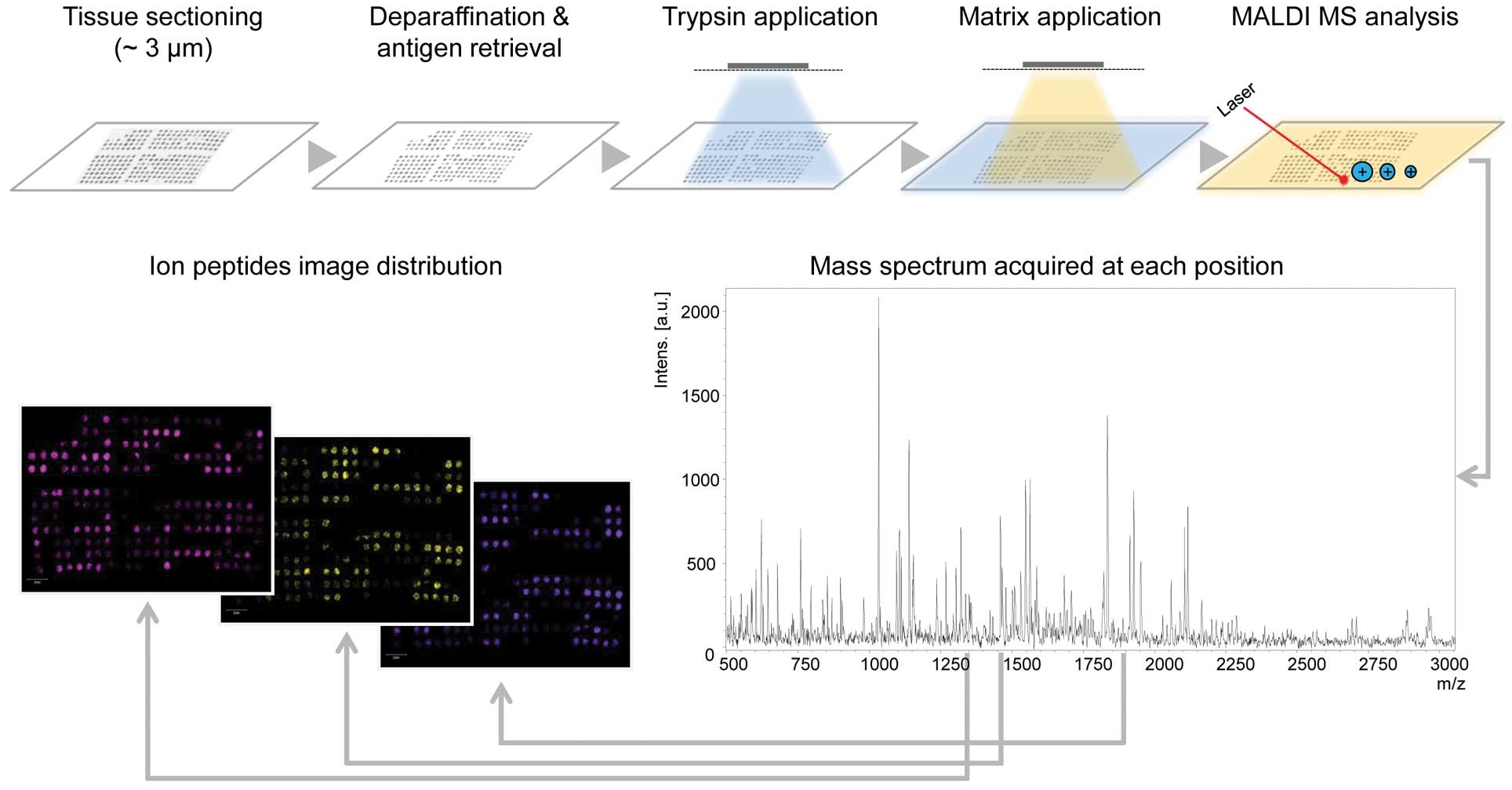
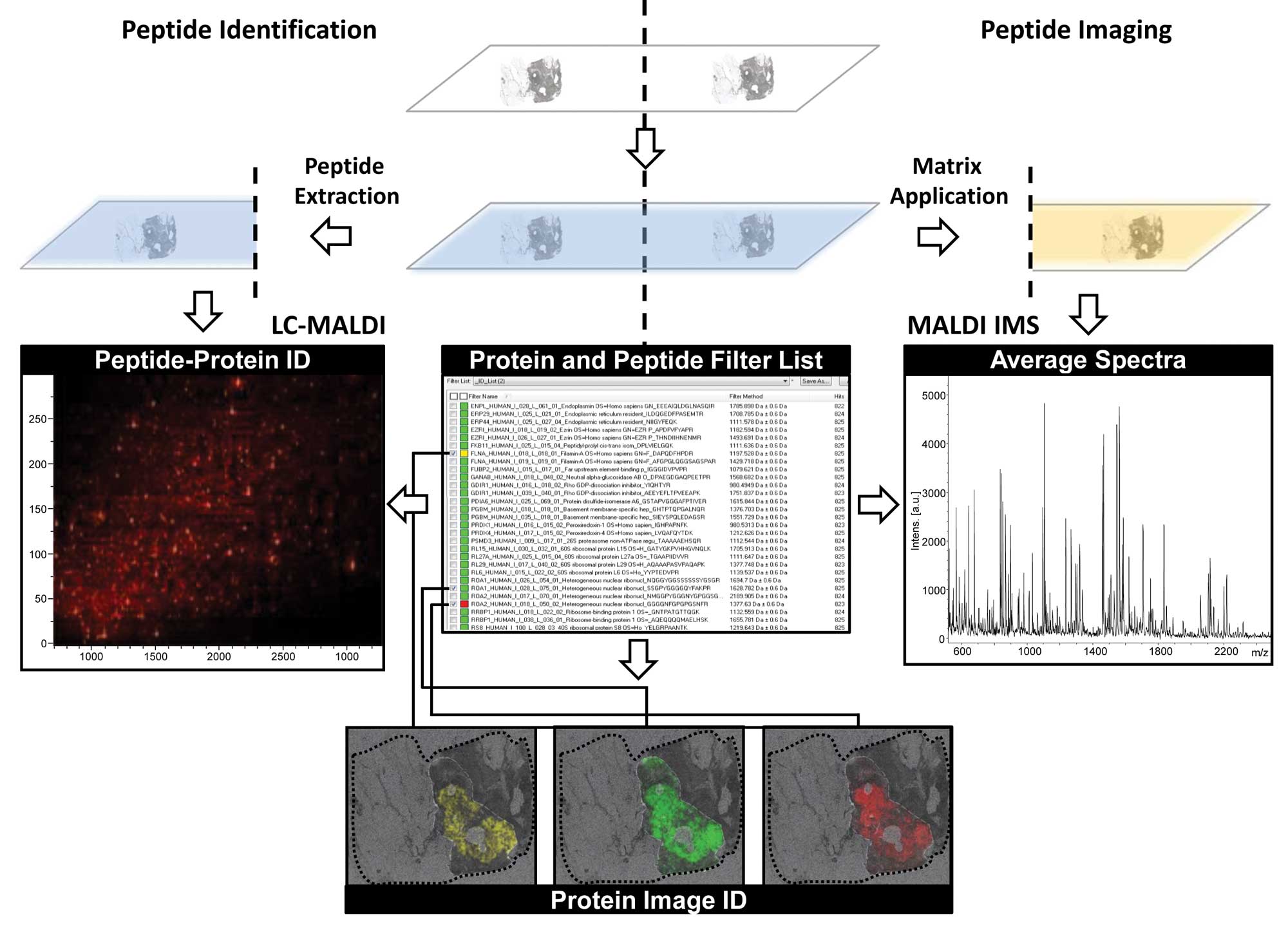
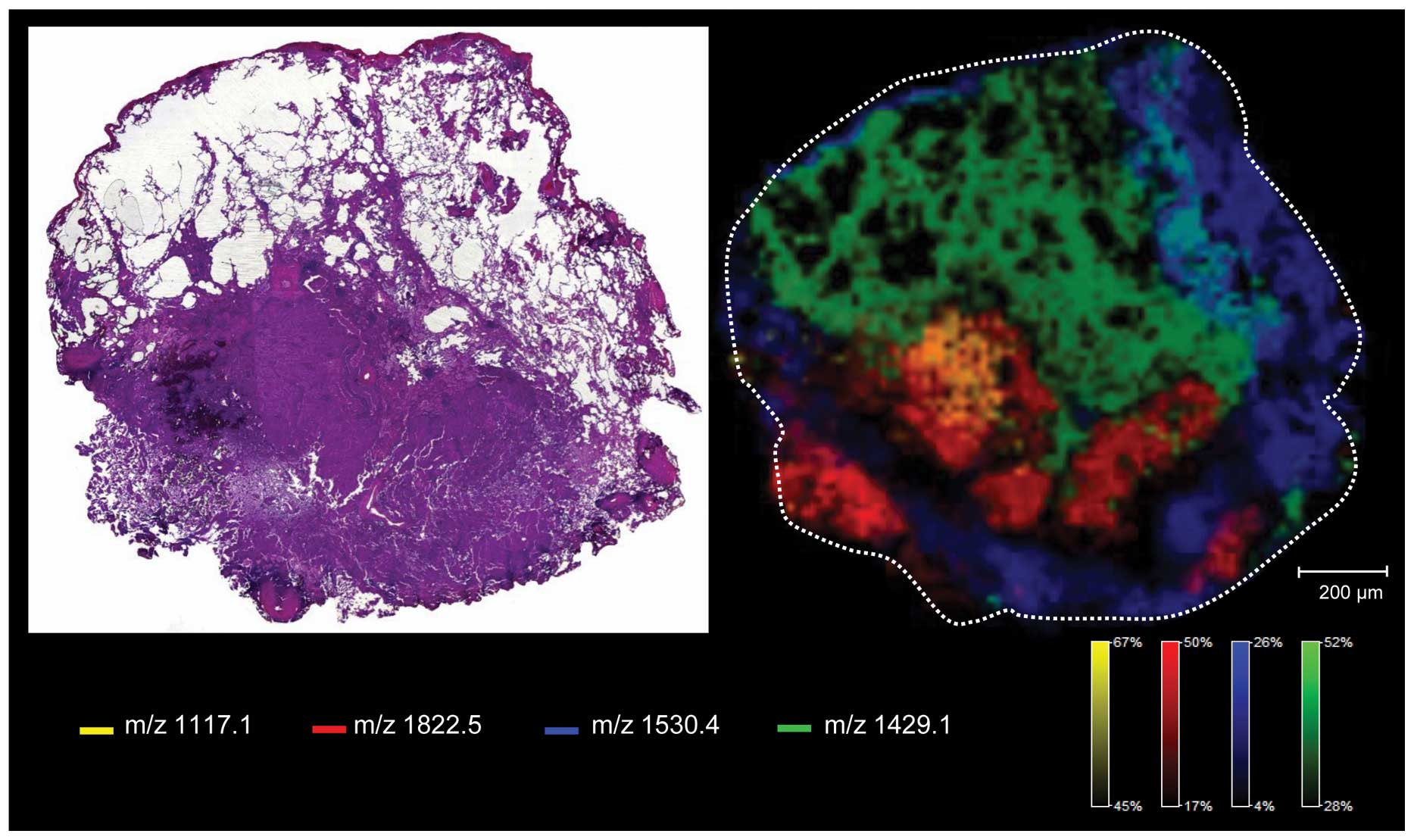
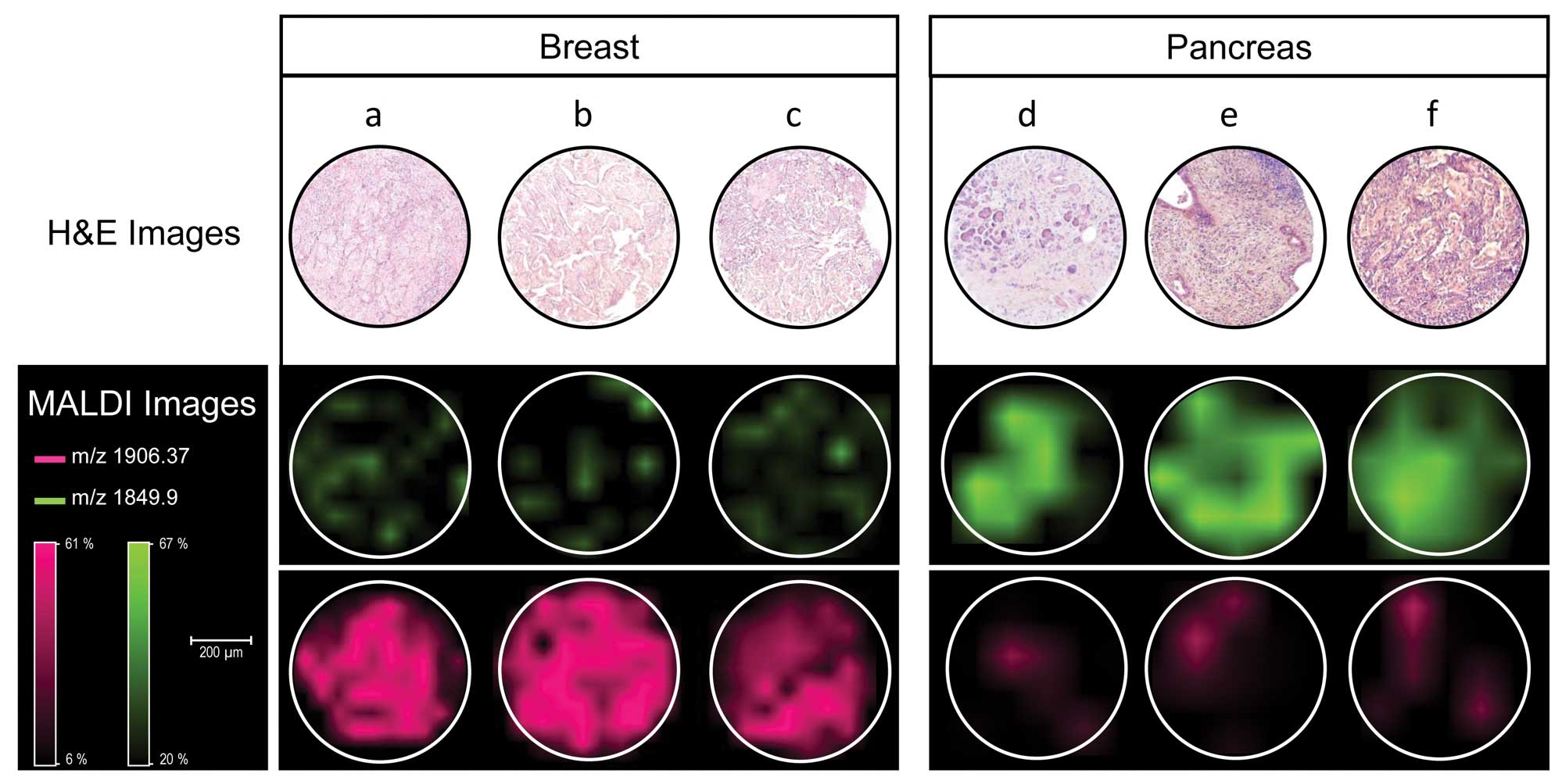
|
1
|
Patel R: MALDI-TOF mass spectrometry:
transformative proteomics for clinical microbiology. Clin Chem.
59:340–342. 2013. View Article : Google Scholar
|
|
2
|
Clark AE, Kaleta EJ, Arora A and Wolk DM:
Matrix-assisted laser desorption ionization-time of flight mass
spectrometry: a fundamental shift in the routine practice of
clinical microbiology. Clin Microbiol Rev. 26:547–603. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Moore JL, Caprioli RM and Skaar EP:
Advanced mass spectrometry technologies for the study of microbial
pathogenesis. Curr Opin Microbiol. 19C:45–51. 2014. View Article : Google Scholar
|
|
4
|
Kueger S, Steinhauser D, Willmitzer L and
Giavalisco P: High-resolution plant metabolomics: from mass
spectral features to metabolites and from whole-cell analysis to
subcellular metabolite distributions. Plant J. 70:39–50. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Schoenian I, Spiteller M, Ghaste M, Wirth
R, Herz H and Spiteller D: Chemical basis of the synergism and
antagonism in microbial communities in the nests of leaf-cutting
ants. Proc Natl Acad Sci USA. 108:1955–1960. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chaurand P, Cornett DS, Angel PM and
Caprioli RM: From whole-body sections down to cellular level,
multiscale imaging of phospholipids by MALDI mass spectrometry. Mol
Cell Proteomics. 10:O110.004259. 2011. View Article : Google Scholar :
|
|
7
|
Trim PJ, Henson CM, Avery JL, et al:
Matrix-assisted laser desorption/ionization-ion mobility
separation-mass spectrometry imaging of vinblastine in whole body
tissue sections. Anal Chem. 80:8628–8634. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Reyzer ML, Chaurand P, Angel PM and
Caprioli RM: Direct molecular analysis of whole-body animal tissue
sections by MALDI imaging mass spectrometry. Methods Mol Biol.
656:285–301. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Remmerbach TW, Maurer K, Janke S, et al:
Oral brush biopsy analysis by matrix assisted laser
desorption/ionisation-time of flight mass spectrometry profiling: a
pilot study. Oral Oncol. 47:278–281. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Schwamborn K, Krieg RC, Uhlig S, Ikenberg
H and Wellmann A: MALDI imaging as a specific diagnostic tool for
routine cervical cytology specimens. Int J Mol Med. 27:417–421.
2011.
|
|
11
|
Kriegsmann M, Seeley EH, Schwarting A, et
al: MALDI MS imaging as a powerful tool for investigating synovial
tissue. Scand J Rheumatol. 41:305–309. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Seeley EH and Caprioli RM: MALDI imaging
mass spectrometry of human tissue: method challenges and clinical
perspectives. Trends Biotechnol. 29:136–143. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Meding S, Nitsche U, Balluff B, et al:
Tumor classification of six common cancer types based on proteomic
profiling by MALDI imaging. J Proteome Res. 11:1996–2003. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nipp M, Elsner M, Balluff B, et al:
S100-A10, thioredoxin, and S100-A6 as biomarkers of papillary
thyroid carcinoma with lymph node metastasis identified by MALDI
imaging. J Mol Med. 90:163–174. 2012. View Article : Google Scholar
|
|
15
|
Steurer S, Seddiqi AS, Singer JM, et al:
MALDI imaging on tissue microarrays identifies molecular features
associated with renal cell cancer phenotype. Anticancer Res.
34:2255–2261. 2014.PubMed/NCBI
|
|
16
|
Morita Y, Ikegami K, Goto-Inoue N, et al:
Imaging mass spectrometry of gastric carcinoma in formalin-fixed
paraffin-embedded tissue microarray. Cancer Sci. 101:267–273. 2010.
View Article : Google Scholar
|
|
17
|
Gode D and Volmer DA: Lipid imaging by
mass spectrometry - a review. Analyst. 138:1289–1315. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Horn PJ and Chapman KD: Lipidomics in
situ: insights into plant lipid metabolism from high resolution
spatial maps of metabolites. Prog Lipid Res. 54:32–52. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sparvero LJ, Amoscato AA, Dixon CE, et al:
Mapping of phospholipids by MALDI imaging (MALDI-MSI): realities
and expectations. Chem Phys Lipids. 165:545–562. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Harvey DJ: Analysis of carbohydrates and
glycoconjugates by matrix-assisted laser desorption/ionization mass
spectrometry: An update for 2009–2010. Mass Spectrom Rev. May
26–2014. View Article : Google Scholar : (Epub ahead of
print).
|
|
21
|
Reyzer ML, Hsieh Y, Ng K, Korfmacher WA
and Caprioli RM: Direct analysis of drug candidates in tissue by
matrix-assisted laser desorption/ionization mass spectrometry. J
Mass Spectrom. 38:1081–1092. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Norris JL and Caprioli RM: Analysis of
tissue specimens by matrix-assisted laser desorption/ionization
imaging mass spectrometry in biological and clinical research. Chem
Rev. 113:2309–2342. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Groseclose MR and Castellino S: A mimetic
tissue model for the quantification of drug distributions by MALDI
imaging mass spectrometry. Anal Chem. 85:10099–10106. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Honisch C, Chen Y, Mortimer C, et al:
Automated comparative sequence analysis by base-specific cleavage
and mass spectrometry for nucleic acid-based microbial typing. Proc
Natl Acad Sci USA. 104:10649–10654. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Okamoto I, Sakai K, Morita S, et al:
Multiplex genomic profiling of non-small cell lung cancers from the
LETS phase III trial of first-line S-1/carboplatin versus
paclitaxel/carboplatin: results of a West Japan Oncology Group
study. Oncotarget. 5:2293–2304. 2014.PubMed/NCBI
|
|
26
|
Castellino S, Groseclose MR and Wagner D:
MALDI imaging mass spectrometry: bridging biology and chemistry in
drug development. Bioanalysis. 3:2427–2441. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Rompp A, Guenther S, Takats Z and Spengler
B: Mass spectrometry imaging with high resolution in mass and space
(HR(2) MSI) for reliable investigation of drug compound
distributions on the cellular level. Anal Bioanal Chem. 401:65–73.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Caprioli RM, Farmer TB and Gile J:
Molecular imaging of biological samples: localization of peptides
and proteins using MALDI-TOF MS. Anal Chem. 69:4751–4760. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Schone C, Hofler H and Walch A: MALDI
imaging mass spectrometry in cancer research: combining proteomic
profiling and histological evaluation. Clin Biochem. 46:539–545.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gessel MM, Norris JL and Caprioli RM:
MALDI imaging mass spectrometry: spatial molecular analysis to
enable a new age of discovery. J Proteomics. 107:71–82. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Thomas A, Lenglet S, Chaurand P, et al:
Mass spectrometry for the evaluation of cardiovascular diseases
based on proteomics and lipidomics. Thromb Haemost. 106:20–33.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
McClure RA, Chumbley CW, Reyzer ML, et al:
Identification of promethazine as an amyloid-binding molecule using
a fluorescence high-throughput assay and MALDI imaging mass
spectrometry. Neuroimage Clin. 2:620–629. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kriegsmann M, Casadonte R, Randau T, et
al: MALDI imaging of predictive ferritin, fibrinogen and proteases
in haemophilic arthropathy. Haemophilia. 20:446–453. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Seeley EH and Caprioli RM: Molecular
imaging of proteins in tissues by mass spectrometry. Proc Natl Acad
Sci USA. 105:18126–18131. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Seeley EH, Washington MK, Caprioli RM and
M’Koma AE: Proteomic patterns of colonic mucosal tissues delineate
Crohn’s colitis and ulcerative colitis. Proteomics Clin Appl.
7:541–549. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lalowski M, Magni F, Mainini V, et al:
Imaging mass spectrometry: a new tool for kidney disease
investigations. Nephrol Dial Transplant. 28:1648–1656. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Mainini V, Pagni F, Ferrario F, et al:
MALDI imaging mass spectrometry in glomerulonephritis: feasibility
study. Histopathology. 64:901–906. 2014. View Article : Google Scholar
|
|
38
|
Xu BJ, Shyr Y, Liang X, et al: Proteomic
patterns and prediction of glomerulosclerosis and its mechanisms. J
Am Soc Nephrol. 16:2967–2975. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lagarrigue M, Becker M, Lavigne R, et al:
Revisiting rat spermatogenesis with MALDI imaging at 20-microm
resolution. Mol Cell Proteomic. 10:M110.005991. 2011. View Article : Google Scholar
|
|
40
|
Willems SM, van Remoortere A, van Zeijl R,
Deelder AM, McDonnell LA and Hogendoorn PC: Imaging mass
spectrometry of myxoid sarcomas identifies proteins and lipids
specific to tumour type and grade, and reveals biochemical
intratumour heterogeneity. J Pathol. 222:400–409. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Schwamborn K, Krieg RC, Reska M, Jakse G,
Knuechel R and Wellmann A: Identifying prostate carcinoma by
MALDI-Imaging. Int J Mol Med. 20:155–159. 2007.PubMed/NCBI
|
|
42
|
Rodrigo MA, Zitka O, Krizkova S, Moulick
A, Adam V and Kizek R: MALDI-TOF MS as evolving cancer diagnostic
tool: a review. J Pharm Biomed Anal. 95:245–255. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Rahman SM, Gonzalez AL, Li M, et al: Lung
cancer diagnosis from proteomic analysis of preinvasive lesions.
Cancer Res. 71:3009–3017. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Oppenheimer SR, Mi D, Sanders ME and
Caprioli RM: Molecular analysis of tumor margins by MALDI mass
spectrometry in renal carcinoma. J Proteome Res. 9:2182–2190. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Jones EE, Powers TW, Neely BA, et al:
MALDI imaging mass spectrometry profiling of proteins and lipids in
clear cell renal cell carcinoma. Proteomics. 14:924–935. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Angel PM and Caprioli RM: Matrix-assisted
laser desorption ionization imaging mass spectrometry: in situ
molecular mapping. Biochemistry. 52:3818–3828. 2013. View Article : Google Scholar
|
|
47
|
Boggio KJ, Obasuyi E, Sugino K, Nelson SB,
Agar NY and Agar JN: Recent advances in single-cell MALDI mass
spectrometry imaging and potential clinical impact. Expert Rev
Proteomics. 8:591–604. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Eriksson C, Masaki N, Yao I, Hayasaka T
and Setou M: MALDI imaging mass spectrometry-A mini review of
methods and recent developments. Mass Spectrom. 2:S00222013.
View Article : Google Scholar
|
|
49
|
Dreisewerd K: Recent methodological
advances in MALDI mass spectrometry. Anal Bioanal Chem.
406:2261–2278. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Fujimura Y and Miura D: MALDI mass
spectrometry imaging for visualizing in situ metabolism of
endogenous metabolites and dietary phytochemicals. Metabolites.
4:319–346. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Gorzolka K and Walch A: MALDI mass
spectrometry imaging of formalin-fixed paraffin-embedded tissues in
clinical research. Histol Histopathol. 29:1365–1376.
2014.PubMed/NCBI
|
|
52
|
Schwamborn K and Caprioli RM: Molecular
imaging by mass spectrometry: looking beyond classical histology.
Nat Rev Cancer. 10:639–646. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Chaurand P: Imaging mass spectrometry of
thin tissue sections: a decade of collective efforts. J Proteomics.
75:4883–4892. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Rompp A and Spengler B: Mass spectrometry
imaging with high resolution in mass and space. Histochem Cell
Biol. 139:759–783. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Goodwin RJ: Sample preparation for mass
spectrometry imaging: small mistakes can lead to big consequences.
J Proteomics. 75:4893–4911. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Kaletas BK, van der Wiel IM, Stauber J, et
al: Sample preparation issues for tissue imaging by imaging MS.
Proteomics. 9:2622–2633. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Stoeckli M, Chaurand P, Hallahan DE and
Caprioli RM: Imaging mass spectrometry: a new technology for the
analysis of protein expression in mammalian tissues. Nat Med.
7:493–496. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
58
|
Cornett DS, Mobley JA, Dias EC, et al: A
novel histology-directed strategy for MALDI-MS tissue profiling
that improves throughput and cellular specificity in human breast
cancer. Mol Cell Proteomics. 5:1975–1983. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Seeley EH, Schwamborn K and Caprioli RM:
Imaging of intact tissue sections: moving beyond the microscope. J
Biol Chem. 286:25459–25466. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Zavalin A, Yang J and Caprioli R: Laser
beam filtration for high spatial resolution MALDI imaging mass
spectrometry. J Am Soc Mass Spectrom. 24:1153–1156. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Caprioli RM: Imaging mass spectrometry:
molecular microscopy for enabling a new age of discovery.
Proteomics. 14:807–809. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Norris JL, Cornett DS, Mobley JA, et al:
Processing MALDI Mass Spectra to improve mass spectral direct
tissue analysis. Int J Mass Spectrom. 260:212–221. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Djidja MC, Claude E, Snel MF, et al: Novel
molecular tumour classification using MALDI-mass spectrometry
imaging of tissue microarray. Anal Bioanal Chem. 397:587–601. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Andersson M, Groseclose MR, Deutch AY and
Caprioli RM: Imaging mass spectrometry of proteins and peptides: 3D
volume reconstruction. Nat Methods. 5:101–108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Crecelius AC, Cornett DS, Caprioli RM,
Williams B, Dawant BM and Bodenheimer B: Three-dimensional
visualization of protein expression in mouse brain structures using
imaging mass spectrometry. J Am Soc Mass Spectrom. 16:1093–1099.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Pagni F, Mainini V, Garancini M, et al:
Proteomics for the diagnosis of thyroid lesions: preliminary
report. Cytopathology. Jul 20–2014. View Article : Google Scholar : (Epub ahead of
print).
|
|
67
|
Groseclose MR, Massion PP, Chaurand P and
Caprioli RM: High-throughput proteomic analysis of formalin-fixed
paraffin-embedded tissue microarrays using MALDI imaging mass
spectrometry. Proteomics. 8:3715–3724. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Gancberg D, Jarvinen T, di Leo A, et al:
Evaluation of HER-2/NEU protein expression in breast cancer by
immunohistochemistry: an interlaboratory study assessing the
reproducibility of HER-2/NEU testing. Breast Cancer Res Treat.
74:113–120. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Thaysen-Andersen M, Wilkinson BL, Payne RJ
and Packer NH: Site-specific characterisation of densely
O-glycosylated mucin-type peptides using electron transfer
dissociation ESI-MS/MS. Electrophoresis. 32:3536–3545. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Ronci M, Sharma S, Chataway T, et al:
MALDI-MS-imaging of whole human lens capsule. J Proteome Res.
10:3522–3529. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Bona A, Papai Z, Maasz G, et al: Mass
spectrometric identification of ancient proteins as potential
molecular biomarkers for a 2000-year-old osteogenic sarcoma. PLoS
One. 9:e872152014. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Morgan TM, Seeley EH, Fadare O, Caprioli
RM and Clark PE: Imaging the clear cell renal cell carcinoma
proteome. J Urol. 189:1097–1103. 2013. View Article : Google Scholar :
|
|
73
|
Dekker TJ, Balluff BD, Jones EA, et al:
Multicenter Matrix-assisted laser desorption/ionization mass
spectrometry imaging (MALDI MSI) identifies proteomic differences
in breast-cancer-associated stroma. J Proteome Res. May
2–2014.(Epub ahead of print). View Article : Google Scholar
|
|
74
|
Rauser S, Marquardt C, Balluff B, et al:
Classification of HER2 receptor status in breast cancer tissues by
MALDI imaging mass spectrometry. J Proteome Res. 9:1854–1863. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Le Faouder J, Laouirem S, Chapelle M, et
al: Imaging mass spectrometry provides fingerprints for
distinguishing hepatocellular carcinoma from cirrhosis. J Proteome
Res. 10:3755–3765. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Yanagisawa K, Shyr Y, Xu BJ, et al:
Proteomic patterns of tumour subsets in non-small-cell lung cancer.
Lancet. 362:433–439. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Gruner BM, Hahne H, Mazur PK, et al: MALDI
imaging mass spectrometry for in situ proteomic analysis of
preneoplastic lesions in pancreatic cancer. PLoS One. 7:e394242012.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Balluff B, Rauser S, Meding S, et al:
MALDI imaging identifies prognostic seven-protein signature of
novel tissue markers in intestinal-type gastric cancer. Am J
Pathol. 179:2720–2729. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Kim HK, Reyzer ML, Choi IJ, et al: Gastric
cancer-specific protein profile identified using endoscopic biopsy
samples via MALDI mass spectrometry. J Proteome Res. 9:4123–4130.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Casadonte R and Caprioli RM: Proteomic
analysis of formalin-fixed paraffin-embedded tissue by MALDI
imaging mass spectrometry. Nat Protoc. 6:1695–1709. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Djidja MC, Chang J, Hadjiprocopis A, et
al: Identification of hypoxia-regulated proteins using MALDI-mass
spectrometry imaging combined with quantitative proteomics. J
Proteome Res. 13:2297–2313. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Steurer S, Borkowski C, Odinga S, et al:
MALDI mass spectrometric imaging based identification of clinically
relevant signals in prostate cancer using large-scale tissue
microarrays. Int J Cancer. 133:920–928. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Casadonte R, Kriegsmann M, Zweynert F, et
al: Imaging mass spectrometry to discriminate breast from
pancreatic cancer metastasis in formalin-fixed paraffin-embedded
tissues. Proteomics. 14:956–964. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Ikeda K, Monden T, Kanoh T, et al:
Extraction and analysis of diagnostically useful proteins from
formalin-fixed, paraffin-embedded tissue sections. J Histochem
Cytochem. 46:397–403. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Becker KF, Schott C, Hipp S, et al:
Quantitative protein analysis from formalin-fixed tissues:
implications for translational clinical research and nanoscale
molecular diagnosis. J Pathol. 211:370–378. 2007. View Article : Google Scholar
|
|
86
|
Chung JY, Lee SJ, Kris Y, Braunschweig T,
Traicoff JL and Hewitt SM: A well-based reverse-phase protein array
applicable to extracts from formalin-fixed paraffin-embedded
tissue. Proteomics Clin Appl. 2:1539–1547. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Nirmalan NJ, Harnden P, Selby PJ and Banks
RE: Development and validation of a novel protein extraction
methodology for quantitation of protein expression in
formalin-fixed paraffin-embedded tissues using western blotting. J
Pathol. 217:497–506. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Crockett DK, Lin Z, Vaughn CP, Lim MS and
Elenitoba-Johnson KS: Identification of proteins from
formalin-fixed paraffin-embedded cells by LC-MS/MS. Lab Invest.
85:1405–1415. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Hood BL, Darfler MM, Guiel TG, et al:
Proteomic analysis of formalin-fixed prostate cancer tissue. Mol
Cell Proteomics. 4:1741–1753. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Palmer-Toy DE, Krastins B, Sarracino DA,
Nadol JB Jr and Merchant SN: Efficient method for the proteomic
analysis of fixed and embedded tissues. J Proteome Res.
4:2404–2411. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Becker KF, Berg D, Malinowsky K, et al:
Update on protein analysis of fixed tissues. Pathologe. 31(Suppl
2): 263–267. 2010. View Article : Google Scholar
|
|
92
|
Shi SR, Liu C, Balgley BM, Lee C and
Taylor CR: Protein extraction from formalin-fixed,
paraffin-embedded tissue sections: quality evaluation by mass
spectrometry. J Histochem Cytochem. 54:739–743. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
McDonnell LA, Walch A, Stoeckli M and
Corthals GL: MSiMass list: a public database of identifications for
protein MALDI MS imaging. J Proteome Res. 13:1138–1142. 2014.
View Article : Google Scholar
|
|
94
|
Maier SK, Hahne H, Gholami AM, et al:
Comprehensive identification of proteins from MALDI imaging. Mol
Cell Proteomics. 12:2901–2910. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Deininger SO, Ebert MP, Futterer A,
Gerhard M and Rocken C: MALDI imaging combined with hierarchical
clustering as a new tool for the interpretation of complex human
cancers. J Proteome Res. 7:5230–5236. 2008. View Article : Google Scholar
|
|
96
|
Caprioli RM: Deciphering protein molecular
signatures in cancer tissues to aid in diagnosis, prognosis, and
therapy. Cancer Res. 65:10642–10645. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Chaurand P, Sanders ME, Jensen RA and
Caprioli RM: Proteomics in diagnostic pathology: profiling and
imaging proteins directly in tissue sections. Am J Pathol.
165:1057–1068. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Kang S, Shim HS, Lee JS, et al: Molecular
proteomics imaging of tumor interfaces by mass spectrometry. J
Proteome Res. 9:1157–1164. 2010. View Article : Google Scholar
|
|
99
|
Han EC, Lee YS, Liao WS, Liu YC, Liao HY
and Jeng LB: Direct tissue analysis by MALDI-TOF mass spectrometry
in human hepatocellular carcinoma. Clin Chim Acta. 412:230–239.
2011. View Article : Google Scholar
|
|
100
|
Eberlin LS, Norton I, Orringer D, et al:
Ambient mass spectrometry for the intraoperative molecular
diagnosis of human brain tumors. Proc Natl Acad Sci USA.
110:1611–1616. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Schwartz SA, Weil RJ, Thompson RC, et al:
Proteomic-based prognosis of brain tumor patients using
direct-tissue matrix-assisted laser desorption ionization mass
spectrometry. Cancer Res. 65:7674–7681. 2005.PubMed/NCBI
|
|
102
|
Lemaire R, Menguellet SA, Stauber J, et
al: Specific MALDI imaging and profiling for biomarker hunting and
validation: fragment of the 11S proteasome activator complex, Reg
alpha fragment, is a new potential ovary cancer biomarker. J
Proteome Res. 6:4127–4134. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Elsner M, Rauser S, Maier S, et al: MALDI
imaging mass spectrometry reveals COX7A2, TAGLN2 and S100-A10 as
novel prognostic markers in Barrett’s adenocarcinoma. J Proteomics.
75:4693–4704. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Flatley B, Malone P and Cramer R: MALDI
mass spectrometry in prostate cancer biomarker discovery. Biochim
Biophys Acta. 1844:940–949. 2014. View Article : Google Scholar
|
|
105
|
Hardesty WM, Kelley MC, Mi D, Low RL and
Caprioli RM: Protein signatures for survival and recurrence in
metastatic melanoma. J Proteomics. 74:1002–1014. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Oezdemir RF, Gaisa NT, Lindemann-Docter K,
et al: Proteomic tissue profiling for the improvement of grading of
noninvasive papillary urothelial neoplasia. Clin Biochem. 45:7–11.
2012. View Article : Google Scholar
|
|
107
|
Chaurand P, Schwartz SA, Billheimer D, Xu
BJ, Crecelius A and Caprioli RM: Integrating histology and imaging
mass spectrometry. Anal Chem. 76:1145–1155. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Schwartz SA, Weil RJ, Johnson MD, Toms SA
and Caprioli RM: Protein profiling in brain tumors using mass
spectrometry: feasibility of a new technique for the analysis of
protein expression. Clin Cancer Res. 10:981–987. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Choi JH, Shin NR, Moon HJ, et al:
Identification of S100A8 and S100A9 as negative regulators for
lymph node metastasis of gastric adenocarcinoma. Histol
Histopathol. 27:1439–1448. 2012.PubMed/NCBI
|
|
110
|
Reyzer ML, Caldwell RL, Dugger TC, et al:
Early changes in protein expression detected by mass spectrometry
predict tumor response to molecular therapeutics. Cancer Res.
64:9093–9100. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Vegvari A, Fehniger TE, Rezeli M, et al:
Experimental models to study drug distributions in tissue using
MALDI mass spectrometry imaging. J Proteome Res. 12:5626–5633.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Sugihara Y, Vegvari A, Welinder C, et al:
A new look at drugs targeting malignant melanoma - an application
for mass spectrometry imaging. Proteomics. 14:1963–1970. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Bianga J, Bouslimani A, Bec N, et al:
Complementarity of MALDI and LA ICP mass spectrometry for platinum
anticancer imaging in human tumor. Metallomics. 6:1382–1386. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Liu X, Ide JL, Norton I, et al: Molecular
imaging of drug transit through the blood-brain barrier with MALDI
mass spectrometry imaging. Sci Rep. 3:28592013.PubMed/NCBI
|
|
115
|
Buck A and Walch A: In situ drug and
metabolite analyzes in biological and clinical research by MALDI MS
imaging. Bioanalysis. 6:1241–1253. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Jirasko R, Holcapek M, Kunes M and Svatos
A: Distribution study of atorvastatin and its metabolites in rat
tissues using combined information from UHPLC/MS and
MALDI-Orbitrap-MS imaging. Anal Bioanal Chem. 406:4601–4610. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Huber K, Aichler M, Sun N, et al: A rapid
ex vivo tissue model for optimising drug detection and ionisation
in MALDI imaging studies. Histochem Cell Biol. 142:361–371. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Yasunaga M, Furuta M, Ogata K, et al: The
significance of microscopic mass spectrometry with high resolution
in the visualisation of drug distribution. Sci Rep. 3:30502013.
View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Salphati L, Shahidi-Latham S, Quiason C,
et al: Distribution of the phosphatidylinositol 3-kinase inhibitors
Pictilisib (GDC-0941) and GNE-317 in U87 and GS2 intracranial
glioblastoma models-assessment by matrix-assisted laser desorption
ionization imaging. Drug Metab Dispos. 42:1110–1116. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Zecchi R, Trevisani M, Pittelli M, et al:
Impact of drug administration route on drug delivery and
distribution into the lung: an imaging mass spectrometry approach.
Eur J Mass Spectrom. 19:475–482. 2013. View Article : Google Scholar
|
|
121
|
Bouamrani A, Ternier J, Ratel D, et al:
Direct-tissue SELDI-TOF mass spectrometry analysis: a new
application for clinical proteomics. Clin Chem. 52:2103–2106. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Agar NY, Malcolm JG, Mohan V, et al:
Imaging of meningioma progression by matrix-assisted laser
desorption ionization time-of-flight mass spectrometry. Anal Chem.
82:2621–2625. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Ronci M, Bonanno E, Colantoni A, et al:
Protein unlocking procedures of formalin-fixed paraffin-embedded
tissues: application to MALDI-TOF imaging MS investigations.
Proteomics. 8:3702–3714. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Qin XJ and Ling BX: Proteomic studies in
breast cancer (Review). Oncol Lett. 3:735–743. 2012.PubMed/NCBI
|
|
125
|
Seeley EH and Caprioli RM: Imaging mass
spectrometry: towards clinical diagnostics. Proteomics Clin Appl.
2:1435–1443. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Walch A, Rauser S, Deininger SO and Hofler
H: MALDI imaging mass spectrometry for direct tissue analysis: a
new frontier for molecular histology. Histochem Cell Biol.
130:421–434. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Amstalden van Hove ER, Blackwell TR,
Klinkert I, Eijkel GB, Heeren RM and Glunde K: Multimodal mass
spectrometric imaging of small molecules reveals distinct
spatio-molecular signatures in differentially metastatic breast
tumor models. Cancer Res. 70:9012–9021. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Chughtai K, Jiang L, Greenwood TR, Glunde
K and Heeren RM: Mass spectrometry images acylcarnitines,
phosphatidylcholines, and sphingomyelin in MDA-MB-231 breast tumor
models. J Lipid Res. 54:333–344. 2013. View Article : Google Scholar :
|
|
129
|
Patel SA, Barnes A, Loftus N, et al:
Imaging mass spectrometry using chemical inkjet printing reveals
differential protein expression in human oral squamous cell
carcinoma. Analyst. 134:301–307. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Thomas A, Patterson NH, Marcinkiewicz MM,
Lazaris A, Metrakos P and Chaurand P: Histology-driven data mining
of lipid signatures from multiple imaging mass spectrometry
analyses: application to human colorectal cancer liver metastasis
biopsies. Anal Chem. 85:2860–2866. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Rahman SM, Shyr Y, Yildiz PB, et al:
Proteomic patterns of preinvasive bronchial lesions. Am J Respir
Crit Care Med. 172:1556–1562. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Lee GK, Lee HS, Park YS, et al: Lipid
MALDI profile classifies non-small cell lung cancers according to
the histologic type. Lung Cancer. 76:197–203. 2012. View Article : Google Scholar
|
|
133
|
Hassanein M, Rahman JS, Chaurand P and
Massion PP: Advances in proteomic strategies toward the early
detection of lung cancer. Proc Am Thorac Soc. 8:183–188. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Schwamborn K, Krieg RC, Jirak P, et al:
Application of MALDI imaging for the diagnosis of classical Hodgkin
lymphoma. J Cancer Res Clin Oncol. 136:1651–1655. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Lazova R, Seeley EH, Keenan M, Gueorguieva
R and Caprioli RM: Imaging mass spectrometry - a new and promising
method to differentiate Spitz nevi from Spitzoid malignant
melanomas. Am J Dermatopathol. 34:82–90. 2012. View Article : Google Scholar :
|
|
136
|
Min KW, Bang JY, Kim KP, et al: Imaging
mass spectrometry in papillary thyroid carcinoma for the
identification and validation of biomarker proteins. J Korean Med
Sci. 29:934–940. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Junker H, Venz S, Zimmermann U, Thiele A,
Scharf C and Walther R: Stage-related alterations in renal cell
carcinoma-comprehensive quantitative analysis by 2D-DIGE and
protein network analysis. PLoS One. 6:e218672011. View Article : Google Scholar
|
|
138
|
Chinello C, Gianazza E, Zoppis I, et al:
Serum biomarkers of renal cell carcinoma assessed using a protein
profiling approach based on ClinProt technique. Urology.
75:842–847. 2010. View Article : Google Scholar
|
|
139
|
Axt J, Murphy AJ, Seeley EH, et al: Race
disparities in Wilms tumor incidence and biology. J Surg Res.
170:112–119. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Steurer S, Singer JM, Rink M, et al: MALDI
imaging-based identification of prognostically relevant signals in
bladder cancer using large-scale tissue microarrays. Urol Oncol.
Aug 14–2014. View Article : Google Scholar : (Epub ahead of
print).
|
|
141
|
Liu Y, Chen Y, Momin A, et al: Elevation
of sulfatides in ovarian cancer: an integrated transcriptomic and
lipidomic analysis including tissue-imaging mass spectrometry. Mol
Cancer. 9:1862010. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Meding S, Martin K, Gustafsson OJ, et al:
Tryptic peptide reference data sets for MALDI imaging mass
spectrometry on formalin-fixed ovarian cancer tissues. J Proteome
Res. 12:308–315. 2013. View Article : Google Scholar
|
|
143
|
Cazares LH, Troyer D, Mendrinos S, et al:
Imaging mass spectrometry of a specific fragment of
mitogen-activated protein kinase/extracellular signal-regulated
kinase kinase kinase 2 discriminates cancer from uninvolved
prostate tissue. Clin Cancer Res. 15:5541–5551. 2009. View Article : Google Scholar : PubMed/NCBI
|