Introduction
Docosahexaenoic acid (DHA) is a 22:6n-3, long-chain
polyunsaturated fatty acid (PUFA) present in fat fish, fish oils
(1) and in marine microalgae
(2). Among numerous beneficial
effects, such as in cardiovascular (3) or metabolic syndromes (4), DHA may play a preventive role in
cancer (5,6). The mechanism by which DHA could
prevent tumorigenesis or directly target cancer cells remains
unclear (7). However, it has been
shown that DHA not only acts as an anti-proliferative agent by
lengthening the cell cycle between the G2/M transition (8), but also it is capable of inducing
apoptosis and reducing the invasive potential of the
triple-negative breast cancer cell line MDA-MB-231 with an optimal
amount of 100 μM (9). DHA can
modify the metastatic phenotype of cancer cells, emphasizing the
anti-cancer potential of the omega-3 (n-3) PUFAs (10,11).
This anti-cancer activity of DHA is promising and could partly
result in a modification of the lipid contents of the plasma
membrane and its fluidity (12).
Metastasis is a multifactorial process involving
extracellular matrix remodeling, extra- and intravasation, and
requiring the involvement of a variety of cell surface proteins.
For example, the urokinase-type plasminogen activator (uPA) and
metalloproteinases (MMPs) are involved in extracellular matrix
disorganization leading to the release of angiogenic factors such
as VEGF or FGF, allowing the sprouting of new blood vessels and
ultimately extra- and intravasation (13–15).
Several studies have shown that lipid rafts in the plasma membrane
can play an important role in cancer cells when n-3 PUFAs were
added, as it altered the cholesterol levels and consequently cell
migration, invasion and angiogenesis (16,17).
The DHA-induced decrease in breast cancer cell invasion may also be
due to inhibition of voltage-gated Na+ channels
(18,19). These voltage-gated channels also
called neonatal Nav1.5 are inhibited in a dose-dependent
manner by DHA and the use of specific blockers, like tetrodoxin,
can reduce the migration of MDA-MB-231 at the same level to that
observed with DHA (18). Thus,
DHA-induced suppression of cellular migration may occur via
downregulation of neonatal Nav1.5 mRNA and functional
protein expression (18). The
localization of voltage-gated Na+ channel in lipid rafts
(20) may be affected by n-3 PUFA
(19).
In addition, DHA may change the biophysical
properties of lipid rafts decreasing the content of cholesterol and
the distribution of key proteins such as EGFR, Src, heterotri-meric
G-protein subunits, or sphingomyelinase. Among these proteins, the
Src kinase might play an important role by regulating the migration
and invasion of the MDA-MB-231 cell line (21). Src was shown to play a role in
cancer and invasiveness (21,22)
and was also related to other molecules such as Keratin, type II
cytoskeletal 1 (KRT1) via integrin β1 (23) or the voltage-gated Na+
channels where one of its subunits can also be considered as an
integrin (24). Together, plasma
membrane and related cytosolic molecules appear to play an
important role in the DHA-induced inhibition of breast cancer cell
invasion.
In this context, we have decided to analyze changes
in the protein content of crude membrane preparations from breast
cancer cells treated with DHA. Two-dimensional electrophoresis
(2-DE) and MALDI-TOF mass spectrometry were used and several
proteins were identified as upregulated by DHA. Increase in the
level of KRT1 was the major change and interestingly functional
in vitro assays have shown its involvement in mediating the
anti-invasive effect of DHA.
Material and methods
Cell culture
The triple-negative breast cancer cell line MDA-
MB-231 was purchased from ATCC (Manassas, VA, USA) and routinely
grown as monolayers at 37°C, in a humidified atmosphere with 5%
CO2, in minimum essential medium (MEM) (Sigma-Aldrich,
Saint-Quentin Fallavier, France) supplemented with 10% fetal calf
serum (FCS) (Gibco; Invitrogen, Cergy Pontoise, France), 20 mM
Hepes, 2 mM L-glutamine, 100 U/ml penicillin/streptomycin
(Sigma-Aldrich).
Crude membrane protein extraction
MDA-MB-231 cells were grown in T75 cm2
flasks until reaching subconfluency. A set of 7×108
cells were treated, or not, with 100 μM DHA (Sigma-Aldrich Chimie
S.a.r.l.) for 24 h. After treatment, cells were rinsed three times
with 10 ml MEM and twice with 20 mM phosphate buffer pH 7.4
containing 150 mM NaCl. Cells were then detached with 2 ml of
Versene (Sigma-Aldrich) and centrifuged at 200 × g for 10 min at
4°C. The supernatant was discarded and the cell pellet stored at
−70°C. The membrane preparations were carried out on ice and at 4°C
according to Venkateswaran et al (25). Briefly, the cell pellets were
defrosted and homogenized in 3 ml buffer A composed with 20 mM
Hepes, 200 mM sucrose and 5 mM EDTA. The suspension was then
transferred in a 7 ml Dounce and cells were disrupted with 40
strikes of pestle. The homogenate was centrifuged at 1,000 × g for
20 min at 4°C in order to pellet the nuclei. The supernatant
containing the membranes was transferred in a tube for
centrifugation. Prior to this step, four volumes of buffer B
composed of 20 mM Hepes, 1 mM CaCl2, 1 mM
MgCl2 (all from Sigma-Aldrich). and 100 mM NaCl were
added to the supernatant to decrease buffer density and to allow
the best ionic environment for membrane proteins. The mixture was
centrifuged at 60,000 × g for 90 min at 4°C in order to obtain
crude membrane pellets. The supernatants were removed and the
pellets suspended in 500 μl buffer B and washed twice in the same
conditions. Aliquots of the pellets were taken for a protein assay
using Bradford’s method (Bio-Rad, Marnes-la-Coquette, France) with
BSA (Sigma-Aldrich) as standard. Aliquots of 100 μg membrane
proteins were stored in Eppendorf tubes and centrifuged at 20,000 ×
g for 60 min at 4°C. The pellets were then solubilized with 50 μl
of a lysis buffer suitable for isoelectric focusing (IEF) (urea 7
M, thiourea 2 M, CHAPS 2%, DTT 40 mM and 0.4% ampholytes 3–10) (GE
Healthcare Europe GmbH, Vélizy-Villacoublay, France) containing 1%
ASB-14 (Sigma-Aldrich), and stored at −70°C.
2-DE samples
A total of 100 μg of membrane proteins prepared as
described above were lysed with 50 μl of lysis buffer as described
above and processed for IEF by incubation for 1 h in 2 μl of 200 mM
tributylphosphine (TBP) (Sigma-Aldrich) followed by incubation for
1.5 h with 5 μl 200 mM iodoacetamide (IAM) (GE Healthcare). At this
stage, samples were loaded for IEF. Three independent experiments
were carried out in duplicate.
IEF and SDS-PAGE
IEF was performed using Ettan IPGphor 3 apparatus
using 7 cm strips with pH 3.0–10.0 (both from GE Healthcare Europe
GmbH). Strips were rehydrated overnight at room temperature
according to the manufacturer’s instructions with DeStreak
Rehydration Solution containing 0.4% ampholytes pH 3.0–10.0 (GE
Healthcare). The samples (100 μg) were cup-loaded near the anode of
the IPG strips and three drops of mineral oil were introduced in
the cups. Then, the tray was filled with mineral oil. The run was
defined as follows: step at 500 V for 500 Vh, gradient to reach
3,000 V for 5,000 Vh, step at 3,000 V for 12,000 Vh, step 1,000 V
for 1,000 Vh. Once the IEF was completed, the strips were processed
for SDS-PAGE after equilibration in urea 6 M, PlusOne Glycerol 30%
w/v, SDS 2% w/v (Bio-Rad), 0.125 M Tris, 0.1 M HCl containing 50 mM
DTT (first equilibration step; Sigma-Aldrich) and 150 mM IAM
(second equilibration step; GE Healthcare), and consisting in two
baths of 20 min each. The strips were placed at the top of 12%
acrylamide-bisacrylamide gels and maintained in position with 2 ml
of stacking gel. The run was performed with a PROTEAN 3 apparatus
(Bio-Rad) at a constant power of 8 W until the Bromophenol Blue
(Merck S.A., Lyon, France) reached the bottom of the gels. Gels
were washed twice for 5 min and stained with Imperial
Blue® (Fisher Scientific, Illkirch-Graffenstaden,
France) according to manufacturer’s instructions. Three independent
experiments were performed in duplicate.
Spot detection and quantification
The 2-D gels were scanned with a GS-800 densitometer
(Bio-Rad). Spot detection, quantification and analysis were
performed with the SameSpots® v4.1 analysis software
(Nonlinear Dynamics, Ltd., Newcastle upon Tyne, UK). Following
linearization towards a reference gel chosen among the experimental
gels, they were grouped either as control or treated. Each group
was the result of three independent experiments performed in
duplicate. Spot detection and quantification were determined and a
difference was considered to be significant, due to the staining
method used, when a 1.5-fold increase or decrease at least was
reached. Statistics using ANOVA were given with the in-built
statistical software.
In-gel digestion of protein
The protein spots differentially expressed were
excised manually and washed five times for 6 min with 100 μl water.
Then the gel spots were soaked in acetonitrile and dried under
vacuum. The gel pieces were rehydrated in a reduction buffer
[ammonium bicarbonate 100 mM (Sigma-Aldrich), DTT 10 mM (GE
Healthcare)] for 1 h at 56°C and 5 min at room temperature. After
removing this buffer, they were incubated with an alkylation buffer
(ammonium bicarbonate 100 mM, IAM 55 mM) for 45 min at room
temperature and protected from light. Then, they were washed in a
25-mM ammonium bicarbonate buffer followed by acetonitrile (Merck
S.A.) and finally dried under vacuum. The gel pieces were
rehydrated in 100 μl of 25 mM ammonium bicarbonate and incubated
with 125 ng of Trypsin Gold (Mass Spectrometry Grade; Promega
France, Charbonnières-les-Bains, France) for 1 h on ice. The
trypsin digestion was performed for 12 h at 37°C after addition of
30 μl of 25 mM ammonium bicarbonate.
Mass spectrometr y analysis
Mass spectrometry analyses were performed using an
Ultra flex™ II MALDI-TOF/TOF instrument (Bruker Daltonics, Bremen,
Germany). MALDI target plate (AnchorChip™; Bruker Daltonics) was
covered with extracted peptides mixed-up with
α-cyano-4-hydroxycinnamic acid matrix (0.3 mg/ml in
acetone:ethanol, 3:6 v/v). The molecular mass measurements were
obtained as previously described (26). Database searches, through Mascot
v.2.2.1 (Matrix Science, Ltd., London, UK), using combined PMF and
PFF datasets were performed against the UnitProt 2013-06 database
(2013-06-17) via ProteinScape 2.1 (Bruker Daltonics). A mass
tolerance of 75 ppm and one missing cleavage site for PMF and MS/MS
tolerance of 0.5 Da and one missing cleavage site for MS/MS search
were allowed. Carbamidomethylation of cysteine and oxidation of
methionine residues were also considered. Relevance of protein
identities was judged according to the probability-based MOWSE
score calculated with a P-value of 0.05 (P≤0.05).
Preparation of siRNA and cell
transfection
A siRNA (Eurogentec S.A., Seraing, Belgium) directed
against KRT1 was used and defined by (GGA-UGU-GGA-UGG-UGC-UUA-U55)
for the forward strand and (AUA-AGC-ACC-AUC-CAC- AUC-C55) for the
reverse. In addition a control siRNA (NEG) provided by the
manufacturer was used. The different siRNA were rehydrated with
ultrapure water to obtain a concentration of 20 μM. In a 24-well
plate, 4×105 living cells/well were seeded. After 12 h,
the medium was removed and rinsed twice with 1 ml/well of Opti-MEM.
Then 950 μl of Opti-MEM were added with 50 μl of a mixture
containing Lipofectamine (Invitrogen) with or without the
appropriate siRNA. To form the mixture, 2.5 μl siRNA at 20 μM were
mixed with 22.5 μl Opti-MEM, and apart, 8.32 μl Lipofectamine were
homogenized with 16.68 μl Opti-MEM. The two solutions were then
mixed. After 10 min of incubation at room temperature the mixture
was transferred to culture wells. Incubation was for 4 h at 37°C
with 5% CO2. The medium was discarded and replaced with
1 ml Opti-MEM containing 5% FCS with or without 100 μM DHA. After
24 h, cells were harvested for invasion assay.
Invasion assays and Hoechst staining
Invasion assays were done in 12-well Boyden
microchambers (Transwell®; Fisher Scientific) with 8-μm
pore membranes. Matrigel® (100 μl; BD Biosciences, Le
Pont de Claix, France) at 10% in MEM were introduced in the upper
chamber and dried overnight at 37°C. Cells treated or not for 24 h
as described in the previous section were dissociated with Versene
and counted by using a Malassez hemocytometer. Living cells
(2×105) treated or not in 400 μl MEM supplemented with
0.5% FCS and 1% BSA were then loaded into the upper chamber. A
volume of 800 μl of MEM with 0.5% FCS, and 1% BSA was introduced
into the lower chamber. After incubating for 24 h, the
Transwell® was rinsed with PBS, and the
Matrigel® was scraped off the upper surfaces of the
membranes. The cells remaining on the underside of the membrane
were fixed for 30 min at −20°C in methanol, then stained with
Hoechst stain (H6024; Sigma-Aldrich), and mounted on glass slides
with glycerol for fluorescence microscopy (Merck S.A.) before
counting (15 fields/membrane) under a UV microscope (Biomed with
fluorescence equipment; Leica, Rueil-Malmaison, France). Light and
fluorescent micrographs were taken with the Lasez software (Leica).
Three independent experiments were performed in duplicate.
Immunocytochemistry
Experiments were performed with 5×104
cells/chamber on a 16-chamber slide (Fisher Scientific) overnight.
Then 200 μl of the Lipofectamine mixture with or without siRNA were
added for 4 h after medium withdrawal and rinsing with 500 μl
Opti-MEM. Then 200 μl Opti-MEM containing 5% SVF with or without
100 μM DHA were added for 24 h. Chambers were rinsed with PBS and
cells fixed with ethanol-methanol-ultrapure water (1:1:2) for 1 h
at −20°C. Cells were treated for endogenous peroxidase with 100 μl
PBS containing 3% H2O2 20 vol, for 10 min at
room temperature. Then the medium was discarded and 200 μl of PBS
containing 5% BSA was added for 1 h followed by an incubation of 2
h with 100 μl mouse monoclonal anti-KRT1 antibody (Mab 191–05;
Diagnostic BioSystems, Inc., Hague, The Netherlands) at 1/500
diluted in PBS with 0.5% BSA. Cells were rinsed three times with
200 μl PBS 0.5% BSA. HRP anti-mouse antibody (Sigma-Aldrich) at
1/200 in PBS 0.5% BSA was added for 1 h. Cells were rinsed three
times with PBS 0.5% BSA and three times with PBS prior HRP
revelation by the adjunction of 100 μl diaminobenzidine (DAB)
prepared in 10 ml water with 20 μl H2O2 20
vol and counterstained with Hoechst staining. Slides were washed
with water and mounted with glycerol for fluorescence microscopy.
For each chamber, 10 randomized fields were photographed and
analyzed with the Quantity One software (Bio-Rad). Staining
intensity in one field was divided by the number of nuclei observed
by Hoechst staining in the same field in order to have the average
value of KRT1 immunoreactivity. Three independent experiments were
performed in duplicate.
Statistics
Statistical analyses for cell culture were performed
using KyPlot® (KyensLab, Inc., Tokyo, Japan) for a one-
way ANOVA followed by a Dunnett’s test to compare untreated or
control cells with the treated one. P-value of <0.01 and
<0.001 respectively, indicates statistically significant result.
In the figures shown as **P<0.01 and
***P<0.001 Statistics for 2-DE are described in the
corresponding paragraph.
Results
Identification of differentially
expressed proteins in DHA-treated cells
MDA-MB-231 cells treated or not with 100 μM DHA for
24 h were processed to obtain membrane extracts. After 2-DE of
membrane proteins (from both DHA-treated and control cells), it
appears that only a few membrane proteins displayed at least a
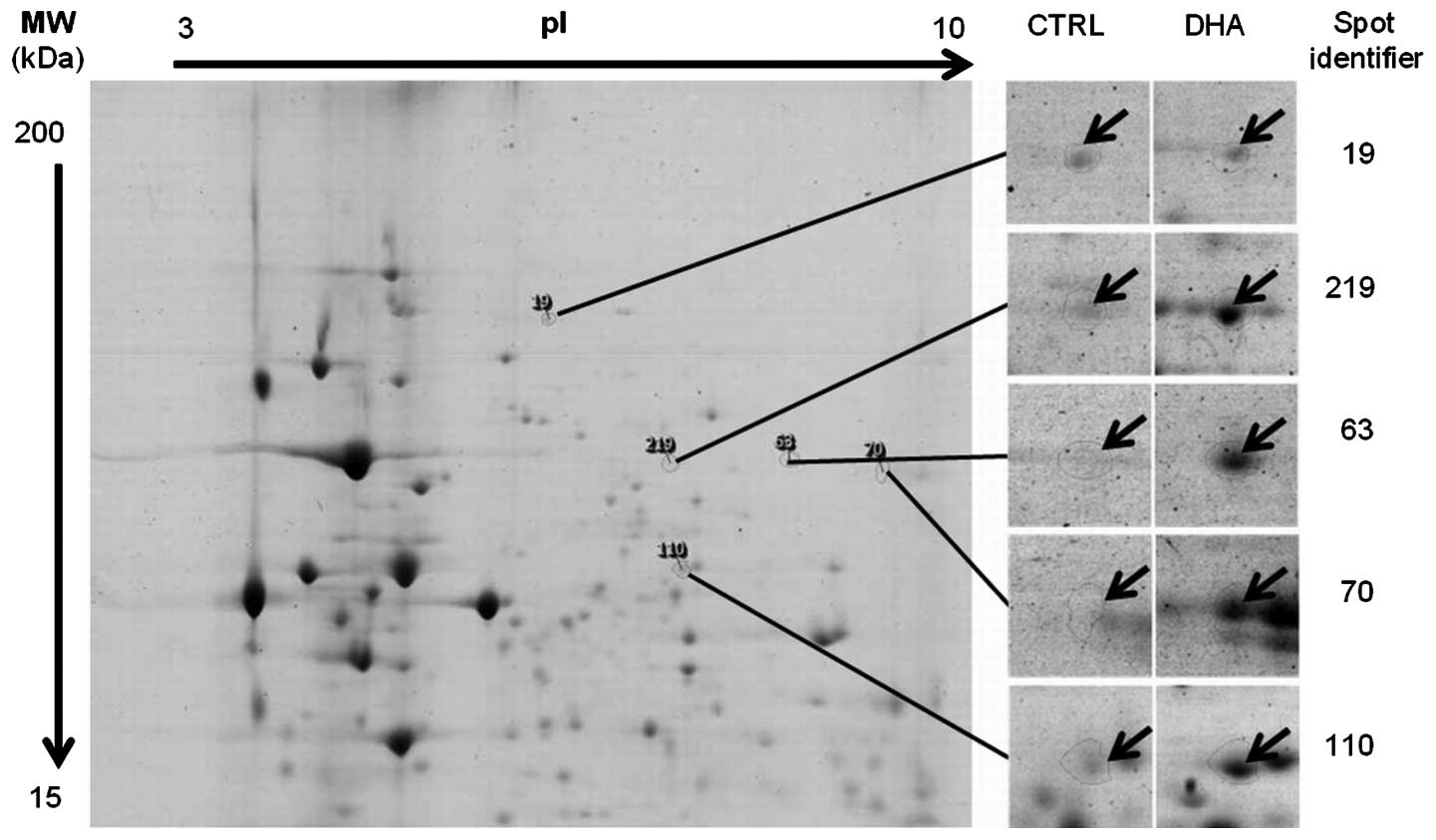
1.5-fold differential expression pattern (Fig. 1). Four proteins were found
upregulated by DHA treatment compared to the control, and one
protein was down-regulated (Table
I). Mass spectrometry for spot no. 19 was not possible due to
insufficient protein quantity and for spot no. 63 it was a mixture
of proteins. Spots nos. 70, 110 and 219 were identified (Fig. 2; Table II) by microsequencing using
coupled mass spectrometry as being KRT1 (UniProt ID P04264),
catalase (UniProt ID P04040) and lamin-A/C (UniProt ID P02545),
respectively.
 | Table IUpregulated proteins in DHA-treated
MDA-MB-231 cells. Cells were treated for 24 h with 100 μM DHA
before membrane protein preparation, 2-DE and identification in
mass spectrometry. |
Table I
Upregulated proteins in DHA-treated
MDA-MB-231 cells. Cells were treated for 24 h with 100 μM DHA
before membrane protein preparation, 2-DE and identification in
mass spectrometry.
| Spot
identifier | Fold | Regulation | ANOVA P-value | Protein name | UniProt ID |
|---|
| 70 | 2.0 | Up | 0.017 | KRT1 | P04264 |
| 110 | 1.7 | Up | 0.008 | Catalase | P04040 |
| 219 | 1.9 | Up | 0.022 | Lamin-A/C | P02545 |
 | Table IICharacteristics of the different
peptides for each protein identified in mass spectrometry. |
Table II
Characteristics of the different
peptides for each protein identified in mass spectrometry.
| UniProt ID | MW (kDa) | pI | Score mascot | MS1a coverage (%) | Peptide sequence
MS2b | Score MS2b | MS2b coverage (%) |
|---|
| P04264 | 65.9 | 8.1 | 65.8 | 31.7 |
WELLQQVDTSTR
THNLEPYFESFINNLR | 62.4 | 4.4 |
| P04040 | 59.7 | 6.9 | 110 | 44.8 | LFAYPDTHR
LGPNYLHIPVNCPYR
AFYVNVLNEEQR | 116.4 | 6.8 |
| P02545 | 74.1 | 6.6 | 107 | 37.5 |
LQEKEDLQELNDR
NSNLVGAAHEELQQSR | 62 | 4.4 |
DHA-induced KRT1 protein upregulation and
inhibition by siRNA
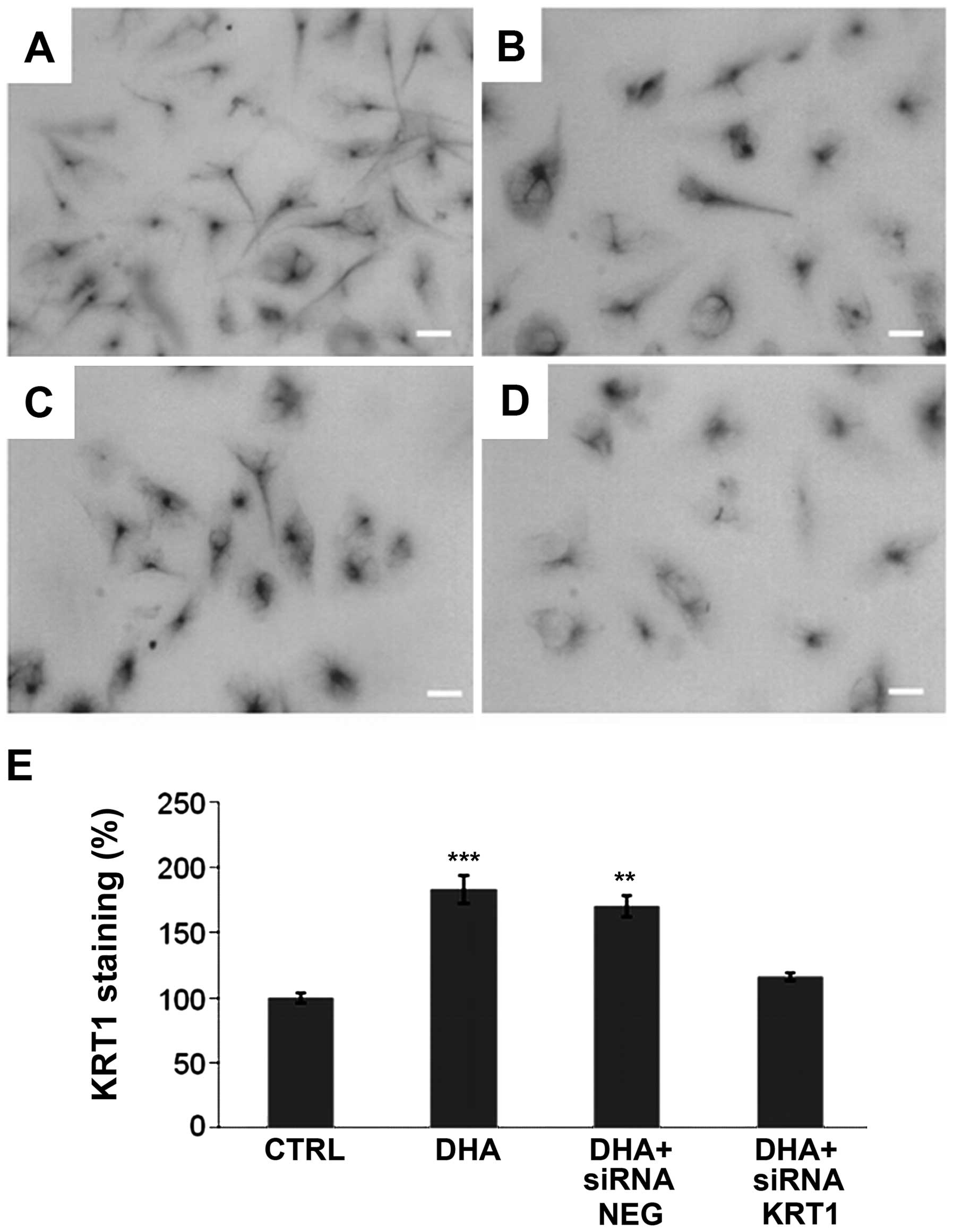
The increase of KRT1 in DHA-treated cells was
confirmed by immunocytochemistry (Fig.
3). KRT1 was present in the cytoplasm of untreated (Fig. 3A) and treated (Fig. 3B–D) cells. KRT1 labeling was
quantified by using Quantity One software (Bio-Rad). The result was
then subtracted by the blank of a similar surface without cells and
divided by the number of nucleus present in the field. Thus, the
average KRT1 quantification corresponded to a single cell expressed
as a percentage (Fig. 3E). The
result indicated that control siRNA (NEG) had no effect on the
level of KRT1 in the DHA-treated cells and in the control. In the
cells treated with DHA and siRNA against KRT1, we observed a level
of KRT1 that was reduced nearly to what was observed in the control
(Fig. 3E).
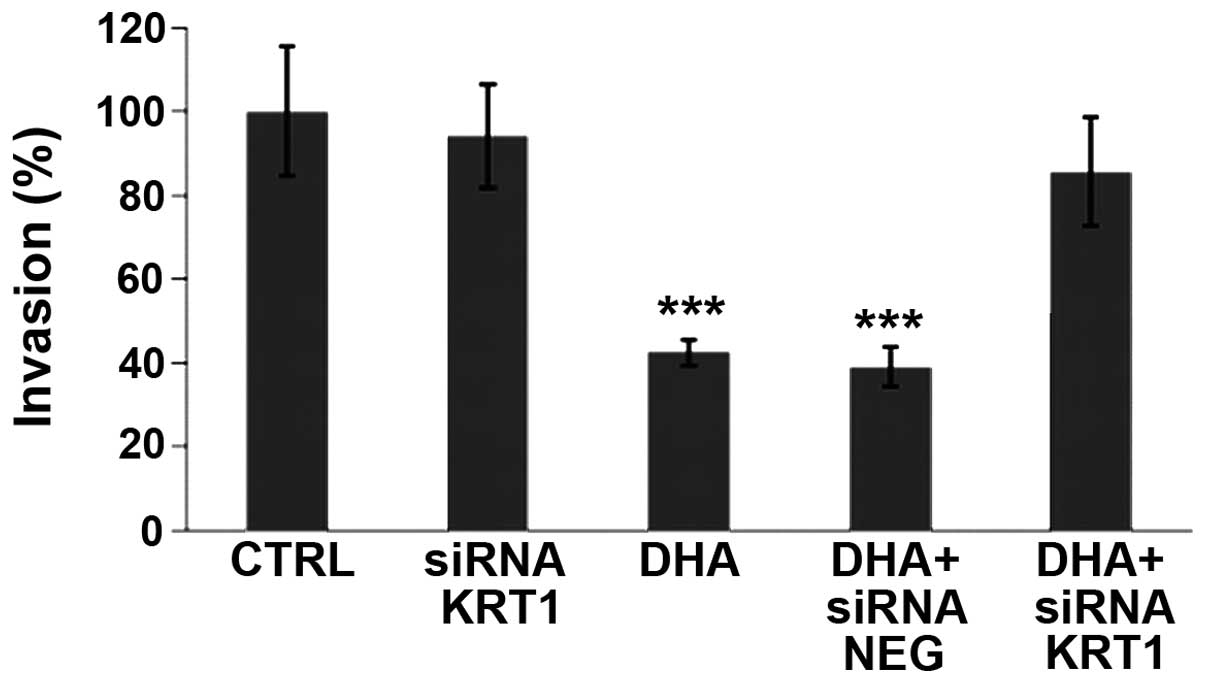
DHA-induced decrease of breast cancer
cell invasiveness is reversed by siRNA against KRT1
Using Matrigel® in Boyden chambers, DHA
was shown to reduce the invasive potential of MDA-MB-231 cells
(Fig. 4). In order to investigate
the role of the differentially expressed KRT1 protein, siRNAs were
used in the invasion assay in presence or absence of DHA. siRNA
against KRT1 was able to restore a percentage of invasive cells
similar to the control level while the control siRNA NEG was
inefficient (Fig. 4). This
indicated the involvement of KRT1 in the DHA-induced decrease of
MBA-MB-231 cell invasiveness.
Discussion
The pro-apoptotic effect of DHA on cancer cells and
especially in breast cancer is well known (11). However, few studies have also shown
the inhibitory effect of DHA on the metastatic and invasive
potential of cancer cells (9,27,28).
In order to identify membrane proteins from MDA-MB-231 that could
be involved, a 2-DE-based proteomic analysis was performed with
crude membrane preparations. This allowed the identification of
three differentially expressed proteins after DHA treatment.
Besides, it appeared that these proteins are well expressed in
differentiated cells and it is possible that the effects observed
could be due to the DHA effect on cell differentiation as reported
by Siddiqui et al (29).
Interestingly, lamin-A/C, a protein of the nuclear envelope, was
present and upregulated in the crude membrane extract from
DHA-treated cells. In stage II and III colon cancer patients, low
expression of lamin-A/C was associated with an increased disease
recurrence (30). In breast
cancer, it has been shown that higher lamin-A/C expression is
associated with: i) early clinical stage; ii) a better clinical
outcomes; and iii) a better overall and disease-free survival,
suggesting a significant role for nuclear and chromosomal stability
in this pathology (31).
Therefore, an increased in lamin-A/C expression appears to be
related to a less aggressive phenotype of breast cancer cells, and
our results are well in range with this notion by showing that the
inhibition of MDA-MB-231 invasiveness induced by DHA is accompanied
by an increase in lamin-A/C.
The enzyme catalase was also upregulated after DHA
treatment, which is in agreement with its protective role against
reactive oxygen species (ROS) and the induction of apoptosis by DHA
(32,33). Moreover, ROS can induce cell
migration and invasion (34,35),
and their impact is well established in the migration process
triggered by growth factors able to activate tyrosine kinase
receptors and MAPK (36). It has
been shown that the lysyl oxidase (LOX) facilitates the invasion of
MDA-MB-231 cells and that the removal of hydrogen peroxide leads to
a dose-dependent loss in Src activation (37). Consequently, LOX was shown to
facilitate migration and cell-matrix adhesion in invasive breast
cancer cells through a hydrogen peroxide-mediated mechanism
involving the FAK/Src signaling pathway (37). It has been shown that an increase
in catalase results in a decreased ROS level close to the plasma
membrane and leads to a reduction of migration and invasion
(38). Consequently, the increased
level of catalase observed in MDA-MB-231 crude membranes is well in
range with other studies and is related to a decreased
invasiveness.
Our study reports that KRT1 is induced upon
stimulation of cancer cells by DHA. siRNA against KRT1 was able to
reduce the de novo expression of KRT1 induced by DHA
treatment in MDA-MB-231 cells, then leading to the reacquisition of
an invasive potential. It has been shown that DHA is able to
selectively alter the subcellular distribution of lipidated
cytosolic proteins, including Ras isoforms, by modifying membrane
lipid composition (39),
indicating that KRT1 can be associated with membrane proteins. In
addition, KRT1 was shown to interact with the tyrosine kinase Src
through binding to integrin β1 (23) and therefore the presence of KRT1 in
a crude membrane preparation is not surprising. It has been shown
that KRT1 level is strongly decreased in breast cancer cells
reaching a metastatic phenotype (40). In addition, a recent study has
shown that KRT1 is decreased in breast tumors (41). In the same study, KRT1 was also
found to be released in sera concomitantly with a 130 kDa
epithelial membrane antigen (EMA) and the EMA/CK1 ratio was
correlated with more aggressive tumor types. Therefore, KRT1
expression is associated with a less aggressive phenotype of breast
cancer and our results suggest a mechanism involving the inhibition
of cancer cell invasiveness.
An indirect interaction between KRT1 and Src was
previously reported (23) as well
as the interaction between Src and the membrane protein
Nav1.5, a sodium ion channel protein encoded by the
SCN5A in humans (24,42). Inactivation of Nav1.5 is
known to induce a loss of invasion capacity in triple-negative
highly metastatic breast cancer cells (18,43).
Then it is conceivable that the overexpression of KRT1 observed
following DHA treatment may lead to KRT1 interaction with Src and
then to Nav1.5, but further experiments are needed to
elucidate this hypothesis and define the precise mechanisms linking
KRT1 and tumor cell invasion.
In conclusion, this proteomics-based study provides
new mechanistic insights into the activity of DHA in breast cancer
cells and in particular identifies KRT1 upregulation as being
involved in the DHA-induced inhibition of breast cancer cell
invasion.
Acknowledgements
We thank Dr S. Duban-Deweer for mass spectrometry
analyses (Centre d’Analyse Protéomique de l’Artois). The mass
spectrometry facility used for this study was funded by the
European Union (FEDER), the Fonds d’Industrialisation du Bassin
Minier (FIBM), the Ministère de l’Enseignement Supérieur et de la
Recherche and l’Université d’Artois. This study was supported by a
Grant from the Mayenne Council.
References
|
1
|
Larsson SC, Kumlin M, Ingelman-Sundberg M
and Wolk A: Dietary long-chain n-3 fatty acids for the prevention
of cancer: A review of potential mechanisms. Am J Clin Nutr.
79:935–945. 2004.PubMed/NCBI
|
|
2
|
Doughman SD, Krupanidhi S and Sanjeevi CB:
Omega-3 fatty acids for nutrition and medicine: Considering
microalgae oil as a vegetarian source of EPA and DHA. Curr Diabetes
Rev. 3:198–203. 2007. View Article : Google Scholar
|
|
3
|
Poudyal H, Panchal SK, Diwan V and Brown
L: Omega-3 fatty acids and metabolic syndrome: Effects and emerging
mechanisms of action. Prog Lipid Res. 50:372–387. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Holub BJ: Docosahexaenoic acid (DHA) and
cardiovascular disease risk factors. Prostaglandins Leukot Essent
Fatty Acids. 81:199–204. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Berquin IM, Edwards IJ and Chen YQ:
Multi-targeted therapy of cancer by omega-3 fatty acids. Cancer
Lett. 269:363–377. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bougnoux P, Hajjaji N, Maheo K, Couet C
and Chevalier S: Fatty acids and breast cancer: Sensitization to
treatments and prevention of metastatic re-growth. Prog Lipid Res.
49:76–86. 2010. View Article : Google Scholar
|
|
7
|
Holmes MD and Willett WC: Does diet affect
breast cancer risk? Breast Cancer Res. 6:170–178. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Barascu A, Besson P, Le Floch O, Bougnoux
P and Jourdan ML: CDK1-cyclin B1 mediates the inhibition of
proliferation induced by omega-3 fatty acids in MDA-MB-231 breast
cancer cells. Int J Biochem Cell Biol. 38:196–208. 2006. View Article : Google Scholar
|
|
9
|
Blanckaert V, Ulmann L, Mimouni V, Antol
J, Brancquart L and Chénais B: Docosahexaenoic acid intake
decreases proliferation, increases apoptosis and decreases the
invasive potential of the human breast carcinoma cell line
MDA-MB-231. Int J Oncol. 36:737–742. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Stoll BA: Breast cancer and the western
diet: Role of fatty acids and antioxidant vitamins. Eur J Cancer.
34:1852–1856. 1998. View Article : Google Scholar
|
|
11
|
Chénais B and Blanckaert V: The janus face
of lipids in human breast cancer: How polyunsaturated fatty acids
affect tumor cell hallmarks. Int J Breast Cancer. 2012:7125362012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Stillwell W and Wassall SR:
Docosahexaenoic acid: Membrane properties of a unique fatty acid.
Chem Phys Lipids. 126:1–27. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Conn EM, Botkjaer KA, Kupriyanova TA,
Andreasen PA, Deryugina EI and Quigley JP: Comparative analysis of
metastasis variants derived from human prostate carcinoma cells:
Roles in intravasation of VEGF-mediated angiogenesis and
uPA-mediated invasion. Am J Pathol. 175:1638–1652. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Brooks SA, Lomax-Browne HJ, Carter TM,
Kinch CE and Hall DM: Molecular interactions in cancer cell
metastasis. Acta Histochem. 112:3–25. 2010. View Article : Google Scholar
|
|
15
|
Buxton IL, Yokdang N and Matz RM:
Purinergic mechanisms in breast cancer support intravasation,
extravasation and angio-genesis. Cancer Lett. 291:131–141. 2010.
View Article : Google Scholar :
|
|
16
|
Schley PD, Brindley DN and Field CJ: (n-3)
PUFA alter raft lipid composition and decrease epidermal growth
factor receptor levels in lipid rafts of human breast cancer cells.
J Nutr. 137:548–553. 2007.PubMed/NCBI
|
|
17
|
Raghu H, Sodadasu PK, Malla RR, Gondi CS,
Estes N and Rao JS: Localization of uPAR and MMP-9 in lipid rafts
is critical for migration, invasion and angiogenesis in human
breast cancer cells. BMC Cancer. 10:647–664. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Isbilen B, Fraser SP and Djamgoz MB:
Docosahexaenoic acid (omega-3) blocks voltage-gated sodium channel
activity and migration of MDA-MB-231 human breast cancer cells. Int
J Biochem Cell Biol. 38:2173–2182. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gillet L, Roger S, Bougnoux P, Le Guennec
JY and Besson P: Beneficial effects of omega-3 long-chain fatty
acids in breast cancer and cardiovascular diseases: Voltage-gated
sodium channels as a common feature? Biochimie. 93:4–6. 2011.
View Article : Google Scholar
|
|
20
|
Maguy A, Hebert TE and Nattel S:
Involvement of lipid rafts and caveolae in cardiac ion channel
function. Cardiovasc Res. 69:798–807. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sánchez-Bailón MP, Calcabrini A,
Gómez-Domínguez D, Morte B, Martín-Forero E, Gómez-López G,
Molinari A, Wagner KU and Martín-Pérez J: Src kinases catalytic
activity regulates proliferation, migration and invasiveness of
MDA-MB-231 breast cancer cells. Cell Signal. 24:1276–1286. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Planas-Silva MD, Bruggeman RD, Grenko RT
and Smith JS: Role of c-Src and focal adhesion kinase in
progression and metastasis of estrogen receptor-positive breast
cancer. Biochem Biophys Res Commun. 341:73–81. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chuang NN and Huang CC: Interaction of
integrin beta1 with cytokeratin 1 in neuroblastoma NMB7 cells.
Biochem Soc Trans. 35:1292–1294. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Andavan B, Shankar G and Lemmens-Gruber R:
Modulation of Nav 1.5 variants by src tyrosine kinase.
Biophys J. 98:310a2010. View Article : Google Scholar
|
|
25
|
Venkateswaran S, Blanckaert V and
Schelling M: Membrane fragments from cultured endothelial cells for
use in screening anti-FGF receptor antibodies. Methods Cell Sci.
14:159–162. 1992.
|
|
26
|
Wilmet JP, Tastet C, Desruelles E,
Ziental-Gelus N, Blanckaert V, Hondermarck H and Le Bourhis X:
Proteome changes induced by overexpression of the p75 neurotrophin
receptor (p75NTR) in breast cancer cells. Int J Dev Biol.
55:801–809. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
D’Eliseo D, Manzi L, Merendino N and
Velotti F: Docosahexaenoic acid inhibits invasion of human RT112
urinary bladder and PT45 pancreatic carcinoma cells via
down-modulation of granzyme B expression. J Nutr Biochem.
23:452–457. 2012. View Article : Google Scholar
|
|
28
|
Mandal CC, Ghosh-Choudhury T, Yoneda T,
Choudhury GG and Ghosh-Choudhury N: Fish oil prevents breast cancer
cell metastasis to bone. Biochem Biophys Res Commun. 402:602–607.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Siddiqui RA, Harvey KA, Walker C,
Altenburg J, Xu Z, Terry C, Camarillo I, Jones-Hall Y and Mariash
C: Characterization of synergistic anti-cancer effects of
docosahexaenoic acid and curcumin on DMBA-induced mammary
tumorigenesis in mice. BMC Cancer. 13:418–434. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Belt EJ, Fijneman RJ, van den Berg EG,
Bril H, Delis-van Diemen PM, Tijssen M, van Essen HF, De Lange-de
Klerk ES, Beliën JA, Stockmann HB, et al: Loss of lamin A/C
expression in stage II and III colon cancer is associated with
disease recurrence. Eur J Cancer. 47:1837–1845. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wazir U, Ahmed MH, Bridger JM, Harvey A,
Jiang WG, Sharma AK and Mokbel K: The clinicopathological
significance of lamin A/C, lamin B1 and lamin B receptor mRNA
expression in human breast cancer. Cell Mol Biol Lett. 18:595–611.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yu L, Wan F, Dutta S, Welsh S, Liu Z,
Freundt E, Baehrecke EH and Lenardo M: Autophagic programmed cell
death by selective catalase degradation. Proc Natl Acad Sci USA.
103:4952–4957. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bechtel W and Bauer G: Catalase protects
tumor cells from apoptosis induction by intercellular ROS
signaling. Anticancer Res. 29:4541–4557. 2009.PubMed/NCBI
|
|
34
|
Richard D, Hollender P and Chénais B:
Butyric acid increases invasiveness of HL-60 leukemia cells: Role
of reactive oxygen species. FEBS Lett. 518:159–163. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Richard D, Hollender P and Chénais B:
Involvement of reactive oxygen species in aclarubicin-induced
differentiation and invasiveness of HL-60 leukemia cells. Int J
Oncol. 21:393–399. 2002.PubMed/NCBI
|
|
36
|
Hurd TR, DeGennaro M and Lehmann R: Redox
regulation of cell migration and adhesion. Trends Cell Biol.
22:107–115. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Payne SL, Fogelgren B, Hess AR, Seftor EA,
Wiley EL, Fong SF, Csiszar K, Hendrix MJ and Kirschmann DA: Lysyl
oxidase regulates breast cancer cell migration and adhesion through
a hydrogen peroxide-mediated mechanism. Cancer Res. 65:11429–11436.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Nikitovic D, Corsini E, Kouretas D,
Tsatsakis A and Tzanakakis G: ROS-major mediators of extracellular
matrix remodeling during tumor progression. Food Chem Toxicol.
61:178–186. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Seo J, Barhoumi R, Johnson AE, Lupton JR
and Chapkin RS: Docosahexaenoic acid selectively inhibits plasma
membrane targeting of lipidated proteins. FASEB J. 20:770–772.
2006.PubMed/NCBI
|
|
40
|
DeAngelis JT, Li Y, Mitchell N, Wilson L,
Kim H and Tollefsbol TO: 2D difference gel electrophoresis analysis
of different time points during the course of neoplastic
transformation of human mammary epithelial cells. J Proteome Res.
10:447–458. 2011. View Article : Google Scholar
|
|
41
|
Attallah AM, El-Far M, Omran MM, Abdallah
SO, El-Desouky MA, El-Dosoky I, Abdelrazek MA, Attallah AA,
Elweresh MA, Abdel Hameed GE, et al: Circulating levels and
clinical implications of epithelial membrane antigen and
cytokeratin-1 in women with breast cancer: Can their ratio improve
the results? Tumour Biol. 35:10737–10745. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Rohl CA, Boeckman FA, Baker C, Scheuer T,
Catterall WA and Klevit RE: Solution structure of the sodium
channel inactivation gate. Biochemistry. 38:855–861. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yang M, Kozminski DJ, Wold LA, Modak R,
Calhoun JD, Isom LL and Brackenbury WJ: Therapeutic potential for
phenytoin: Targeting Na(v)1.5 sodium channels to reduce migration
and invasion in metastatic breast cancer. Breast Cancer Res Treat.
134:603–615. 2012. View Article : Google Scholar : PubMed/NCBI
|