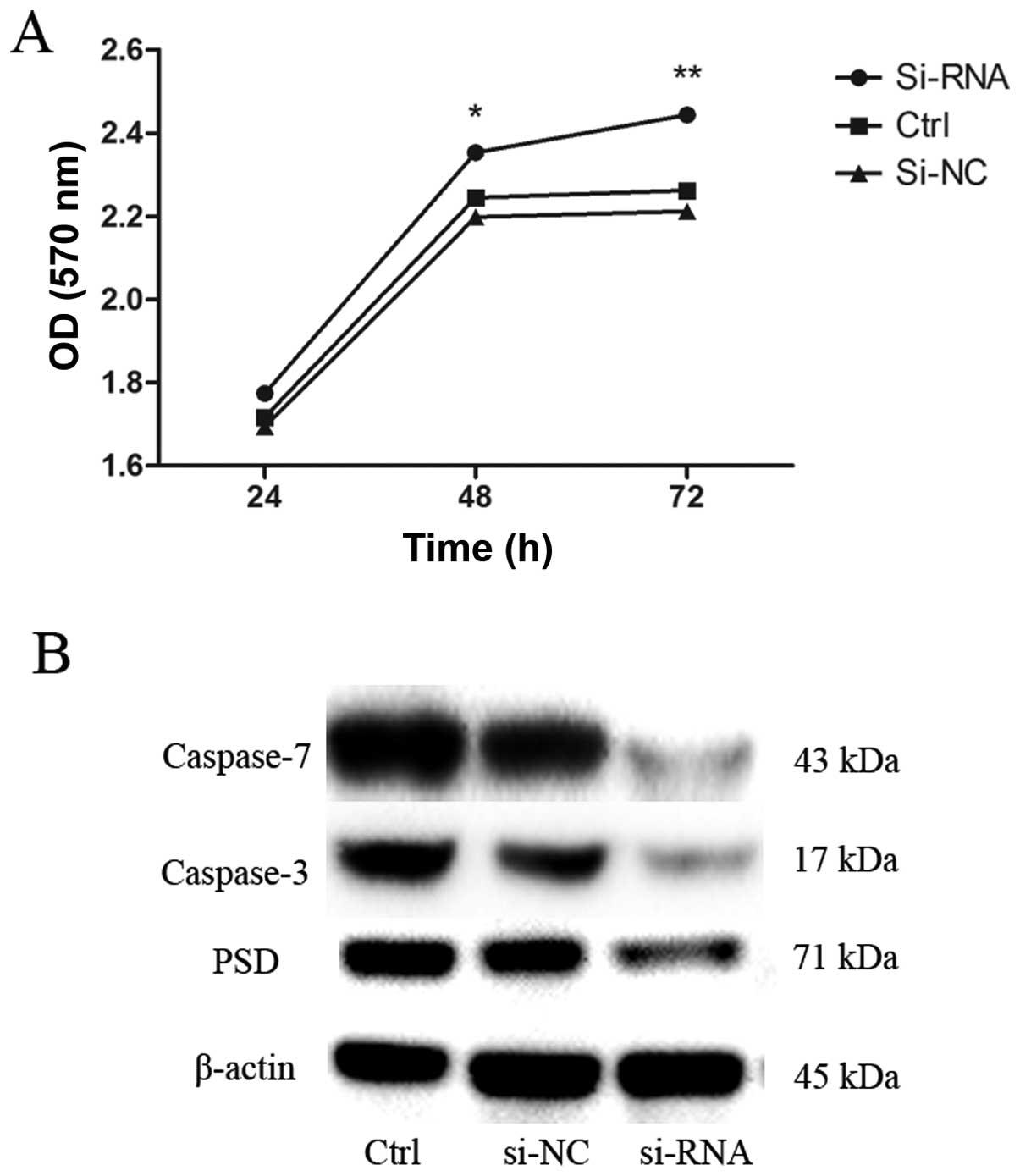
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
2
|
Park YS, Jin MY, Kim YJ, Yook JH, Kim BS
and Jang SJ: The global histone modificaton pattern correlates with
cancer recurrence and overall survival in gastric adenocarcinoma.
Ann Surg Oncol. 15:1968–1976. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Meng CF, Zhu XJ, Peng G and Dai DQ: Role
of histone modifications and DNA methylation in the regulation of
O6-methylguanine-DNA methyltransferase gene expression
in human stomach cancer cells. Cancer Invest. 28:331–339. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kim M, Jang HR, Kim JH, et al: Epigenetic
inactivation of protein kinase D1 in gastric cancer andits role in
gastric cancer cell migration and invasion. Carcinogenesis.
29:629–637. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jaenisch R and Bird A: Epigenetic
regulation of gene expression: how the genome integrates intrinsic
and environmental signals. Nat Genet. 33:245–254. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sharma S, Kelly TK and Jones PA:
Epigenetics in cancer. Carcinogenesis. 31:27–36. 2010. View Article : Google Scholar
|
|
7
|
Nan X, Ng HH, Johnson CA, Laherty CD,
Turner BM, Eisenman RN and Bird A: Transcriptional repression by
the methyl-CpG-binding protein MeCP2 involves a histone deacetylase
complex. Nature. 393:386–389. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hendrich B and Bird A: Identification and
characterization of a family of mammalian methyl-CpG binding
proteins. Mol Cell Biol. 18:6538–6547. 1998.PubMed/NCBI
|
|
9
|
Bird A: DNA methylation patterns and
epigenetic memory. Genes Dev. 16:6–21. 2002. View Article : Google Scholar
|
|
10
|
Tariq M, Saze H, Probst AV, Lichota J,
Habu Y and Paszkowski J: Erasure of CpG methylation in arabidopsis
alters patterns of histone H3 methylation in heterochromatin. Proc
Natl Acad Sci USA. 100:8823–8827. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Espada J, Ballestar E, Fraga MF, et al:
Human DNA methyltransferase 1 is required for maintenance of the
histone H3 modification pattern. J Biol Chem. 279:37175–37184.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Meng CF, Zhu XJ, Peng G and Dai DQ:
Promoter histone H3 lysine 9 di-methylation is associated with DNA
methylation and aberrant expression of p16 in gastric cancer cells.
Oncol Rep. 22:1221–1227. 2009.PubMed/NCBI
|
|
13
|
Kondo Y, Shen L, Yan PS, Huang TH and Issa
JP: Chromatin immunoprecipitation microarrays for identification of
genes silenced by histone H3 lysine 9 methylation. Proc Natl Acad
Sci USA. 101:7398–7403. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rubinek T1, Shulman M, Israeli S, et al:
Epigenetic silencing of the tumor suppressor klotho in human breast
cancer. Breast Cancer Res Treat. 133:649–657. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gulbins E, Coggeshall KM, Brenner B,
Schlottmann K, Linderkamp O and Lang F: Fas-induced apoptosis is
mediated by activation of a Ras and Rac protein-regulated signaling
pathway. J Biol Chem. 271:26389–26394. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Esteve P, Embade N, Perona R, et al:
Rho-regulated signals induce apoptosis in vitro and in vivo by a
p53-independent, but Bcl2 dependent pathway. Oncogene.
17:1855–1869. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Perletti L, Talarico D, Trecca D,
Ronchetti D and Fracchiolla NS: Identification of a novel gene,
PSD, adjacent to NFKB2/lyt-10, which contains Sec7 and
pleckstrin-homologydomains. Genomics. 46:251–259. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Okada S, Suzuki K, Takaharu K, et al:
Aberrant methylation of the Pleckstrin and Sec7 domain-containing
gene is implicated in ulcerative colitis-associated carcinogenesis
throughits inhibitory effect on apoptosis. Int J Oncol. 40:686–694.
2012.
|
|
19
|
Fahrner JA, Eguchi S, Herman JG and Baylin
SB: Dependence of histone modifications and gene expression on DNA
hypermethylation in cancer. Cancer Res. 62:7213–7218.
2002.PubMed/NCBI
|
|
20
|
Cameron EE, Bachman KE, Myohane S, Herman
JG and Baylin SB: Synergy of demethylation and histone deacetylase
inhibition in the re-expression of genes silenced in cancer. Nat
Genet. 21:103–107. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li LC and Dahiya R: MethPrimer: designing
primers for methylation PCRs. Bioinformatics. 18:1427–1431. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fuks F: DNA methylation and histone
modifications: teaming up to silence genes. Curr Opin Genet Dev.
15:490–495. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wu J, Wang SH, Potter D, et al: Diverse
histone modifications on histone 3 lysine 9 andtheir relation to
DNA methylation in specifying gene silencing. BMC Genomics.
8:1312007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu J, Zhu X, Xu X and Dai D: DNA promoter
and histone H3 methylation downregulate NGX6 in gastric cancer
cells. Med Oncol. 31:8172014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kato T, Suzuki K, Okada S, et al: Aberrant
methylation of PSD disturbs. Rac1-mediated immune responses
governing neutrophil chemotaxis and apoptosis in ulcerative
colitis-associated carcinogenesis. Int J Oncol. 40:942–950.
2012.
|
|
26
|
Saez R, Chan AM, Miki T and Aaronson SA:
Oncogenic activation of human R-ras by point mutations analogous to
those of prototype H-ras oncogenes. Oncogene. 9:2977–2982.
1994.PubMed/NCBI
|
|
27
|
Nishida K, Kaziro Y and Satoh T:
Anti-apoptotic function of Rac in hematopoietic cells. Oncogene.
18:407–415. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Eom YW, Yoo MH, Woo CH, et al: Implication
of the small GTPase Rac1 in the apoptosis induced by UV in Rat-2
fibroblasts. Biochem Biophys Res Commun. 285:825–829. 2001.
View Article : Google Scholar : PubMed/NCBI
|