–843. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Leeksma OC, Van Achterberg TA, Tsumura Y,
et al: Human sprouty 4, a new ras antagonist on 5q31, interacts
with the dual specificity kinase TESK1. Eur J Biochem.
269:2546–2556. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Russo A, Ficili B, Candido S, et al:
Emerging targeted therapies for melanoma treatment (review). Int J
Oncol. 45:516–524. 2014.PubMed/NCBI
|
|
58
|
Wu CF, Tan GH, Ma CC and Li L: The
non-coding RNA llme23 drives the malignant property of human
melanoma cells. J Genet Genomics. 40:179–188. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Patton JG, Porro EB, Galceran J, Tempst P
and Nadal-Ginard B: Cloning and characterization of PSF, a novel
pre-mRNA splicing factor. Genes Dev. 7:393–406. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Song X, Sun Y and Garen A: Roles of PSF
protein and VL30 RNA in reversible gene regulation. Proc Natl Acad
Sci USA. 102:12189–12193. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wang G, Cui Y, Zhang G, Garen A and Song
X: Regulation of proto-oncogene transcription, cell proliferation,
and tumori-genesis in mice by PSF protein and a VL30 noncoding RNA.
Proc Natl Acad Sci USA. 106:16794–16798. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Song X, Wang B, Bromberg M, Hu Z,
Konigsberg W and Garen A: Retroviral-mediated transmission of a
mouse VL30 RNA to human melanoma cells promotes metastasis in an
immunodeficient mouse model. Proc Natl Acad Sci USA. 99:6269–6273.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Tsukahara T, Matsuda Y and Haniu H: PSF
knockdown enhances apoptosis via downregulation of LC3B in human
colon cancer cells. Biomed Res Int. 2013:2049732013. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Ren S, She M, Li M, et al: The
RNA/DNA-binding protein PSF relocates to cell membrane and
contributes cells’ sensitivity to antitumor drug, doxorubicin.
Cytometry A. 85:231–241. 2014.PubMed/NCBI
|
|
65
|
Davies H, Bignell GR, Cox C, et al:
Mutations of the BRAF gene in human cancer. Nature. 417:949–954.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Flaherty KT and McArthur G: BRAF, a target
in melanoma: implications for solid tumor drug development. Cancer.
116:4902–4913. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Bamford S, Dawson E, Forbes S, et al: The
COSMIC (Catalogue of Somatic Mutations in Cancer) database and
website. Br J Cancer. 91:355–358. 2004.PubMed/NCBI
|
|
68
|
Jarkowski A 3rd and Khushalani NI: BRAF
and beyond: tailoring strategies for the individual melanoma
patient. J Carcinog. 13:12014. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Flockhart RJ, Webster DE, Qu K, et al:
BRAFV600E remodels the melanocyte transcriptome and induces BANCR
to regulate melanoma cell migration. Genome Res. 22:1006–1014.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Berger MF, Levin JZ, Vijayendran K, et al:
Integrative analysis of the melanoma transcriptome. Genome Res.
20:413–427. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Kong L, Zhang Y, Ye ZQ, et al: CPC: assess
the protein-coding potential of transcripts using sequence features
and support vector machine. Nucleic Acids Res. 35:W345–W349. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Kawada K, Sonoshita M, Sakashita H, et al:
Pivotal role of CXCR3 in melanoma cell metastasis to lymph nodes.
Cancer Res. 64:4010–4017. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Fu Y, Dominissini D, Rechavi G and He C:
Gene expression regulation mediated through reversible
m6A RNA methylation. Nat Rev Genet. 15:293–306. 2014.
View Article : Google Scholar : PubMed/NCBI
|