Introduction
Breast cancer is the most frequent malignant disease
and the second cause of death from cancer in US women (1). Most of the patients die of
metastases, rather than their primary tumors. Although successful
treatment of the primary malignancy is achieved as a result of
early diagnosis by mammographic screening and implementation of
systemic adjuvant therapy (2),
relapse and consequent metastatic growth of cancer cells can still
occur at distant sites, including bone, lung, liver and brain
(3,4). It was reported that metastasis to
distant sites accounts for >90% of breast cancer-related
mortality (5). However, metastasis
remains the most insidious aspect of breast cancer.
How tumors spread and kill their host organism
remains an enigma. Over the past decade, research effort on
metastatic disease has been focused on the biological processes
that influence the establishment of metastases (6,7). It
has been well established that tumor metastasis is a complex
multistep process that requires migration, invasion and
angiogenesis (7). Development of
new blood vessels, is a very critical event in formation of
metastases. This process is orchestrated by a large number of
cytokines and associated receptors and proteinases, such as
vascular endothelial growth factor (VEGF), fibroblast growth
factor, interleukin-8 and matrix metallopeptidases (MMPs) (8). Breakdown of the extracellular matrix
(ECM) is another crucial step in the metastatic cascade. Tumor
cells degrade surrounding ECM and basement membrane to facilitate
invasion and metastasis by secreting several proteases. MMPs play
an important role in this process. It was found that these enzymes
participate in proteolysis of ECM, modulation of cell adhesion,
migration (9,10), and epithelial to mesenchymal
transition (EMT) (11), processing
of growth factors, and tumor-induced angiogenesis. Collectively,
metastasis comprises multiple consecutive steps, and control one of
these processes represent promising therapeutic targets for cancer
therapy.
Extensive evidence shows that Runx2 maybe a
potential target for inhibition of metastatic growth of breast
cancer cells (7). Runx2, also
named PEBP2αA/AMl3/Cbfa1, is a transcription factor which is one of
members in runx gene family encoding proteins homologous to
Drosophila Runt (12).
Originally, it is found that this transcription factor plays a
crucial role in the formation of the skeleton (13–15).
Studies have demonstrated that atypical expression and function of
Runx2 are associated with the formation of bone metastasis in
breast cancer (16). In addition,
Runx2 is ectopically expressed in metastatic cancer cells but not
in non-metastatic cancer cells. Several genes required for bone
development and turnover, such as opn, bsp are major targets of
Runx2 (17). Additionally, Runx2
is involved in metastatic foci formation. Accumulating evidence
suggests that Runx2 is also a ‘master’ transcriptional factor in
ECs. It is reportedly involved in proliferation (18,19),
apoptotic resistance (18),
migration and invasion (20) of
ECs.
EMD is a natural anthraquinone compound extracted
from the root and rhizome of Rheum palmatum L. (Fig. 1). This traditional Chinese
medicinal herb was widely used for treatment of various ailments
and the anti-cancer activity of EMD was demonstrated, and the
ability to inhibit metastasis and angiogenesis was also shown
(21,22). However, clear information how EMD
affects angiogenesis and metastasis in human breast cancer is still
requied. To further evaluate its potential mechanisms in treatment
of metastatic breast cancer, we evaluated the anti-metastatic and
anti-angiogenic properties and its underlying mechanisms in this
study. We found that EMD inhibited tumor-induced angiogenesis and
metastasis in vitro and in vivo, and that the primary
action of EMD in breast cancer cells is throught a Runx2-induced
inhibitive mechanism.
Materials and methods
Materials
EMD for research use was from Sigma-Aldrich (cat.
no. E7881, Beijing, China), with thepurity of ≥90%, as determined
by high-performance liquid chromatography. A stock solution (1 mM)
was prepared by dissolving EMD in dimethyl sulfoxide (DMSO).
Recombinant human VEGF165 (cat. no. 293-VE) is a product
of R&D Systems (Minneapolis, MN, USA). A potent inhibitor of
VEGFR-2, ZD6474 (Selleck Chemicals Inc., cat. no. S1046, Shanghai,
China), and a broad spectrum MMP inhibitor, Batimastat (Bat; Santa
Cruz Biotechnology Inc., cat. no. SC-203833, Santa Cruz, CA, USA)
were used in this study and served as positive controls. In
addition, L-sulforaphane (SFP, Sigma-Aldrich, cat. no. S6317,
Beijing, China) was used as a positive control in tube formation
assay.
Cell culture
The human umbilical vein cell line, EA.hy 926, was
purchased from cell bank of Shanghai Institute for Biological
Sciences, CAS. EA.hy 926 cells were maintained in Dulbecco’s
modified Eagle’s medium supplemented with 10% FBS, 100 IU/ml
penicillin and 100 μg/ml streptomycin (Invitrogen, Beijing, China)
in a humidified incubator containing 5% CO2 at 37°C.
Prior to performing each assay, the ECs were serum starved for 4 h.
MDA-MB-231 human breast cancer cells, purchased from Cell Resource
Center, Institute of Basic Medical Sciences, Chinese Academy of
Medical Sciences, were maintained in Dulbecco’s modified Eagle’s
medium (DMEM) supplemented with 10% fetal bovine serum (FBS; Gibco,
Inc.), 100 IU/ml penicillin and 100 μg/ml streptomycin (Invitrogen,
Inc.) in a humidified incubator containing 5% CO2 at
37°C.
Animals and ethics statement
Five- to-six-week-old female NOD/SCID mice were
purchased from Vital River Laboratories (Beijing, China) to
establish orthotopic and experimental lung metastatic xenograft
model. Eight- to-nine-week-old male Sprague Dawley (SD) rats were
also purchased from Vital River Laboratories to obtain thoracic
aorta in the rat aortic ring angiogenesis assay. The animal
experimental protocol complied with the Animal Management Rules of
the Chinese Ministry of Health (document no. 55, 2001), and was
approved by the Animal Ethics Committees of Jiangxi University of
Traditional Chinese Medicine. All of the animal experiments were
conducted in strict accordance with the requirements listed in this
protocol. All surgery was performed under sodium pentobarbital
anesthesia, and all efforts were made to minimize suffering.
In vivo growth assay
For the growth assay, MDA-MB-231 cell xenografts
were established by injection of 1×107 cells at the
mammary fat pad in NOD/SCID mice. After 3 weeks of growth, the
tumors were removed and 1×1×1 mm tumor pieces were then implanted
at the mammary fat pad of mice. The mice bearing tumor chunks were
randomly divided into three groups: control, EMD (40 mg/kg per day)
treatment group, and EMD (80 mg/kg per day) treatment group.
Forty-eight hours later, EMD was administered by gavage on a
regimen of 6-day dosing per week for 5 weeks. Tumor growth was
assessed by measuring volumes of tumors with electronic calipers
every 3–4 days continuously. All of the mice were sacrificed 5
weeks after inoculation of the cancer cells and the tumors were
collected and weighed.
In vivo experimental metastasis assay and
survival assay
To establish experimental lung metastasis xenograft
model, 2×106 MDA-MB-231 cells in 200 μl normal saline
were injected through tail vein of NOD/SCID mice. The mice were
divided into control, EMD (40 mg/kg per day) treatment group, and
EMD (80 mg/kg per day) treatment group. EMD treatment was began 24
h after tumor injection. After 8-week treatment the formation of
metastatic foci in lung tissues was measured by nest
reverse-transcription polymerase chain reaction (RT-PCR). The mice
were euthanized and dissected, and lungs were snap frozen in liquid
nitrogen. Total RNA was isolated from each lung using the
QIAshredder and RNeasy Protect Mini kit (Qiagen) for detection of
human cytokeratin 19 (ck19) by nest PCR. For the survival assay,
the experimental lung metastasis xenograft model was established
using the same method as in vivo experimental metastasis
assay, but the observation period was 120 days.
RNA isolation and nest RT-PCR
Nest RT-PCR was used to detect expression of the
human ck19 gene in lung tissues of tumor-bearing mice. Total RNA
was isolated from each lung using the QIAshredder and RNeasy
Protect Mini kit (Qiagen). Ck19 was reverse transcribed in the
presence of 2 μl enzyme mix, 10 μl RNA and outer primer for 30 min
at 50°C, according to the manufacturer’s instructions (Qiagen
OneStep RT-PCR kit, Qiagen). After Taq polymerase activation for 15
min at 95°C, samples were amplified for 37 cycles at 94°C for 45
sec, 58°C for 45 sec and 72°C for 90 sec. Target primers for
amplifying ck19 (outer primer) were designed using Primer Designer
(Scientific & Educational Software Version 2.0). The forward
primer for ck19 (GenBank no. BC010409) was 5′-cca cgt cgt cct tcg
gag gcc-3′ (64–84 bp) and reverse primer was 5′-gttc cgt ctc aaa
ctt ggt tc-3′ (529–549 bp). After a final extension for 10 min at
72°C, the RT-PCR product was further subjected to nest PCR using
the Qiagen Multiplex PCR kit (Qiagen). The inner primers were
forward 5′-tac agc cac tac tac acg acc atc c-3′ (432–456 bp) and
reverse 5′-gga caa tcc tgg agt tct caa tg-3′ (488–510 bp). The nest
PCR profile was as follows: 3 min at 94°C, followed by 35
three-step cycles of 45 sec at 94°C, 45 sec at 60°C and 1 min at
72°C. PCR reactions were subjected to final extension at 72°C for
10 min. Nest RT-PCR analysis was performed using the Mastercycler
gradient (Eppendorf). The β-actin gene was used as an internal
control for standardization and the primers, Tm, and cycles are the
same as reported (23). The nest
PCR product was separated by 4% agarose (UltraPure™ Agarose,
Invitrogen) gel electrophoresis, and the gels were viewed by UV
transillumination and photographed by the UVP EC3 gel imaging
system.
Immunohistochemistry for tumor
tissues
MVD was determined by CD34 staining against ECs in
tumor tissues. Firstly, 4-μm-thick sections from paraffin-embedded
formalin-fixed MDA-MB-231 tumor tissues were made. After dewaxing,
and hydration, the slides were incubated with Proteinase K at 37°C
for 15 min to retrieve antigen, and then washed in PBS (0.01 mol/l,
5 min, three times). In order to block endogenous peroxidase
activity, the sections were treated with 3%
H2O2 in methanol for 10 min. Followed by
blocking with 10% normal goat serum (Cell Signaling, Danvers, MA,
USA) and washing in PBS, the slides were incubated with anti-CD34
antibody (1:50; cat. no. 3569; Cell Signaling) or PBS (0.01 mol/l)
at 4°C overnight. The slices were incubated with second antibody
(HRP-labeled goat anti-mouse antibody) at room temperature for 60
min, and then peroxidase activity was detected by
SignalStain® DAB substrate kit (Cell Signaling). All the
slides were checked under light microscopy (Olympus, BX-63), and
images were analyzed by Image Pro plus software 5.0 (Media
Cybernetics Inc. Silver Spring, MD, USA).
Tube formation assay
The tube formation assay was used to investigate the
effects of EMD on angiogenesis in vitro. Briefly, 80 μl of
liquid growth factor-reduced Matrigel (BD Biosciences, San Jose,
CA, USA) were added to each well of a 96-well plate. After 45-min
incubation at 37°C, 3×104 EA.hy 926 cells per well in
100 μl complete culture medium containing vehicle, ZD6474 or EMD
was seeded in each well. Then, 100 μl serum-free medium containing
VEGF165 (final concentration 2 ng/ml) was added. After
24-h incubation at 37°C and 5% CO2, the images of each
well were recorded by an inverted microscope (Leica, DMI 3000B,
Germany).
Rat aortic ring angiogenesis assay
In the present study, an ex vivo tube
formation system was used to evaluate the anti-angiogenic effect of
EMD. Male SD rats (8–9-week-old) were euthanized and thoracic
aortas were retrieved. After rinsing with 1% antibiotic/antimycotic
cocktail in 1X PBS (100 U/ml penicillin, 100 μg/ml streptomycin and
0.25 μg/ml amphotericin B; Invitrogen), the surrounding
fibro-adipose tissue of thoracic aorta was completely removed with
fine microdissection scissors and the thoracic aorta was cut into 1
mm thick rings with a scalpel blade. Then, individual ring was
implanted on a Matrigel-coated 96-well microtiter plate. Matrigel
was added again to embed and fix rings. After 30-min incubation in
5% CO2 at 37°C, the aorta rings were incubated in human
endothelial serum-free medium (Gibco, Carlsbad, CA, USA)
supplemented with 2% FBS, 50 U/ml penicillin and 50 μg/ml
streptomycin (Invitrogen) for 24 h and then treated with different
doses of EMD for 13 days. Finally, 8 μg/ml Calcein AM (BD
Biosciences) was added to stain microvessels. Photographs of the
microvessels were obtained using an inverted fluorescence
microscope (Leica, DMI 3000B, Germany).
In vitro invasion assay
Effects of EMD on invasion were measured by a
48-well microchemotaxis system (AP 48, Neuro Probe; Gaithersburg,
MD, USA). Briefly, 5 μg of fibronectin in a volume of 50 μl was
applied on the rough (lower) surface of the polycarbonate membrane
and 5 μg/filter Matrigel was plated to the smooth (upper) surface.
The lower chambers of the plates were then filled with 30 μl medium
containing 0.1% BSA. Log-phase cells were harvested and
re-suspended in culture medium with 0.1% BSA. Cell suspensions (100
μl containing 1×105 cells) were added to the upper
compartment and incubated for 16 h at 37°C in a 5% CO2
atmosphere. After incubation, the filters were fixed with methanol
and stained with 0.5% crystal violet for 60 min. The cells on the
upper surface of the filters were removed by wiping with cotton
swabs. The cells invading to the lower surface of the filter
through Matrigel and filter were quantified with the Image Pro plus
software 5.0 (Media Cybernetics Inc.), and representative results
are illustrated in the figures. Each assay was performed in
triplicate.
In vitro migratory assay
In vitro migration of MDA-MB-231 cells was
measured using the AP 48 chamber (Neuro Probe), similar to the
in vitro invasive assay, but without Matrigel pre-coating on
the smooth (upper) surface of filters. Briefly, the underside of
the polycarbonate membrane was coated with 10 μg/ml fibronectin
overnight at 4°C. Thirty microliters of DMEM (with 10% FBS and 10
μg/ml Collagen I) was added to the lower chamber, and the chamber
was covered by the filter. MDA-MB-231 cells were trypsinized and
washed with FBS-free DMEM, and 100 μl of cell suspensions (in
FBS-free DMEM, containing 2×105 cells) with or without
EMD were added to the upper chambers and incubated for 14 h in an
incubator containing 5% CO2 at 37°C. Determination of
migrated cells was conducted as described for the in vitro
invasive assay.
Immunocytochemistry
Log-phase cells EA.hy 926 cells were harvested,
re-suspended and seeded on the 8 wells Nunc™ Lab-Tek™ II Chamber
Slide™ system. The cells were grown at 37°C in a humidified
CO2 incubator until they were 50–70% confluent. The
culture medium was aspirated from each well and the cells gently
rinse twice in PBS at room temperature. Then, the cells were
exposed to EMD and/or vehicle for 24 h. After exposure, the
chambers of the Chamber Slide system were removed gently and the
slides were rinsed twice in PBS. Then cells were fixed by
incubation in 4% (v/v) paraformaldehyde in PBS for 20 min at room
temperature. The slides were heated in antigen retrieval buffer
[100 mM Tris, 5% (w/v) urea, pH 9.5] at 95°C for 10 min. After
rinsed in PBS, cells were blocked by incubating the cells in 0.1%
Triton X-100 in PBS for 15 min at room temperature. Then, cells
were incubated with the primary antibodies at 4°C overnight. After
rinsing in PBS, the slices were incubated with second antibody
(HRP-labeled goat anti-mouse antibody) at room temperature for 60
min, and then peroxidase activity was detected by
SignalStain® DAB substrate kit (Cell Signaling). All the
slides were checked under a light microscope (Olympus, BX-63), and
images were analyzed by Image Pro plus software 5.0 (Media
Cybernetics Inc.).
FRET-based MMP activity assay
The activity of MMPs was measured by the
SensoLyte® 570 Generic MMP assay kit (AnaSpec, Fremont,
CA, USA). This kit provides a FRET-based method to detect the
activity of a variety of MMPs including MMP-1, 2, 3, 7, 8, 9, 10,
11, 12, 13 and 14. It uses 5-FAM (fluorophore) and QXL520™
(quencher) labeled FRET peptide substrates for continuous
measurement of MMP activity. In an intact FRET peptide, the
fluorescence of 5-FAM is quenched by SensoLyte. Upon the cleavage
of FRET peptide by MMPs, the fluorescence of 5-FAM is recovered.
Analyses were performed according to the manufacturer’s
instructions. Briefly, supernatants of MDA-MB-231 cells were
collected after incubation with or without EMD for 12 h. MMPs in
the supernatants were activated by incubation with
4-aminophenylmercuric acetate for 1 h at 37°C. Fifty micro-liter
MMPs-containing samples and 50 μl MMP substrate solution were added
into a 96-well plate. The reagents were mixed by shaking the plate
gently for 30 sec. After a 50-min incubation period at 37°C, the
counter Victor3™, Perkin-Elmer (Waltham, MA, USA) was applied at
Ex/Em=540 nm/575 and then the action was stopped by adding stop
solution, and fluorescence intensity was measured.
Runx2 transcription factor assay
Runx2 activity in MDA-MB-231 and EA.hy 926 nuclear
extracts was detected using the TransAM™ AMl-3/Runx2 kit (Active
Motif North America, Carlsbad, CA, USA) following the
manufacturer’s instructions. Cell extracts were prepared using the
nuclear extract kit (Active Motif). Then, 20 μl of extracts diluted
in complete lysis buffer and containing 15 μg nuclear extract were
added into a 96-well plate. This plate immobilizes oligonucleotides
containing Runx2 consensus binding sites. Saos-2 nuclear extract
served as a positive control for Runx2 activation and 20 μl
complete lysis buffer served as the blank. The wild-type consensus
oligonucleotide was provided as a competitor for Runx2 binding to
monitor the specificity of the assay. After 1-h incubation at room
temperature, the plate was washed three times with washing buffer.
Diluted primary antibody (100 μl) was added into wells and
incubated for 1 h at room temperature without agitation. After
three washes, HRP-labeled secondary antibody was added and
incubated for 1 h at room temperature. Then, 100 μl developing
solution was added to initiate the color reaction. After 100 μl
stop solution was added, the absorbance was measured within 5 min
at 450 nm with a reference wavelength of 655 nm using an ELx800
microplate reader (BioTek, Winooski, VT, USA).
Western blot analysis
For western blot analysis, ECs were washed twice
with PBS and then lysed by the addition of 1 ml lysis buffer (10
mmol/l Tris, pH 7.6, 150 mmol/l NaCl, 5 mmol/l EDTA, pH 8.0, 10
ml/l Triton X-100, 1 mmol/l DTT) containing 0.1 mmol/l PMSF. After
30 min on ice, lysates were collected and clarified by
centrifugation at 15,000 g for 10 min at 4°C. Aliquots of whole
cell lysates were subjected to 10% SDS-PAGE and then transferred to
Hybond nitro-blotting membranes. The membranes were blocked with 3%
bovine serum albumin in Tris-buffered saline containing 0.5 ml/l
Tween-20 (TTBS) and then incubated with a 1:500–1,000 dilution of
the indicated primary antibodies, followed by incubation with
horseradish peroxidase (HRP)-conjugated secondary antibodies.
Immunoreactive proteins were detected using an enhanced
chemiluminescence kit (Millipore). β-actin (Santa Cruz, SC-130301)
served as an internal control.
Statistical analysis
The mean values were obtained from at least three
independent tests. The data are presented as mean ± SD and analyzed
with the GraphPad Prism 5.0 software program (La Jolla, CA, USA).
Comparison among different groups was carried out by analysis of
variance (one-way ANOVA). Differences between means were considered
statistically significant at p<0.05.
Results
EMD inhibits tumor growth and development
of metastasis in vivo
Initially the study was to determine the inhibitory
effects of EMD on breast cancer in vivo. The growth
inhibition in MDA-MB-231 cell line-xenografts was determined first.
Tumor-bearing mice were treated with various doses of EMD for 5
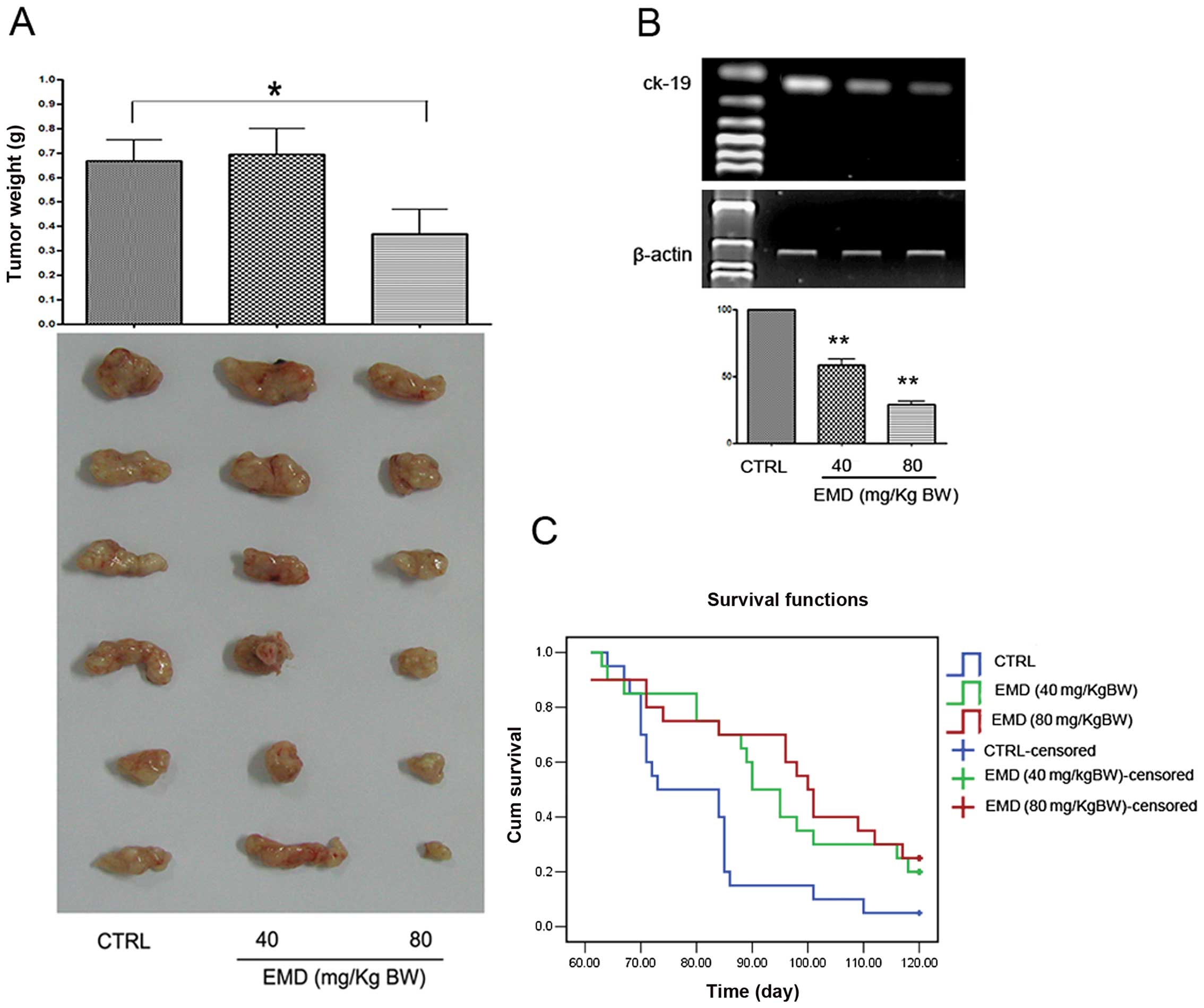
weeks. A significant suppression on tumor weight was found when the
mice were treated with 80 mg/kg b.w. of EMD (Fig. 2A). These results indicated that EMD
may contribute to breast cancer. Considering the clinical
importance of metastasis in breast cancer patients, the role of EMD
in inhibition of metastasis was evaluated in vivo.
Experimental lung metastasis models were established by injecting
human breast cancer MDA-MB-231 cells through tail vein of NOD/SCID
mice and the tumor cells invaded into lung tissues were detected by
amplification of human ck19 gene with nest RT-PCR assay. As shown
in Fig. 2B, EMD at 80 mg/kg b.w.
daily for 8 weeks reduced >70% ck19 expression compared to the
vehicle control. To determine whether EMD influence survival of the
tumor cell injected mice, overall survival assay was performed. Our
data showed that EMD lengthen survival time of NOD/SCID mice in a
dose-dependent manner (Fig.
2C).
EMD inhibits invasion and migration of
MDA-MB-231 cells in vitro
Invasion and migration are the critical steps for
the spread of tumor cells to distant organs. To determine whether
inhibition of lung tumor formation by EMD in vivo was due to
the ability of EMD to influence tumor cell migration and/or
invasion, we next determined the effects of EMD on cell invasion
and migration in vitro. As shown in Fig. 3A, EMD blocked trans-membrane
invasion of MDA-MB-231 cells significantly, when MDA-MB-231 cells
were incubated with 10, 20 and 40 μM EMD for 14 h. We next
evaluated the effects of EMD on cell migration with similar
methods. Here, we also found that migration of MDA-MB-231 cells was
significantly blocked by EMD (Fig.
3B) in a dose-dependent manner. The data suggested that the
inhibitory effects of EMD on metastasis may be associated with its
significant inhibition on cell invasion and migration.
EMD inhibits the activity of MMPs in
MDA-MB-231 cells in vitro
Breakdown of the extracellular matrix by MMPs in
surrounding tissues is a fundamental step of invasion and migration
in tumor cell metastasis. Therefore, we examined the effects of EMD
on MMPs in MDA-MB-231 cells. Fig.
4 shows the effects of EMD on MMPs in MDA-MB-231 cells. Using a
fluorescence resonance energy transfer (FRET)-based analysis, we
found that EMD exhibited significant suppression of FRET substrate
cleavage of MMPs in a dose-dependent manner (Fig. 4A). To further determine whether EMD
inhibited the functional activity or expression of MMPs, we
analyzed the expression of MMP-9 and MMP-13 in MDA-MB-231 cells
treated with EMD. As shown in Fig.
4B, EMD significantly decreased MMP-9 and MMP-13 expression of
MDA-MB-231 in a dose-dependent manner. These data suggested that
the anti-metastatic properties of EMD may be due to downregulation
of expression of MMP-9 and MMP-13 in MDA-MB-231 cells.
EMD inhibits the development of
angiogenesis in vivo
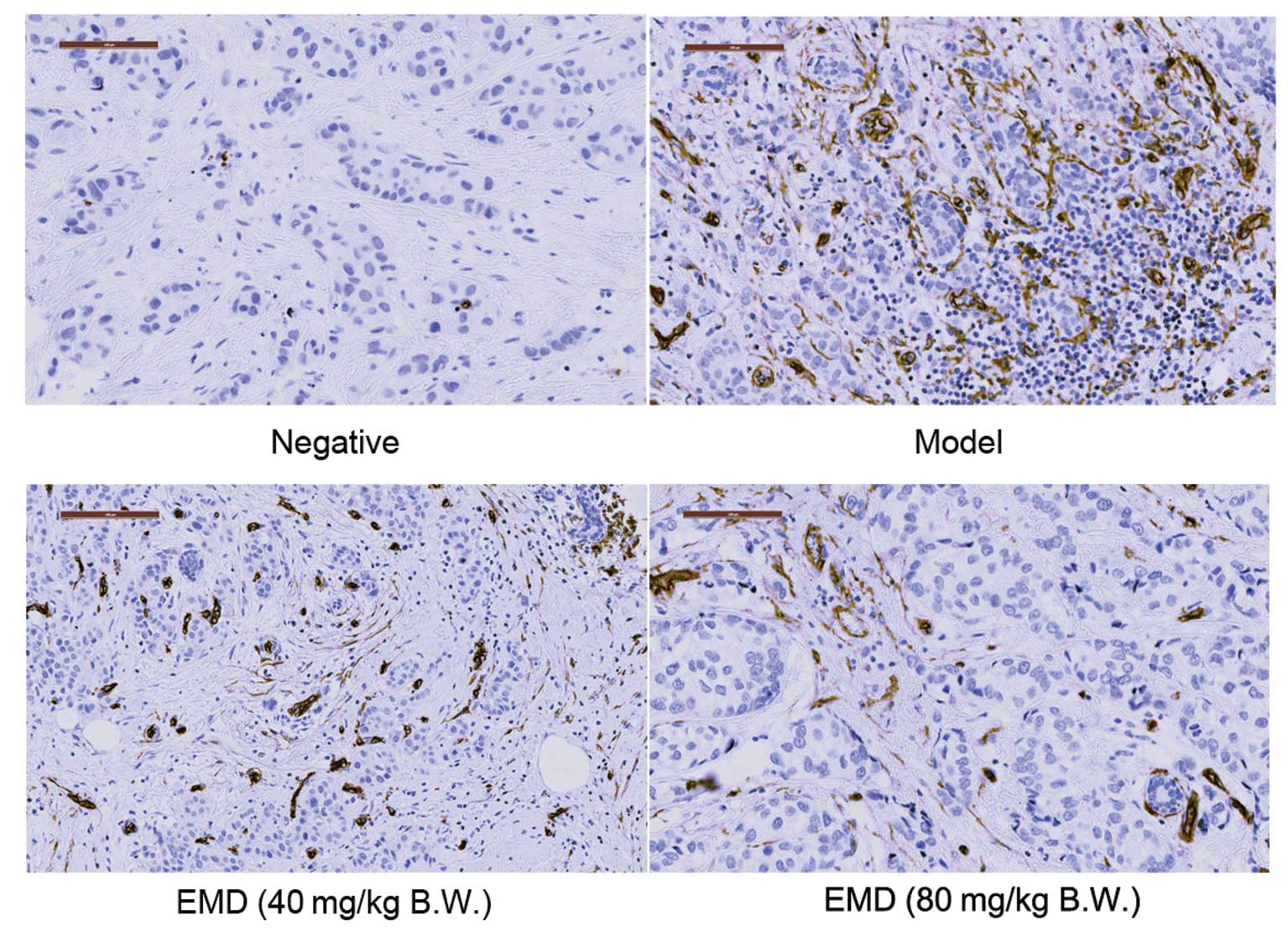
To determine whether tumor growth and metastasis
inhibition by EMD was associated with inhibition of tumor vessel
formation, we next measured effects of EMD on microvessel density
(MVD) in MDA-MB-231 tumor blocks by IHC assay. Representative
results are illustrated in Fig. 5
and semi-quantitative analysis of CD34 in these tumor blocks was
shown in Table I. A 42.49%
(p<0.01 compared with CTRL) and 78.91% (p<0.01 compared with
CTRL) reduction of positive area of CD34 staining were observed
when the nude mice were treated with 40 and 80 mg/kg/day EMD,
respectively.
 | Table ISemi-quantitative analysis of CD34 in
MDA-MB-231 tumor tissues. |
Table I
Semi-quantitative analysis of CD34 in
MDA-MB-231 tumor tissues.
| Group | Relative area of
CD34 immunostaining (%) | Inhibition rate
(%) |
|---|
| Negative | 0.87±0.22 | - |
| CTRL | 10.05±2.47 | - |
| EMD 40
mg/kg/day | 5.78±2.23a | 42.49 |
| EMD 80
mg/kg/day | 2.12±0.81a | 78.91 |
EMD shows direct inhibitory effects on
angiogenesis by targeting endothelial cell activation
To determine whether inhibition of tumor induced
angiogenesis in vivo by EMD was due to a direct inhibition
targeting ECs, we used the tube formation assay to evaluate the
effects of EMD on angiogenesis. As shown in Fig. 6A, when EA.hy 926 cells were seeded
on Matrigel, capillary-like structures with a lumen were formed.
After exposure to EMD solution at various concentrations for 24 h,
these structures were destroyed in a dose-dependent manner. In
addition, rat aortic ring angiogenesis assay was used in this study
to verify the anti-angiogenic effects of EMD. Similarly, new blood
vessels, triggered by the injury of the dissection procedure and
mediated by growth factors produced from aortic ring were
demolished by EMD in a dose-dependent manner (Fig. 6B).
EMD inhibits VRGFR-2 activation of
endothelial cells in vitro
VEGFR-2 is a crucial regulator in all aspects of
normal and pathological vascular endothelial cell biology. In order
to understand whether VEGFR-2 is involved in the inhibition of
angiogenesis induced by EMD, we next evaluated the effects of EMD
on VEGFR-2 phosphorylation in EA.hy 926 cells. To this end,
immunocytochemistry assay was employed and the results are shown in
Fig. 7. From these data, it was
found that EMD produced a dose-dependent decrease in
phospho-VEGFR-2 expression. Collectively, these findings indicated
that EMD inhibited angiogenesis by directly inhibiting activation
of VEGFR-2 in human ECs.
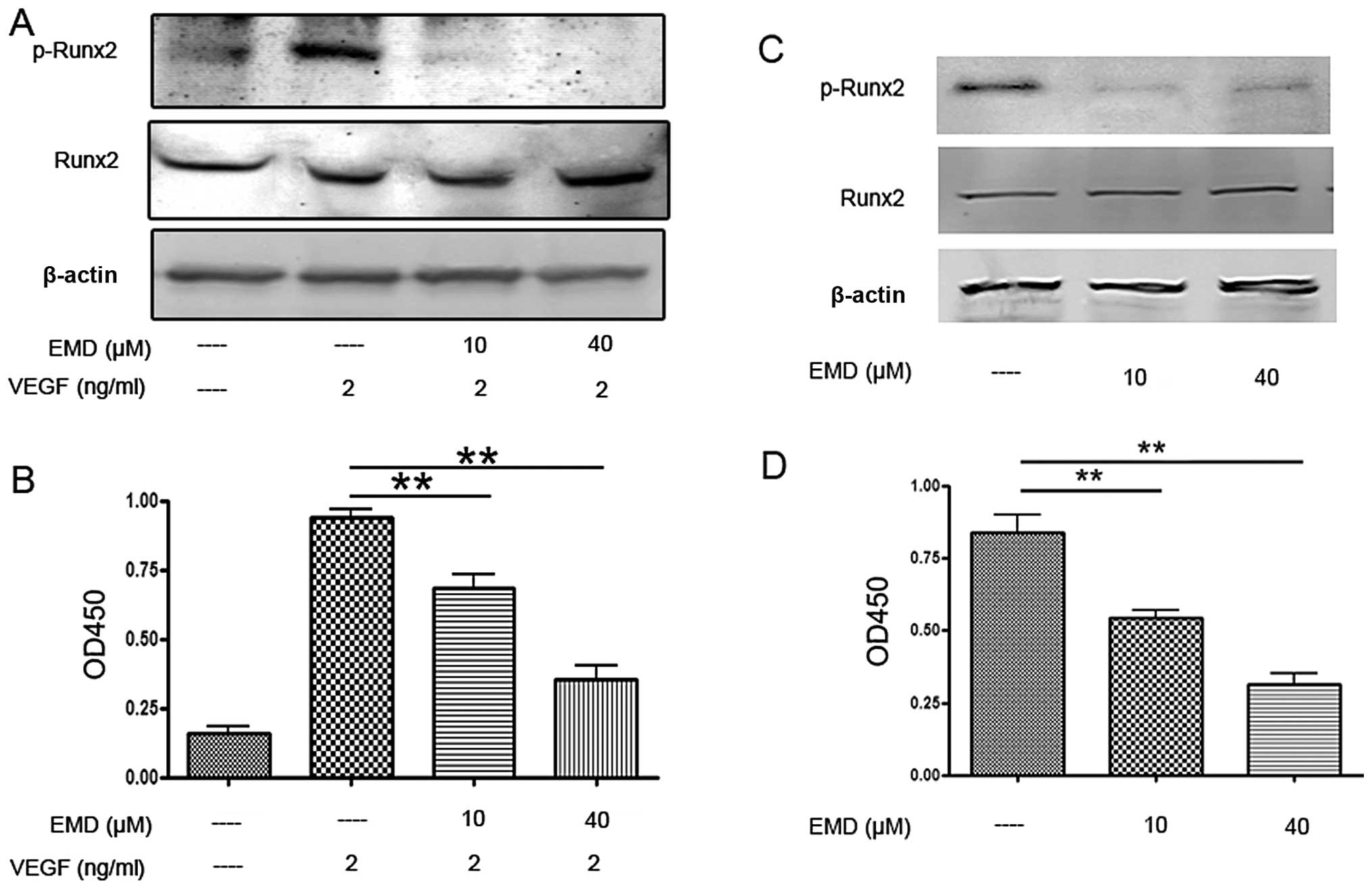
EMD inhibits activation of Runx2 in
endothelial and MDA-MB-231 cells
Runx2 is a transcriptional factor of metastatic
growth of breast cancer cells. Several genes required for the
formation of metastatic foci, including MMP-9, MMP-13, bone
sialoprotein, osteopontin, VEGF, are targets of this
transcriptional factor. Therefore, next we examined Runx2
activities in ECs and MDA-MB-231 cells. As shown in Fig. 8A and C, EMD had no effect on total
Runx2 expression, but caused a significant decrease of
phospho-Runx2 expression. To confirm the western blot results,
Runx2 transcription factor assay was employed. Here, an ELISA-based
kit for the Runx2 transcription factor was used to analyze the
effects of EMD on Runx2. Nuclear extracts incubated with EMD or
vehicle were prepared and the binding activity between Runx2 with
its target sequence was determined. The results showed that EMD
significant decreased the binding activity of Runx2 to its target
sequences in EA.hy 926 and MDA-MB-231 cells (Fig. 8B and D). Together, these findings
suggested that the inhibitory effects of EMD on angiogenesis and
metastasis were caused by inhibition of MMPs and VEGFR-2, which may
be associated with suppression of Runx2 phosphorylation.
Discussion
Emodin is an anthraquinone derivative of the root
and rhizome of Rheum palmatum L. and also found in other
plants (24). This active compound
has been reported to show anti-bacterial, antitumor, diuretic and
vasorelaxant effects (24). EMD
reportedly inhibits tumor-induced angiogenesis and metastasis by
blocking VEGFR signaling in human colon cancer cells and inhibiting
MMP expression in human neuroblastoma cells (25). However, there is no clear
information how EMD affects angiogenesis and metastasis in human
breast cancer. Therefore, we evaluated the inhibitory effects of
EMD on angiogenesis and metastasis in breast cancer. We found that
this anthraquinone derivative showed significant antitumor
properties and improvement of overall survival in human breast
cancer. It was found that EMD attenuated tumor cell-induced
angiogenesis and metastasis both in vitro and in
vivo. Furthermore, these inhibitory effects of EMD were caused
by MMPs and VEGFR-2 inhibition, which may be associated with
downregulation of Runx2 transcriptional activity in breast
cancer.
Migration and invasion of tumor cells are critical
events in the metastatic processes. In addition, breakdown of the
extracellular matrix by MMPs in surrounding tissues is a
fundamental step of invasion and migration in tumor cell
metastasis. Thus, in the present study, we examined the
anti-metastatic properties of EMD in vitro and in
vivo. It was demonstrated that EMD decreased tumor foci
formation in experimental metastasis in vivo and inhibited
tumor cells invasion and migration in vitro. FRET-based MMPs
analysis and western blot assay indicated that EMD attenuated MMP
activity and expression of MMP-9 and MMP-13. MMPs are a family of
structurally and functionally related zinc-dependent
endopeptidases. To date, 23 human MMPs, including 17 soluble,
secreted enzymes and six membrane-associated enzymes, have been
identified (10). These enzymes
are involved in a wide range of physiological and pathological
processes, such as embryonic development, wound healing, tumor
growth, invasion and metastasis (9), and participate in the proteolysis of
the ECM, modulation of cell adhesion, migration, and EMT,
processing of growth factors, and tumor-induced angiogenesis. In
breast cancer, reported data suggested critical roles for MMPs in
both breast cancer initiation and progression (9,11,26).
These data suggested EMD attenuation of breast cancer metastatic
properties may be associated with inhibition of MMPs.
It is well recognized that the growth of both
primary and metastatic tumors depends on adequate vascular support.
The increase in vasculature also increases the ability of tumor
cells to invade, enter the circulation to reach distant organs and
give rise to metastasis (27). In
order to understand the anti-growth and anti-metastatic properties
of EMD in breast cancer, we also evaluated its anti-angiogenic
effects. Our data showed that EMD inhibited the development of
angiogenesis in MDA-MB-231 breast cancer. Then, we used tube
formation assay of ECs and rat aortic ring angiogenesis assay to
evaluate the anti-angiogenic effects of EMD. It was revealed that
mesh-like structure formation on Matrigel was significantly
impaired by EMD both in tube formation assay and in rat aortic ring
angiogenesis assay, when 2 ng/ml of VEGF was added into the culture
system. Because of the important role of VEGFR signaling in
tumor-induced angiogenesis, we next determined the activity of
VEGFR-2 in ECs. Immunocytochemistry assay indicated that EMD
suppressed expression of phospho-VEGFR-2 in EA.hy 926 cells
significantly. These data suggested that the direct effects of EMD
on ECs may also contribute its inhibitory properties on tumor
growth and metastasis.
Runx2, also named PEBP2αA/AMl3/Cbfa1, is a critical
transcription factor for osteoblastic differentiation and skeletal
morphogenesis. This protein belongs to the Runx family encoding
proteins homologous to Drosophila Runt and has a conserved
Runt DNA-binding domain. Originally, Runx2 was found to act as a
master regulatory factor in skeletal development (15). To date, extensive evidence shows a
close association between Runx2 and breast cancer metastasis, and
this transcriptional factor is becoming a potential target of novel
antimetastatic agents and diagnostic approaches to breast cancer
control (28–30). Jiménez et al first found
that MMP-13, also named Collagenase-3, was highly expressed in
MDA-MB-231 cells, and that it was one of target genes of Runx2
(31). These observations were
demonstrated by Selvamurugan and colleagues (32,33).
Studies showed that the runt domain (RD) binding site and Runx2
were required for maximal constitutive and basal expression of
MMP-13 in MDA-MB-231 cells. ChIP assay confirmed two Runx2 binding
sites in the MMP-13 promoter, and these sites are occupied by
Runx2. Pratap et al investigated the role of Runx2 in the
regulation of the promoter of MMP-9, in MDA-MB-231 and MCF-7 cells
(34). Collectively, Runx2 acts a
‘master’ transcription factor of MMPs expression. Therefore, we
wondered whether the inhibitory effects of EMD on MMP-9 and MMP-13
were associated with the downregulation of Runx2 activities in
breast cancer cells. Our findings from western blotting
demonstrated that EMD significantly inhibited the expression of
p-Runx2 indicating that the transcriptional ability of Runx2 is
impaired by EMD. To confirm these findings, we used an ELISA-based
Runx2 transcription factor assay, and the results revealed that the
interaction between Runx2 and its target sequence sequences was
significantly inhibited by EMD. We also reported here that EMD
impaired activity of Runx2 in ECs. Numerous evidence supports that
VEGF is one of the target genes of Runx2, and VEGF-VEGFRs signaling
is controlled by Runx2 activity (18,19,35).
However, the direct evidence of the relationship between Runx2 and
VEGFR-2 is still lacking. Therefore, on-going research is directed
to exact the mechanisms of EMD on Runx2 and VEGFR-2 to clarify
their causal relation.
In conclusion, we report that EMD, an anthraquinone
derivative, impaired the metastatic and angiogenic potential of
breast cancer, and the inhibitory effects were due to its ability
to reduce the expression of MMP-9 and MMP-13 in breast cancer cells
and the activation of VEGFR-2 in ECs. These effects may be
associated with inhibition of transcriptional activity of
Runx2.
Acknowledgements
This study was supported by grants from the National
Natural Science Foundation of China (grant no. 81160530 and
81260656), National Basic Research Program of China (‘973’ Program)
(grant no. 2010CB530603), Key Research Project from the Ministry of
Education of China (grant no. 211091), and the Natural Science
Foundation of Jiangxi Province (grant no. 2010GQY0147).
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
Statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Weigelt B, Peterse JL and van ‘t Veer LJ:
Breast cancer metastasis: markers and models. Nat Rev Cancer.
5:591–602. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Leong SP, Cady B, Jablons DM, et al:
Clinical patterns of metastasis. Cancer Metastasis Rev. 25:221–232.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Stevanovic A, Lee P and Wilcken N:
Metastatic breast cancer. Aust Fam Physician. 35:309–312.
2006.PubMed/NCBI
|
|
5
|
Liu S, Goldstein RH, Scepansky EM, et al:
Inhibition of rho-associated kinase signaling prevents breast
cancer metastasis to human bone. Cancer Res. 69:8742–8751. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chiang AC and Massagué J: Molecular basis
of metastasis. N Engl J Med. 359:2814–2823. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gupta GP and Massagué J: Cancer
metastasis: building a framework. Cell. 127:679–695. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Adams RH and Alitalo K: Molecular
regulation of angiogenesis and lymphangiogenesis. Nat Rev Mol Cell
Biol. 8:464–478. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Duffy MJ, Maguire TM, Hill A, et al:
Metalloproteinases: role in breast carcinogenesis, invasion and
metastasis. Breast Cancer Res. 2:252–257. 2000. View Article : Google Scholar
|
|
10
|
Roy R, Yang J and Moses MA: Matrix
metalloproteinases as novel biomarkers and potential therapeutic
targets in human cancer. J Clin Oncol. 27:5287–5297. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Radisky ES and Radisky DC: Matrix
metalloproteinase-induced epithelial-mesenchymal transition in
breast cancer. J Mammary Gland Biol Neoplasia. 15:201–212. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lian JB, Stein JL, Stein GS, et al:
Runx2/Cbfa1 functions: diverse regulation of gene transcription by
chromatin remodeling and co-regulatory protein interactions.
Connect Tissue Res. 44(Suppl 1): S141–S148. 2003. View Article : Google Scholar
|
|
13
|
Karsenty G: Role of Cbfa1 in osteoblast
differentiation and function. Semin Cell Dev Biol. 11:343–346.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Komori T: Runx2, a multifunctional
transcription factor in skeletal development. J Cell Biochem.
87:1–8. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Stein GS, Lian JB, van Wijnen AJ, et al:
Runx2 control of organization, assembly and activity of the
regulatory machinery for skeletal gene expression. Oncogene.
23:4315–4329. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Harada S and Rodan GA: Control of
osteoblast function and regulation of bone mass. Nature.
423:349–355. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shore P: A role for Runx2 in normal
mammary gland and breast cancer bone metastasis. J Cell Biochem.
96:484–489. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sun L, Vitolo MI, Qiao M, et al:
Regulation of TGFbeta1-mediated growth inhibition and apoptosis by
RUNX2 isoforms in endothelial cells. Oncogene. 23:4722–4734. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Qiao M, Shapiro P, Fosbrink M, et al: Cell
cycle-dependent phosphorylation of the RUNX2 transcription factor
by cdc2 regulates endothelial cell proliferation. J Biol Chem.
281:7118–7128. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sun L, Vitolo MI and Passaniti A:
Runt-related gene 2 in endothelial cells: inducible expression and
specific regulation of cell migration and invasion. Cancer Res.
61:4994–5001. 2001.PubMed/NCBI
|
|
21
|
Liu A, Chen H, Wei W, et al:
Antiproliferative and antimetastatic effects of emodin on human
pancreatic cancer. Oncol Rep. 26:81–89. 2011.PubMed/NCBI
|
|
22
|
Kaneshiro T, Morioka T, Inamine M, et al:
Anthraquinone derivative emodin inhibits tumor-associated
angiogenesis through inhibition of extracellular signal-regulated
kinase 1/2 phosphorylation. Eur J Pharmacol. 553:46–53. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fu J, Ding Y, Huang D, et al: The retinoid
X receptor-selective ligand, LGD1069, inhibits tumor-induced
angiogenesis via suppression of VEGF in human non-small cell lung
cancer. Cancer Lett. 248:153–163. 2007. View Article : Google Scholar
|
|
24
|
Hsu S-C and Chung J-G: Anticancer
potential of emodin. BioMedicine. 2:108–116. 2012. View Article : Google Scholar
|
|
25
|
Lu HF, Lai KC, Hsu SC, et al: Involvement
of matrix metalloproteinases on the inhibition of cells invasion
and migration by emodin in human neuroblastoma SH-SY5Y cells.
Neurochem Res. 34:1575–1583. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chambers AF and Matrisian LM: Changing
views of the role of matrix metalloproteinases in metastasis. J
Natl Cancer Inst. 89:1260–1270. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fidler IJ: Angiogenesis and cancer
metastasis. Cancer J. 6(Suppl 2): S134–S141. 2000.PubMed/NCBI
|
|
28
|
Inman CK, Li N and Shore P: Oct-1
counteracts autoinhibition of Runx2 DNA binding to form a novel
Runx2/Oct-1 complex on the promoter of the mammary gland-specific
gene beta-casein. Mol Cell Biol. 25:3182–3193. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Barnes GL, Javed A, Waller SM, et al:
Osteoblast-related transcription factors Runx2 (Cbfa1/AMl3) and
MSX2 mediate the expression of bone sialoprotein in human
metastatic breast cancer cells. Cancer Res. 63:2631–2637.
2003.PubMed/NCBI
|
|
30
|
Inman CK and Shore P: The osteoblast
transcription factor Runx2 is expressed in mammary epithelial cells
and mediates osteopontin expression. J Biol Chem. 278:48684–48689.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jiménez MJ, Balbín M, López JM, et al:
Collagenase 3 is a target of Cbfa1, a transcription factor of the
runt gene family involved in bone formation. Mol Cell Biol.
19:4431–4442. 1999.PubMed/NCBI
|
|
32
|
Selvamurugan N and Partridge NC:
Constitutive expression and regulation of collagenase-3 in human
breast cancer cells. Mol Cell Biol Res Commun. 3:218–223. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Selvamurugan N, Kwok S and Partridge NC:
Smad3 interacts with JunB and Cbfa1/Runx2 for transforming growth
factor-beta1-stimulated collagenase-3 expression in human breast
cancer cells. J Biol Chem. 279:27764–27773. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pratap J, Javed A, Languino LR, et al: The
Runx2 osteogenic transcription factor regulates matrix
metalloproteinase 9 in bone metastatic cancer cells and controls
cell invasion. Mol Cell Biol. 25:8581–8591. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kwon TG, Zhao X, Yang Q, et al: Physical
and functional interactions between Runx2 and HIF-1α induce
vascular endothelial growth factor gene expression. J Cell Biochem.
112:3582–3593. 2011. View Article : Google Scholar : PubMed/NCBI
|