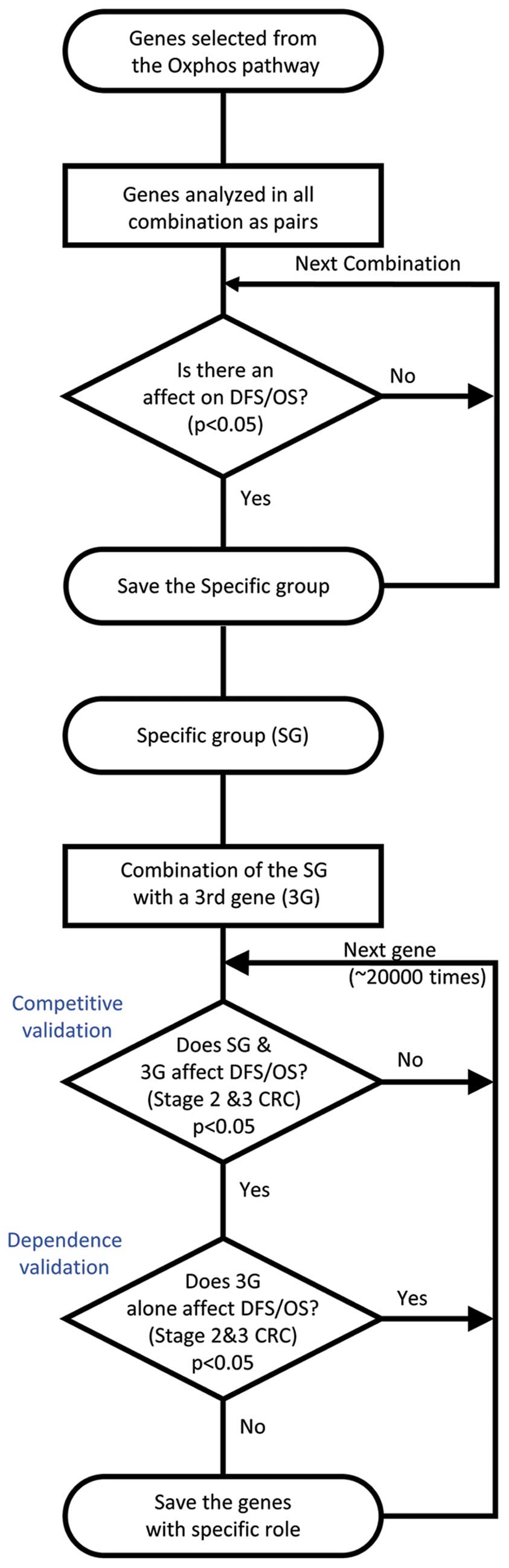
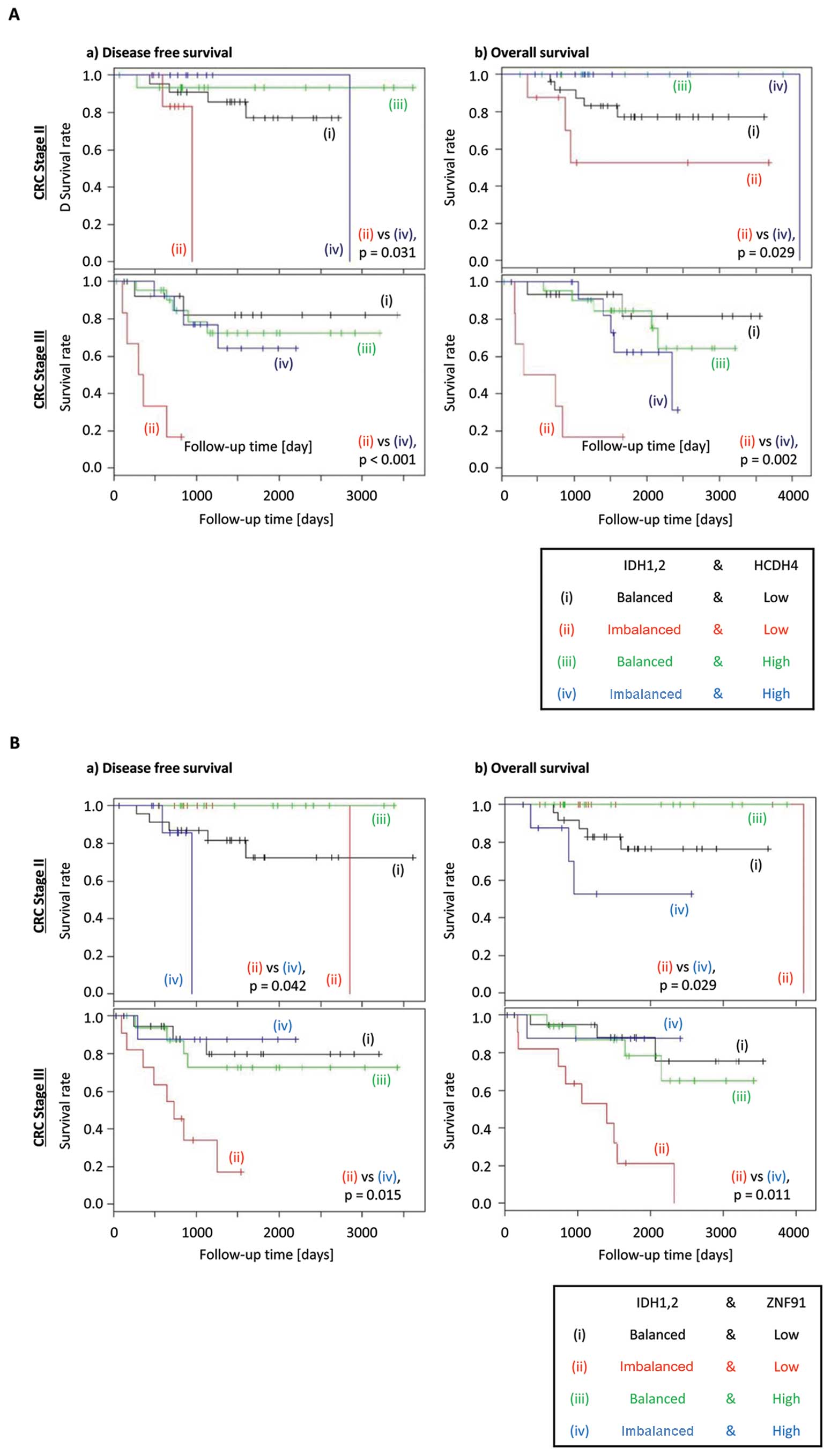
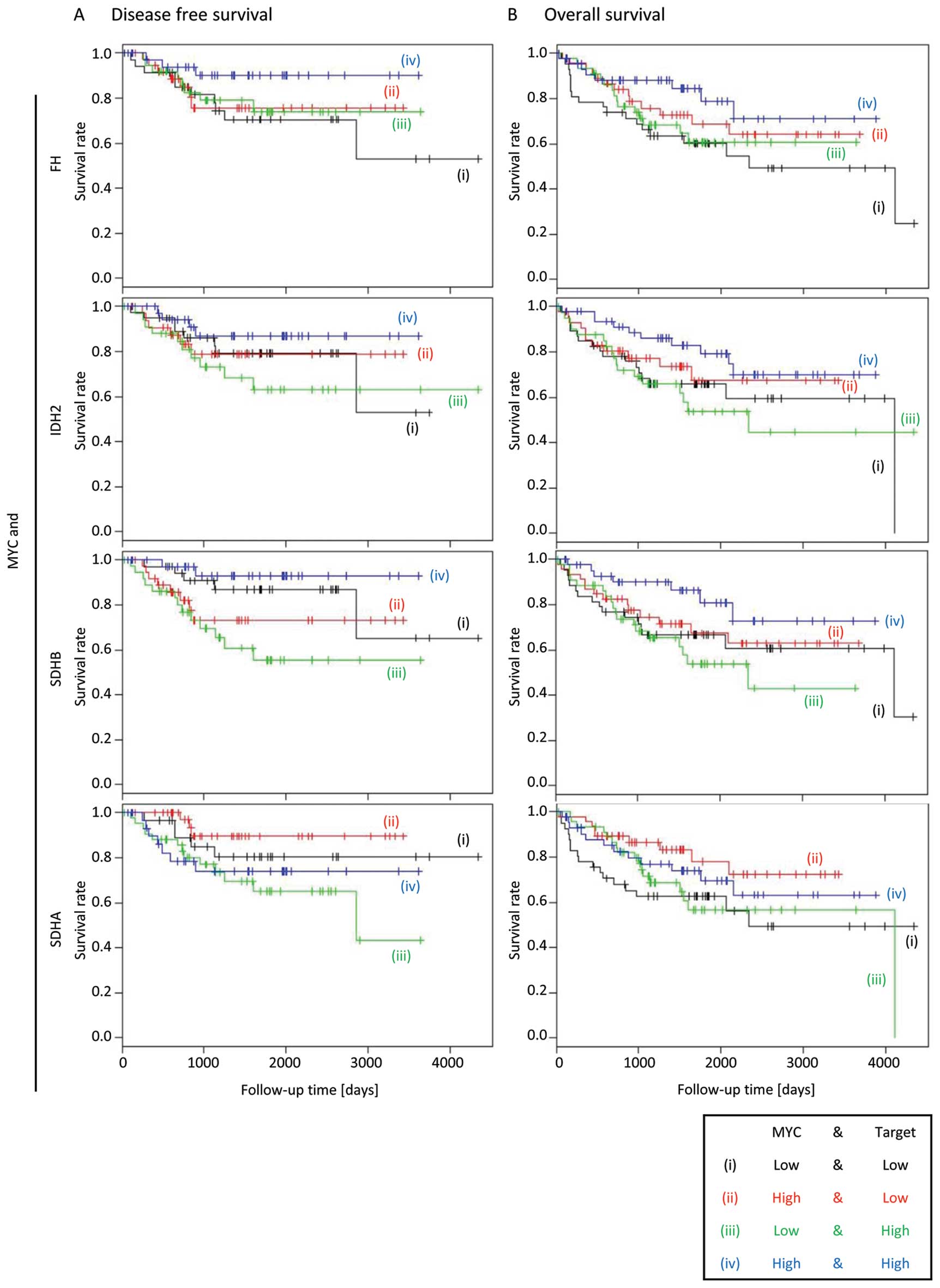
|
1
|
Chena M and Hofestädtb R: A medical
bioinformatics approach for metabolic disorders: Biomedical data
prediction, modeling, and systematic analysis. J Biomed Inform.
39:147–159. 2006. View Article : Google Scholar
|
|
2
|
Reya T, Morrison SJ, Clarke MF and
Weissman IL: Stem cells, cancer, and cancer stem cells. Nature.
414:105–111. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Meacham CE and Morrison SJ: Tumour
heterogeneity and cancer cell plasticity. Nature. 501:328–337.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nowell PC and Croce CM: Chromosomes,
genes, and cancer. Am J Pathol. 125:8–15. 1986.
|
|
5
|
Kouzarides T: Chromatin modifications and
their function. Cell. 128:693–705. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yaniv M: Chromatin remodeling: from
transcription to cancer. Cancer Genet. Mar 21–2014.(Epub ahead of
print). View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fujimoto A, Totoki Y, Abe T, et al:
Whole-genome sequencing of liver cancers identifies etiological
influences on mutation patterns and recurrent mutations in
chromatin regulators. Nat Genet. 44:760–764. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Parsons DW, Jones S, Zhang X, et al: An
integrated genomic analysis of human glioblastoma multiforme.
Science. 321:1807–1812. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mardis ER, Ding L, Dooling DJ, et al:
Recurring mutations found by sequencing an acute myeloid leukemia
genome. N Engl J Med. 361:1058–1066. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yan H, Parsons DW, Jin G, et al: IDH1 and
IDH2 mutations in gliomas. N Engl J Med. 360:765–773. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cairns RA and Mak TW: Oncogenic isocitrate
dehydrogenase mutations: mechanisms, models, and clinical
opportunities. Cancer Discov. 3:730–741. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dang L, White DW, Gross S, et al:
Cancer-associated IDH1 mutations produce 2-hydroxyglutarate.
Nature. 462:739–744. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ward PS, Patel J, Wise DR, et al: The
common feature of leukemia-associated IDH1 and IDH2 mutations is a
neomorphic enzyme activity converting alpha-ketoglutarate to
2-hydroxyglutarate. Cancer Cell. 17:225–234. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ponnaluri VK, Maciejewski JP and Mukherji
M: A mechanistic overview of TET-mediated 5-methylcytosine
oxidation. Biochem Biophys Res Commun. 436:115–120. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Smith JJ, Deane NG, Wu F, Merchant NB, et
al: Experimentally derived metastasis gene expression profile
predicts recurrence and death in patients with colon cancer.
Gastroenterology. 138:958–968. 2010. View Article : Google Scholar
|
|
16
|
Takatsuno Y, Mimori K, Yamamoto K, et al:
The rs6983267 SNP is associated with MYC transcription efficiency,
which promotes progression and worsens prognosis of colorectal
cancer. Ann Surg Oncol. 20:1395–1402. 2013. View Article : Google Scholar
|
|
17
|
Therneau TM and Grambsch PM: Modeling
Survival Data: Extending the Cox Model. Springer; NY: 2000
|
|
18
|
Team RC: A language and environment for
statistical computing. R Foundation for Statistical Computing;
Vienna: 2013, http://www.R-project.org/.
|
|
19
|
Unoki M, Okutsu J and Nakamura Y:
Identification of a novel human gene, ZFP91, involved in acute
myelogenous leukemia. Int J Oncol. 22:1217–1223. 2003.PubMed/NCBI
|
|
20
|
Micci F, Skotheim RI, Haugom L, et al:
Array-CGH analysis of microdissected chromosome 19 markers in
ovarian carcinoma identifies candidate target genes. Genes
Chromosomes Cancer. 49:1046–1053. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Suzuki Y, Jiang LL, Souri M, et al:
D-3-hydroxyacyl-CoA dehydratase/D-3-hydroxyacyl-CoA dehydrogenase
bifunctional protein deficiency: a newly identified peroxisomal
disorder. Am J Hum Genet. 61:1153–1162. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bian YS, Osterheld MC, Fontolliet C,
Bosman FT and Benhattar J: p16 inactivation by methylation of the
CDKN2A promoter occurs early during neoplastic progression in
Barrett’s esophagus. Gastroenterology. 122:1113–1121. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Alder H, Taccioli C, Chen H, et al:
Dysregulation of miR-31 and miR-21 induced by zinc deficiency
promotes esophageal cancer. Carcinogenesis. 33:1736–1744. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Frisch MJ, Trucks GW, Schlegel HB, et al:
Gaussian 09. Gaussian, Inc; Wallingford, CT: 2009
|
|
25
|
Terunuma A, Putluri N, Mishra P, et al:
MYC-driven accumulation of 2-hydroxyglutarate is associated with
breast cancer prognosis. J Clin Invest. 124:398–412. 2014.
View Article : Google Scholar :
|
|
26
|
Kats LM, Reschke M, Taulli R, et al:
Proto-oncogenic role of mutant IDH2 in leukemia initiation and
maintenance. Cell Stem Cell. 14:329–341. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Montrose DC, Zhou XK, Kopelovich L, et al:
Metabolic profiling, a noninvasive approach for the detection of
experimental colorectal neoplasia. Cancer Prev Res Phila.
5:1358–1367. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Haffner MC, Chaux A, Meeker AK, et al:
Global 5-hydroxy-methylcytosine content is significantly reduced in
tissue stem/ progenitor cell compartments and in human cancers.
Oncotarget. 2:627–637. 2011.PubMed/NCBI
|
|
29
|
Smith DR and Goh HS: Overexpression of the
c-myc proto-oncogene in colorectal carcinoma is associated with a
reduced mortality that is abrogated by point mutation of the p53
tumor suppressor gene. Clin Cancer Res. 2:1049–1053.
1996.PubMed/NCBI
|
|
30
|
Struys EA, Salomons GS, Achouri Y, et al:
Mutations in the D-2-hydroxyglutarate dehydrogenase gene cause
D-2-hydroxy-glutaric aciduria. Am J Hum Genet. 76:358–360. 2005.
View Article : Google Scholar :
|