Introduction
Hepatocellular carcinoma (HCC) is the most common
primary malignancy of the liver and the third most common cause of
cancer-related death worldwide (1,2).
Development of HCC is considered as a discriminative event because
it occurs in chronically damaged tissue due to chronic hepatitis
and liver cirrhosis, whereas other common malignancies develop on
otherwise healthy tissue (3–5).
Because of the accumulated genome instability and numerous
epigenetic alterations induced by the microenvironment of the
background liver, HCC is a more heterogeneous disease (3).
Aberrant DNA methylation is one of the most common
epigenetic alterations in malignancies and is specific to
individual organs and diseases (6–8).
Furthermore, several studies have shown that aberrant DNA
methylation contributes to the initiation and progression of
malignant tumors through inactivation of tumor suppressors
(9,10). Therefore, identification of novel
methylated genes is important for the development of both
diagnostic markers and therapeutic targets, such as demethylation
agents.
Kallmann syndrome-1 gene (KAL1), also named
anosmin-1, encodes an extracellular matrix (ECM) related protein
with a role in cellular adhesion. KAL1 contains a WAP domain and
three FnIII domains, and promotes the migration of
gonadotropin-releasing hormone neurons from the olfactory placode
to the hypothalamus during development (11–13).
KAL1 also induces neurite outgrowth and cell migration through
fibroblast growth factor receptor 1 (FGFR1) pathways (14,15).
Studies have demonstrated that ECM proteins play a vital role in
proliferation and invasion of tumor cells (16). However, to date, conflicting
results have been reported regarding the oncological role of KAL1.
Decreased KAL1 expression is observed in colon, lung, and ovarian
cancers compared with corresponding adjacent normal tissues
(17). Conversely, KAL1
overexpression promotes brain tumor malignancy through integrin
signal pathways and facilitates colon cancer cell migration and
anti-apoptotic capacity (15).
These studies indicate that KAL1 exhibits diverse functions in
cancer initiation and progression. To the best of our knowledge,
there have been no studies of expression analysis of KAL1 in HCC.
Moreover, although loss-of-function mutations of the KAL1 gene have
been known to underlie Kallmann syndrome (18,19),
the significance of the methylation status of the KAL1 gene has yet
to be determined.
In our previous microarray project exploring
HCC-related tumor suppressors, we found that KAL1 was downregulated
in HCC tissues (Log2 ratio: −2.1) (16,20–22).
Accordingly, we hypothesized that KAL1 might act as a putative
tumor suppressor and mediate tumorigenesis of HCC. To
systematically address this idea, we examined the expression and
methylation status of KAL1 in HCC.
Materials and methods
Sample collection
We purchased nine HCC cell lines from the American
Type Culture Collection (Manassas, VA, USA) and cultured cells as
previously described (23).
Primary HCC and adjacent liver tissues were collected from 144
patients who underwent hepatectomy for HCC at Nagoya University
Hospital between January 1998 and January 2012. The ages of the 144
patients ranged from 34 to 84 years (median, 65.5 years), and the
male-to-female ratio was 121:23. The median duration of patient
follow-up was 40.1 months (range, 2.3–145 months). Thirty-seven
were infected with hepatitis B and 80 patients were infected with
hepatitis C virus. Ten patients had normal liver, 82 patients had
chronic hepatitis, and 52 patients showed cirrhosis. Ninety, 37,
and 17 patients were in stages I, II, and III, respectively.
Tissue samples were frozen immediately
after resection and stored at −80°C until use
Genomic DNA and total RNA was extracted from both
HCC and adjacent noncancerous tissues approximately 5
mm2 in diameter, avoiding necrotic areas. Specimens were
classified histologically according to the 7th edition of the Union
for International Cancer Control (24). Written informed consent for the use
of clinical samples and data was obtained from all enrolled
patients as required by the Institutional Review Board of Nagoya
University, Japan.
Analysis of the KAL1 promoter region
Nucleotide sequencing was used to determine the
presence of CpG islands in the KAL1 promoter region, defined as
follows: ≥200 bp region with GC content >50% and an observed
CpG/expected CpG ratio ≥0.6 (25).
CpG Island Searcher software (http://cpgis-lands.usc.edu/) was used to determine the
locations of CpG islands (26).
Quantitative real-time reverse
transcription-polymerase chain reaction (qRT-PCR)
KAL1 mRNA levels were determined using qRT-PCR.
Total RNA (10 μg) was isolated from nine HCC cell lines, 144
primary HCCs, and adjacent non-cancerous tissues and used as a
template for complementary DNA synthesis.
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA (TaqMan,
GAPDH control reagents, Applied Biosystems, Foster City, CA, USA)
was quantified in each sample for standardization. Specific primers
and annealing temperatures are listed in Table I. qRT-PCR was performed using the
SYBR Green PCR Core Reagents kit (Applied Biosystems) as follows:
one cycle at 95°C for 10 min, 40 cycles at 95°C for 5 sec, and 60°C
for 60 sec. Real-time detection of SYBR Green fluorescence was
conducted using an ABI StepOnePlus Real-Time PCR System (Applied
Biosystems). All samples were analyzed in triplicate. The
expression level of each sample is shown as the value of the KAL1
amplicon divided by that of GAPDH (27).
 | Table IPrimers and annealing
temperatures. |
Table I
Primers and annealing
temperatures.
| Gene | Experiment | Type | Sequence
(5′-3′) | Product size
(bp) | Annealing
temperature (°C) |
|---|
| KAL1 | qRT-PCR | Forward |
AACAATGGTTCCCTGGTTTG | 110 | 60 |
| Reverse |
TCACAAAAGCTTTGGCACTG |
| MSP | Forward |
GTGCGAACGGGAGAGGC | 109 | 68 |
| Reverse |
GTCAACTACGAACCCGAACG |
| U-MSP | Forward |
AAAACCCATAAACCAATCTCA | 126 | 58 |
| Reverse |
TGAATGGGAGAGGTGTTTGT |
| Bisulfite
sequencing | Forward |
TATTGGGAGGGAGTTTGGGA | 411 | 66 |
| Reverse | TAC TCC CCA CCC TCA
AAC TA |
| EZR | qRT-PCR | Forward |
GATAGTCGTGTTTTCGGGGA | 91 | 60 |
| Reverse |
CTCTGCATCCATGGTGGTAA |
| FAK | qRT-PCR | Forward |
GCCAAAAGGATTTCTAAACCAG | 110 | 64 |
| Reverse |
CCTGGTCCACTTGATCAGCTA |
| SRC | qRT-PCR | Forward |
CTGACCGCATGGACCGT | 107 | 58 |
| Reverse |
AAGCCAACCTGTCACTTGGTA |
| DPYSL3 | qRT-PCR | Forward |
AGAAGAAGGAGGGAGGGAGC | 110 | 60 |
| Reverse |
CTCCCTTGATAAGGAGACGG |
| GAPDH | qRT-PCR | Forward |
GAAGGTGAAGGTCGGAGTC | 226 | 60 |
| Probe |
CAAGCTTCCCGTTCTCAGCC |
| Reverse |
GAAGATGGTGATGGGATTTC | |
Methylation-specific PCR (MSP) and
bisulfite sequence analysis
Genomic DNA samples from nine HCC cell lines and 144
HCC tissues were subjected to bisulfite treatment. MSP was
conducted to determine the presence or absence of promoter
hypermethylation of KAL1 gene. Bisulfite DNA from HCC cell lines
was sequenced to determine the reliability of MSP results. Primer
sequences are shown in Table
I.
5-Aza-2′-deoxycytidine (5-aza-dC)
treatment
To assess the relation of promoter hypermethylation
to KAL1 transcription, HCC cell lines were treated with the DNA
methylation inhibitor 5-aza-dC (Sigma-Aldrich, St. Louis, MO, USA)
as previously described (10,28).
Expression of genes that encode cell
adhesion factors
To identify cell adhesion proteins that may interact
with KAL1, expression levels of Ezrin (EZR), focal adhesion kinase
(FAK), cellular SRC (SRC) and dihydropyrimidinase-like 3 (DPYSL3)
genes were determined by qRT-PCR in HCC cell lines (29,30).
Primers specific for EZR, FAK, SRC and DPYSL3 are listed in
Table I.
Immunohistochemical (IHC) staining
KAL1 protein localization was determined by IHC
using 64 representative formalin-fixed and paraffin-embedded
sections of well-preserved HCC tissue using a rabbit polyclonal
antibody against KAL1 (ABN486, Millipore, Darmstadt, Germany)
diluted 1:150 in antibody diluent (Dako, Glostrup, Denmark) as
previously described (7,31). Samples were then washed with
phosphate-buffered saline, followed by 10 min incubation with a
biotinylated secondary antibody (Histofine SAB-PO(R), Nichirei,
Tokyo, Japan). Sections were subsequently developed for 3 min using
3,3′-diaminobenzidine as substrate (Nichirei) and analyzed. To
avoid bias, specimens were randomized, coded, and then analyzed by
two independent observers who were uninformed of the identities of
the samples.
Statistical analysis
The qualitative χ2 test and quantitative
Mann-Whitney test were used to compare two groups. Correlations
between mRNA levels of KAL1 and those of EZR, FAK, SRC, or DPYSL3
as well as tumor size and preoperative serum protein induced by
vitamin K antagonists II (PIVKA-II) level were analyzed using the
Spearman rank correlation test. Overall and disease-free survival
rates were calculated using the Kaplan-Meier method, and the
difference in survival curves was evaluated using the log rank
test. A P-value <0.05 was considered statistically significant.
All statistical analysis was performed using JMP 10®
software (SAS Institute Inc., Cary, NC, USA).
Results
KAL1 mRNA expression and methylation
status in HCC cell lines
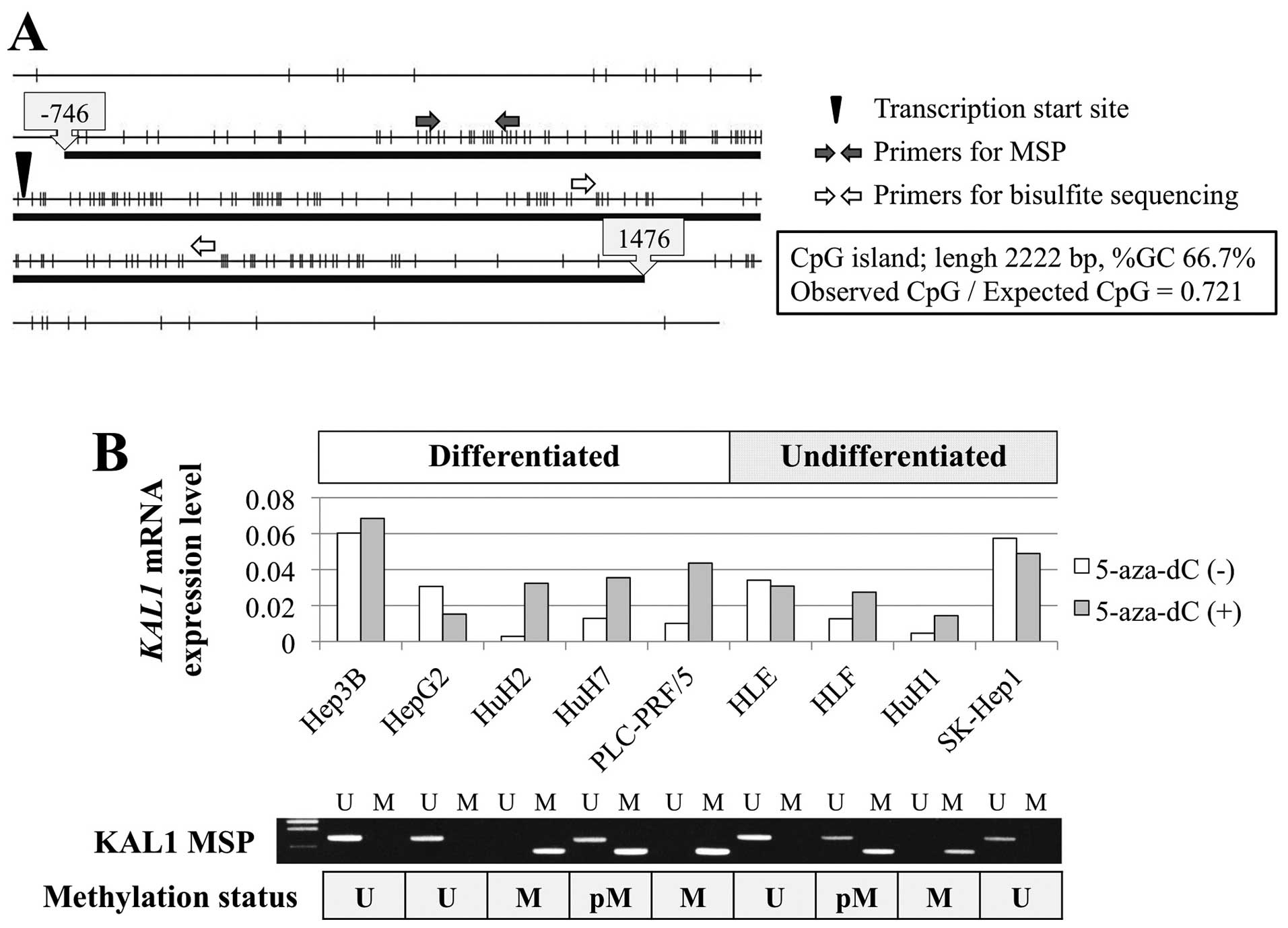
The KAL1 gene harbors a CpG island around the
promoter region (Fig. 1A),
suggesting that hypermethylation of the CpG island may regulate
KAL1 transcription. KAL1 mRNA expression levels were heterogeneous
among nine HCC cell lines, regardless of differentiation (Fig. 1B). MSP revealed methylation in HLF,
HuH1, HuH2, HuH7 and PLC/PRF/5 cells. When comparing the levels of
KAL1 mRNA in HCC cell lines before and after demethylation by
5-aza-dC treatment, reactivation of KAL1 mRNA expression was
observed in cells with promoter hypermethylation of the KAL1 gene
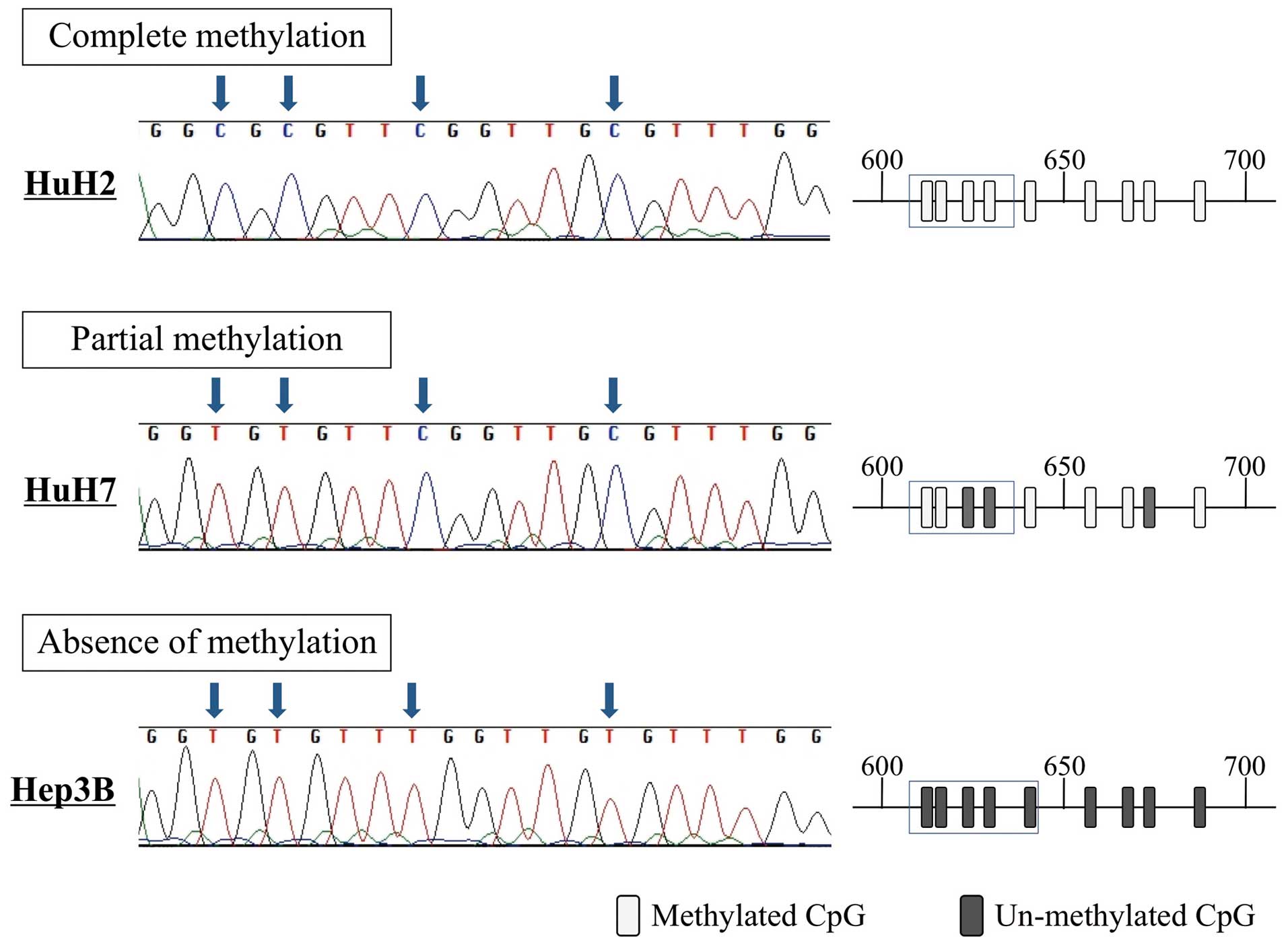
(Fig. 1B). Direct sequence
analysis revealed that all CpG sites in HuH2 cells (complete
methylation) were CG (cytosine and guanine), whereas the
corresponding positions in Hep3B cells (absence of methylation)
were TG (thymine and guanine) (Fig.
2). These results confirm the accuracy of the MSP results.
Expression analysis of KAL1 and genes
encoding putative functional partners in HCC cell lines
We next evaluated the expression levels of genes
encoding other cell adhesion factors that could potentially
functionally interact with KAL1. The relative expression levels of
EZR, FAK, SRC, DPYSL3, and KAL1 mRNAs in HCC cell lines are shown
in Fig. 3A. The results showed
that KAL1 mRNA levels inversely correlated with those of EZR
(correlation coefficient −0.667, P=0.049; Fig. 3B).
KAL1 status in surgically-resected
tissues
We next examined KAL1 mRNA levels in 144 HCC tissues
compared with the corresponding noncancerous liver tissues. Results
showed that KAL1 mRNA levels were lower in HCC tissues compared
with the corresponding noncancerous liver tissues in 106 (74%) of
144 patients. We next evaluated the association between expression
levels of KAL1 mRNA and protein. Results of IHC, qPCR and MSP in
representative patients are shown in Fig. 4A and B. One patient with reduced
KAL1 mRNA levels showed reduced expression of KAL1 protein in the
cytoplasm of HCC cells accompanied with promoter hypermethylation
(Fig. 4A). Equivalent expression
of KAL1 in cancer and normal cells was detected in a patient
without downregulation of KAL1 mRNA and methylation (Fig. 4B). The expression patterns of KAL1
in 64 patients correlated significantly with those of KAL1 mRNA
(P=0.023, Fig. 4C).
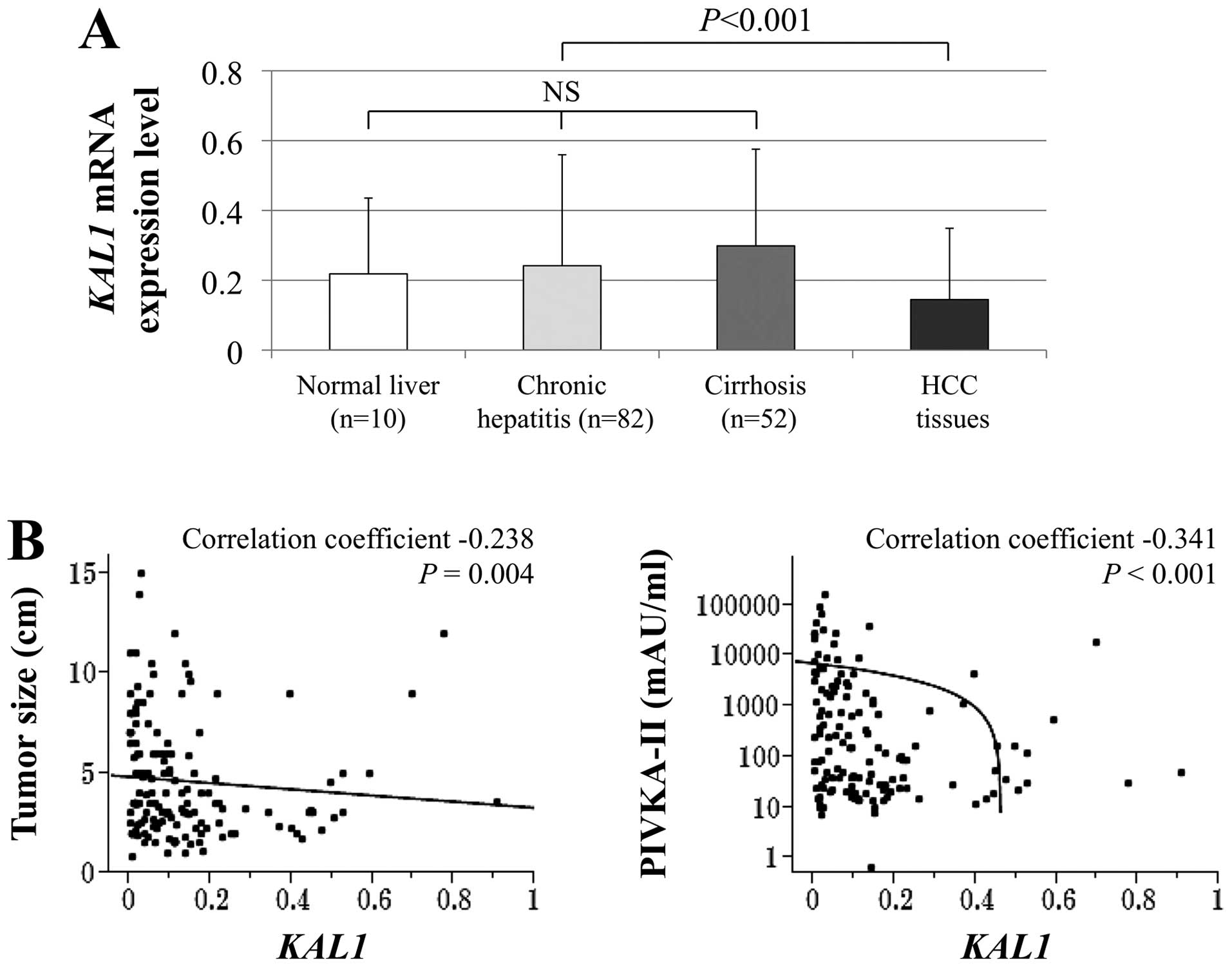
There were no significant differences in KAL1 mRNA
levels between normal liver, chronic hepatitis, and cirrhosis in
noncancerous liver tissues. In contrast, HCC tissues showed
significantly decreased KAL1 mRNA levels compared with the
corresponding noncancerous liver tissues (Fig. 5A). The KAL1 mRNA levels in HCCs
correlated inversely with tumor size and preoperative serum
PIVKA-II level (Fig. 5B). In 62
patients, KAL1 mRNA expression level in HCC was less than half of
that in the corresponding noncancerous liver tissue, and these
patients were categorized into the ‘downregulation of KAL1’ group
for the following analyses. Downregulation of KAL1 was
significantly associated with α-fetoprotein >20 ng/ml, PIVKA-II
>40 mAU/ml, tumor size ≥3.0 cm, moderate to poor
differentiation, formation of a capsule, vascular invasion, and
hypermethylation of KAL1 (Table
II).
 | Table IIAssociation between expression levels
of KAL1 mRNA and clinicopathological parameters in 144
patients with hepato-cellular carcinoma (HCC). |
Table II
Association between expression levels
of KAL1 mRNA and clinicopathological parameters in 144
patients with hepato-cellular carcinoma (HCC).
| Clinicopathological
parameters | Downregulation of
KAL1 mRNA in HCCs (n=62) | Others (n=82) | P-value |
|---|
| Age | | | 0.312 |
| <65 year | 25 | 40 | |
| ≥65 year | 37 | 42 | |
| Gender | | | 0.067 |
| Male | 56 | 65 | |
| Female | 6 | 17 | |
| Background
liver | | | 0.679 |
| Normal liver | 3 | 7 | |
| Chronic
hepatitis | 36 | 46 | |
| Cirrhosis | 23 | 29 | |
| Pugh-Child’s
classification | | | 0.075 |
| A | 55 | 79 | |
| B | 7 | 3 | |
| Hepatitis
virus | | | 0.329 |
| Absent | 15 | 12 | |
| HBV | 14 | 23 | |
| HCV | 33 | 47 | |
| AFP (ng/ml) | | | 0.004a |
| ≤20 | 25 | 53 | |
| >20 | 37 | 29 | |
| PIVKA II
(mAU/ml) | | | 0.002a |
| ≤40 | 16 | 42 | |
| >40 | 46 | 40 | |
| Tumor
multiplicity | | | 0.928 |
| Solitary | 48 | 64 | |
| Multiple | 14 | 18 | |
| Tumor size | | | 0.004a |
| <3.0 cm | 12 | 34 | |
| ≥3.0 cm | 50 | 48 | |
|
Differentiation | | | 0.015a |
| Well | 9 | 26 | |
| Moderate to
poor | 53 | 56 | |
| Growth type | | | 0.126 |
| Expansive
growth | 55 | 65 | |
| Invasive
growth | 7 | 17 | |
| Serosal
infiltration | | | 0.450 |
| Absent | 45 | 64 | |
| Present | 17 | 18 | |
| Formation of
capsule | | | 0.003a |
| Absent | 12 | 35 | |
| Present | 50 | 47 | |
| Infiltration to
capsule | | | 0.066 |
| Absent | 23 | 43 | |
| Present | 39 | 39 | |
| Septum
formation | | | 0.370 |
| Absent | 19 | 31 | |
| Present | 43 | 51 | |
| Vascular
invasion | | | 0.033a |
| Absent | 41 | 67 | |
| Present | 21 | 15 | |
| Hypermethylation of
KAL1 in HCCs | | | 0.019a |
| Absent | 32 | 58 | |
| Present | 30 | 24 | |
| UICC pathological
stage | | | 0.062 |
| I | 33 | 57 | |
| II | 22 | 15 | |
| III | 7 | 10 | |
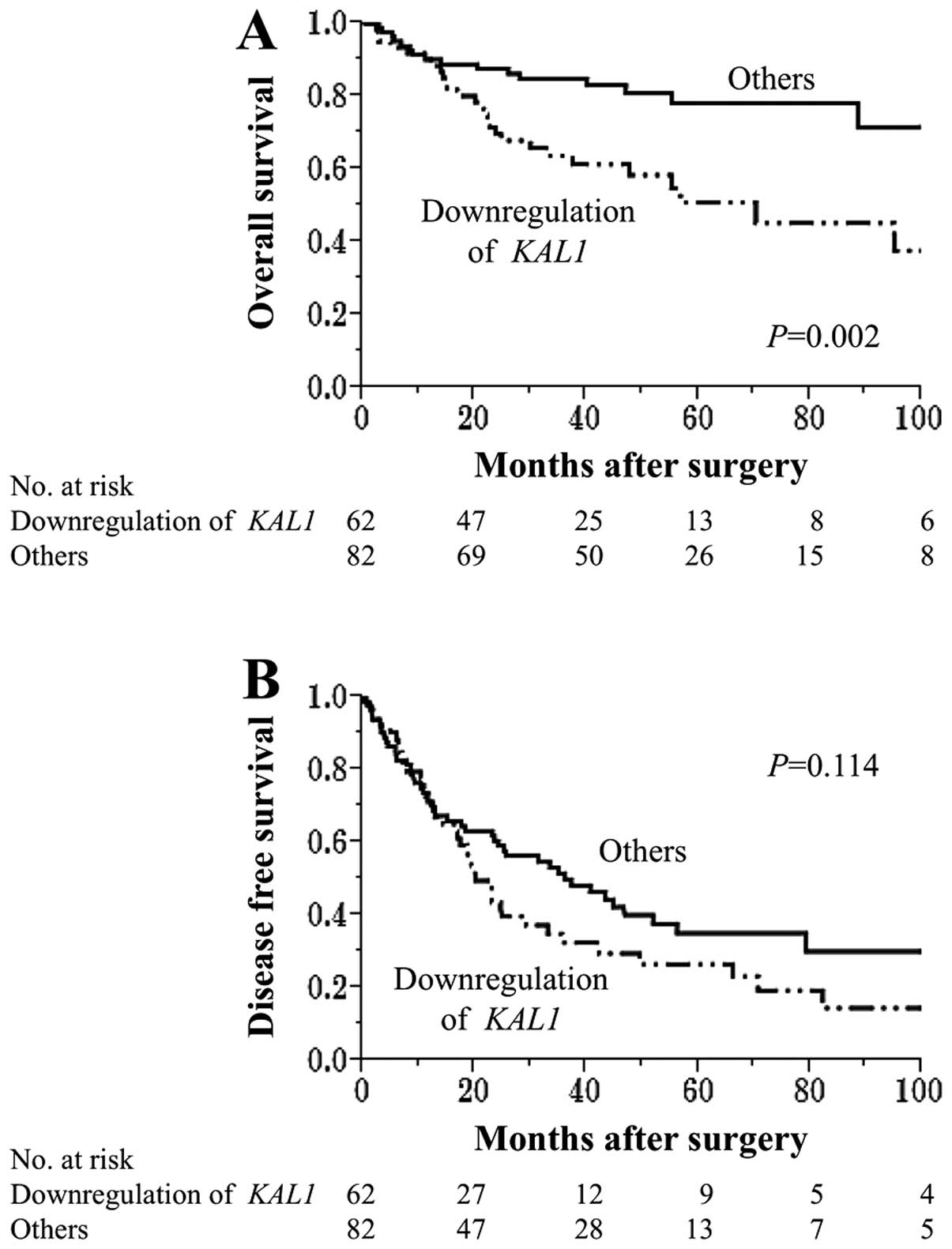
Impact of KAL1 mRNA expression on patient
outcome
Patients with downregulation of KAL1 were more
likely to have a shorter overall survival than other patients
(5-year survival rates 51% and 78%, respectively, P=0.002)
(Fig. 6A). In multivariate
analysis, downregulation of KAL1 was identified as an independent
prognostic factor (hazard ratio 2.04, 95% confidence interval
1.11–3.90, P=0.022; Table III).
Additionally, patients with downregulation of KAL1 tended to have a
shorter disease-free survival compared with other patients,
although it did not reach statistical significance (3-year survival
rates 32% and 50%, respectively, P=0.014) (Fig. 6B).
 | Table IIIPrognostic factors of 144 patients
with hepatocellular carcinoma (HCC) for overall survival. |
Table III
Prognostic factors of 144 patients
with hepatocellular carcinoma (HCC) for overall survival.
| | Univariate | Multivariate |
|---|
| |
|
|
|---|
| Variable | n | Hazard ratio | 95% CI | P-value | Hazard ratio | 95% CI | P-value |
|---|
| Age (≥65) | 79 | 1.75 | 0.96–3.30 | 0.068 | | | |
| Gender (male) | 121 | 1.82 | 0.78–5.29 | 0.178 | | | |
| Background liver
(cirrhosis) | 52 | 1.53 | 0.84–2.75 | 0.161 | | | |
| Pugh-Child’s
classification (B) | 10 | 1.68 | 0.50–4.19 | 0.360 | | | |
| AFP (>20
ng/ml) | 66 | 1.96 | 1.09–3.58 | 0.024a | 1.49 | 0.81–2.78 | 0.196 |
| PIVKA II (>40
mAU/ml) | 86 | 1.90 | 1.03–3.71 | 0.041a | 1.04 | 0.50–2.06 | 0.909 |
| Tumor multiplicity
(multiple) | 32 | 1.83 | 0.94–3.38 | 0.073 | | | |
| Tumor size (≥3.0
cm) | 98 | 2.84 | 1.38–6.64 | 0.004a | 1.95 | 0.88–4.86 | 0.103 |
| Tumor
differentiation (well) | 35 | 0.72 | 0.34–1.41 | 0.349 | | | |
| Growth type
(invasive growth) | 24 | 1.71 | 0.84–3.26 | 0.136 | | | |
| Serosal
infiltration | 35 | 2.23 | 1.16–4.11 | 0.017a | 1.70 | 0.87–3.18 | 0.115 |
| Formation of
capsule | 97 | 0.95 | 0.52–1.81 | 0.861 | | | |
| Infiltration to
capsule | 78 | 1.24 | 0.69–2.29 | 0.478 | | | |
| Septum
formation | 94 | 0.77 | 0.43–1.43 | 0.402 | | | |
| Vascular
invasion | 36 | 3.75 | 2.05–6.78 | <0.001a | 2.48 | 1.30–4.71 | 0.006a |
| Hypermethylation of
KAL1 in HCCs | 54 | 1.23 | 0.67–2.33 | 0.511 | | | |
| Downregulation of
KAL1 mRNA | 62 | 2.53 | 1.40–4.73 | 0.002a | 2.04 | 1.11–3.90 | 0.022a |
Discussion
Impaired expression of genes encoding ECM proteins
plays an important role in the initiation and progression of HCC
(16,32). KAL1, one of the ECM-related
proteins, has been reported to have diverse oncological functions
(15,17). In the present study, the clinical
significance of the expression and methylation status of KAL1 was
evaluated in HCC.
Consistent with earlier studies in colon, lung and
breast cancer (17), our results
showed that expression levels of KAL1 were reduced in HCC tissues
compared to adjacent noncancerous liver tissues. Furthermore, KAL1
expression was independent of chronic inflammation or fibrosis of
the background liver, suggesting that downregulation of KAL1 is a
specific event in hepatocarcinogenesis or at later stages.
Loss-of-function mutations in the KAL1 gene are responsible for
Kallmann syndrome, a developmental disorder characterized by the
association of hypogonadotropic hypogonadism and anosmia (14,18,19).
However, no studies have investigated the regulatory mechanisms of
KAL1 expression in malignancies. Since a CpG island was found in
the promoter region of the KAL1 gene, we focused on aberrant DNA
methylation, which is an important mechanism for inactivation of
tumor suppressors (33,34). Our results showed that HCC cell
lines with profoundly suppressed KAL1 expression also harbored
promoter hypermethylation of the KAL1 gene, and expression levels
of KAL1 were restored by demethylation. Additionally, there was a
significant association between downregulation of KAL1 mRNA and
hypermethylation of KAL1 gene in surgically resected HCC tissues.
These findings implicated that aberrant methylation is a pivotal
regulatory mechanism for KAL1 expression in HCC. Promoter
hypermethylation of the KAL1 gene has the potential for becoming a
novel biomarker of HCC as well as a therapeutic target for specific
demethylation agents (34,35).
We also investigated the levels of other important
ECM-related proteins, and found that the expression level of KAL1
had a significant inverse association with EZR expression. EZR is a
cytoplasmic peripheral membrane protein that functions as a
substrate of protein tyrosine kinases, regulates cellular survival,
adhesion, migration, and invasion. Importantly, EZR is also one of
the key factors involved in tumor progression and metastasis in HCC
(36–39). Our finding supports the notion that
KAL1 may function through tumor suppressor mechanisms and led us to
speculate that KAL1 may interact with EZR and mediate tumorigenesis
of HCC.
The significant correlation between the IHC and
qRT-PCR data allowed us to evaluate the prognostic significance of
KAL1 mRNA levels in a quantitative manner. Downregulation of KAL1
mRNA in HCC was significantly associated with factors reflecting
the malignant potential of HCC and consequently deteriorated
patient outcomes after curative hepatectomy. In contrast to the
previous study showing that KAL1 overexpression promotes brain
tumor malignancy (15), our
results instead support a tumor suppressive role for KAL1 in
HCC.
KAL1 was first identified through its function in
the development of gonadotropin-releasing hormone neurons. Previous
studies demonstrated that the expression of KAL1 is modulated by
FGFR1 and hypoxia inducible factor-1α (HIF-1α) (11,14).
FGFR-1 expression was reported as low in normal liver epithelium
and high in human liver cancer epithelium (40,41).
FGFR-1 protein may be important in regulating cytoskeletal dynamics
and function in cancer cell invasion and metastatic behavior
(42). HIF-1α is an important
transcription factor in essential adaptive responses to hypoxia,
and plays a major role in the development of characteristic tumor
phenotypes, including growth rate, angiogenesis, invasiveness, and
metastasis, via activation of target genes by binding to
hypoxia-responsive elements in the gene regulatory sequences
(43–45). The interactions with these major
oncogenic pathways might provide a mechanism(s) underlying the
correlation between KAL1 expression and malignant phenotype of HCC.
Future studies, including pathway analysis in hepatocarcinogenesis,
hypoxic stress and functional analysis, are required to elucidate
the molecular mechanisms that underlie the biological function of
KAL1 in HCC.
Taken together, our results indicate that KAL1 acts
as a putative tumor suppressor in HCC that is inactivated by
promoter hypermethylation. Our findings suggest that KAL1 may serve
as a promising biomarker of malignant phenotype of HCC.
References
|
1
|
Siegel R, Ward E, Brawley O and Jemal A:
Cancer statistics, 2011: The impact of eliminating socioeconomic
and racial disparities on premature cancer deaths. CA Cancer J
Clin. 61:212–236. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Galuppo R, Ramaiah D, Ponte OM and Gedaly
R: Molecular therapies in hepatocellular carcinoma: What can we
target? Dig Dis Sci. 59:1688–1697. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Giannelli G, Rani B, Dituri F, Cao Y and
Palasciano G: Moving towards personalised therapy in patients with
hepatocellular carcinoma: The role of the microenvironment. Gut.
63:1668–1676. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kanda M, Nomoto S, Nishikawa Y, Sugimoto
H, Kanazumi N, Takeda S and Nakao A: Correlations of the expression
of vascular endothelial growth factor B and its isoforms in
hepatocellular carcinoma with clinicopathological parameters. J
Surg Oncol. 98:190–196. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
El-Serag HB: Hepatocellular carcinoma. N
Engl J Med. 365:1118–1127. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sawan C, Vaissière T, Murr R and Herceg Z:
Epigenetic drivers and genetic passengers on the road to cancer.
Mutat Res. 642:1–13. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kanda M, Sugimoto H, Nomoto S, Oya H,
Hibino S, Shimizu D, Takami H, Hashimoto R, Okamura Y, Yamada S, et
al: B-cell translocation gene 1 serves as a novel prognostic
indicator of hepatocellular carcinoma. Int J Oncol. 46:641–648.
2015.
|
|
8
|
Hernandez-Gea V, Toffanin S, Friedman SL
and Llovet JM: Role of the microenvironment in the pathogenesis and
treatment of hepatocellular carcinoma. Gastroenterology.
144:512–527. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Arzumanyan A, Reis HM and Feitelson MA:
Pathogenic mechanisms in HBV- and HCV-associated hepatocellular
carcinoma. Nat Rev Cancer. 13:123–135. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kanda M, Sugimoto H, Nomoto S, Oya H,
Shimizu D, Takami H, Hashimoto R, Sonohara F, Okamura Y, Yamada S,
et al: Clinical utility of PDSS2 expression to stratify patients at
risk for recurrence of hepatocellular carcinoma. Int J Oncol.
45:2005–2012. 2014.PubMed/NCBI
|
|
11
|
Soussi-Yanicostas N, de Castro F, Julliard
AK, Perfettini I, Chédotal A and Petit C: Anosmin-1, defective in
the X-linked form of Kallmann syndrome, promotes axonal branch
formation from olfactory bulb output neurons. Cell. 109:217–228.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Di Schiavi E and Andrenacci D:
Invertebrate models of kallmann syndrome: Molecular pathogenesis
and new disease genes. Curr Genomics. 14:2–10. 2013.PubMed/NCBI
|
|
13
|
Liu J, Cao W, Chen W, Xu L and Zhang C:
Decreased expression of Kallmann syndrome 1 sequence gene (KAL1)
contributes to oral squamous cell carcinoma progression and
significantly correlates with poorly differentiated grade. J Oral
Pathol Med. 44:109–114. 2015. View Article : Google Scholar
|
|
14
|
González-Martínez D, Kim SH, Hu Y, Guimond
S, Schofield J, Winyard P, Vannelli GB, Turnbull J and Bouloux PM:
Anosmin-1 modulates fibroblast growth factor receptor 1 signaling
in human gonadotropin-releasing hormone olfactory neuroblasts
through a heparan sulfate-dependent mechanism. J Neurosci.
24:10384–10392. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Choy CT, Kim H, Lee JY, Williams DM,
Palethorpe D, Fellows G, Wright AJ, Laing K, Bridges LR, Howe FA,
et al: Anosmin-1 contributes to brain tumor malignancy through
integrin signal pathways. Endocr Relat Cancer. 21:85–99. 2014.
View Article : Google Scholar :
|
|
16
|
Kanda M, Nomoto S, Okamura Y, Hayashi M,
Hishida M, Fujii T, Nishikawa Y, Sugimoto H, Takeda S and Nakao A:
Promoter hypermethylation of fibulin 1 gene is associated with
tumor progression in hepatocellular carcinoma. Mol Carcinog.
50:571–579. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jian B, Nagineni CN, Meleth S, Grizzle W,
Bland K, Chaudry I and Raju R: Anosmin-1 involved in neuronal cell
migration is hypoxia inducible and cancer regulated. Cell Cycle.
8:3770–3776. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Guioli S, Incerti B, Zanaria E, Bardoni B,
Franco B, Taylor K, Ballabio A and Camerino G: Kallmann syndrome
due to a translocation resulting in an X/Y fusion gene. Nat Genet.
1:337–340. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hardelin JP, Levilliers J, del Castillo I,
Cohen-Salmon M, Legouis R, Blanchard S, Compain S, Bouloux P, Kirk
J, Moraine C, et al: X chromosome-linked Kallmann syndrome: Stop
mutations validate the candidate gene. Proc Natl Acad Sci USA.
89:8190–8194. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Nomoto S, Kanda M, Okamura Y, Nishikawa Y,
Qiyong L, Fujii T, Sugimoto H, Takeda S and Nakao A: Epidermal
growth factor-containing fibulin-like extracellular matrix protein
1, EFEMP1, a novel tumor-suppressor gene detected in
hepato-cellular carcinoma using double combination array analysis.
Ann Surg Oncol. 17:923–932. 2010. View Article : Google Scholar
|
|
21
|
Kanda M, Nomoto S, Oya H, Takami H, Hibino
S, Hishida M, Suenaga M, Yamada S, Inokawa Y, Nishikawa Y, et al:
Downregulation of DENND2D by promoter hypermethylation is
associated with early recurrence of hepatocellular carcinoma. Int J
Oncol. 44:44–52. 2014.
|
|
22
|
Shimizu D, Kanda M, Nomoto S, Oya H,
Takami H, Hibino S, Suenaga M, Inokawa Y, Hishida M, Takano N, et
al: Identification of intragenic methylation in the TUSC1 gene as a
novel prognostic marker of hepatocellular carcinoma. Oncol Rep.
31:1305–1313. 2014.
|
|
23
|
Takami H, Kanda M, Oya H, Hibino S,
Sugimoto H, Suenaga M, Yamada S, Nishikawa Y, Asai M, Fujii T, et
al: Evaluation of MAGE-D4 expression in hepatocellular carcinoma in
Japanese patients. J Surg Oncol. 108:557–562. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sobin LH, Gospodarowicz MK and Wittekind
C: International Union Against Cancer, TNM Classification of
Malignant Tumors. 7th edition. Wiley-Blackwell; New York: 2009
|
|
25
|
Kanda M, Nomoto S, Oya H, Hashimoto R,
Takami H, Shimizu D, Sonohara F, Kobayashi D, Tanaka C, Yamada S,
et al: Decreased expression of prenyl diphosphate synthase subunit
2 correlates with reduced survival of patients with gastric cancer.
J Exp Clin Cancer Res. 33:882014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Takai D and Jones PA: The CpG island
searcher: A new WWW resource. In Silico Biol. 3:235–240.
2003.PubMed/NCBI
|
|
27
|
Kanda M, Nomoto S, Oya H, Shimizu D,
Takami H, Hibino S, Hashimoto R, Kobayashi D, Tanaka C, Yamada S,
et al: Dihydropyrimidinase-like 3 facilitates malignant behavior of
gastric cancer. J Exp Clin Cancer Res. 33:662014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kanda M, Shimizu D, Nomoto S, Hibino S,
Oya H, Takami H, Kobayashi D, Yamada S, Inokawa Y, Tanaka C, et al:
Clinical significance of expression and epigenetic profiling of
TUSC1 in gastric cancer. J Surg Oncol. 110:136–144. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kawahara T, Hotta N, Ozawa Y, Kato S, Kano
K, Yokoyama Y, Nagino M, Takahashi T and Yanagisawa K: Quantitative
proteomic profiling identifies DPYSL3 as pancreatic ductal
adenocarcinoma-associated molecule that regulates cell adhesion and
migration by stabilization of focal adhesion complex. PLoS One.
8:e796542013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Oya H, Kanda M, Sugimoto H, Shimizu D,
Takami H, Hibino S, Hashimoto R, Okamura Y, Yamada S, Fujii T, et
al: Dihydropyrimidinase-like 3 is a putative hepatocellular
carcinoma tumor suppressor. J Gastroenterol. Aug 31–2014.(Epub
ahead of print). PubMed/NCBI
|
|
31
|
Kanda M, Shimizu D, Nomoto S, Takami H,
Hibino S, Oya H, Hashimoto R, Suenaga M, Inokawa Y, Kobayashi D, et
al: Prognostic impact of expression and methylation status of
DENN/MADD domain-containing protein 2D in gastric cancer. Gastric
Cancer. 18:288–296. 2015. View Article : Google Scholar
|
|
32
|
Yam JW, Tse EY and Ng IO: Role and
significance of focal adhesion proteins in hepatocellular
carcinoma. J Gastroenterol Hepatol. 24:520–530. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Maunakea AK, Nagarajan RP, Bilenky M,
Ballinger TJ, D’Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C,
Zhao Y, et al: Conserved role of intragenic DNA methylation in
regulating alternative promoters. Nature. 466:253–257. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Khare S, Zhang Q and Ibdah JA: Epigenetics
of hepatocellular carcinoma: Role of microRNA. World J
Gastroenterol. 19:5439–5445. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Miki D, Ochi H, Hayes CN, Aikata H and
Chayama K: Hepato-cellular carcinoma: Towards personalized
medicine. Cancer Sci. 103:846–850. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kang YK, Hong SW, Lee H and Kim WH:
Prognostic implications of ezrin expression in human hepatocellular
carcinoma. Mol Carcinog. 49:798–804. 2010.PubMed/NCBI
|
|
37
|
Chen Y, Wang D, Guo Z, Zhao J, Wu B, Deng
H, Zhou T, Xiang H, Gao F, Yu X, et al: Rho kinase phosphorylation
promotes ezrin-mediated metastasis in hepatocellular carcinoma.
Cancer Res. 71:1721–1729. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ghaffari A, Hoskin V, Szeto A, Hum M,
Liaghati N, Nakatsu K, Madarnas Y, Sengupta S and Elliott BE: A
novel role for ezrin in breast cancer angio/lymphangiogenesis.
Breast Cancer Res. 16:4382014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Piao J and Liu S, Xu Y, Wang C, Lin Z, Qin
Y and Liu S: Ezrin protein overexpression predicts the poor
prognosis of pancreatic ductal adenocarcinomas. Exp Mol Pathol.
98:1–6. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ogasawara S, Yano H, Iemura A, Hisaka T
and Kojiro M: Expressions of basic fibroblast growth factor and its
receptors and their relationship to proliferation of human
hepatocellular carcinoma cell lines. Hepatology. 24:198–205. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Huang X, Yu C, Jin C, Kobayashi M, Bowles
CA, Wang F and McKeehan WL: Ectopic activity of fibroblast growth
factor receptor 1 in hepatocytes accelerates hepatocarcinogenesis
by driving proliferation and vascular endothelial growth
factor-induced angiogenesis. Cancer Res. 66:1481–1490. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang J, Li J, Wang X, Zheng C and Ma W:
Downregulation of microRNA-214 and overexpression of FGFR-1
contribute to hepatocellular carcinoma metastasis. Biochem Biophys
Res Commun. 439:47–53. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zheng SS, Chen XH, Yin X and Zhang BH:
Prognostic significance of HIF-1α expression in hepatocellular
carcinoma: A meta-analysis. PLoS One. 8:e657532013. View Article : Google Scholar
|
|
44
|
Wu L, Fu Z, Zhou S, Gong J, Liu CA, Qiao Z
and Li S: HIF-1α and HIF-2α: Siblings in promoting angiogenesis of
residual hepatocellular carcinoma after high-intensity focused
ultrasound ablation. PLoS One. 9:e889132014. View Article : Google Scholar
|
|
45
|
Yamada S, Utsunomiya T, Morine Y, Imura S,
Ikemoto T, Arakawa Y, Kanamoto M, Iwahashi S, Saito Y, Takasu C, et
al: Expressions of hypoxia-inducible factor-1 and epithelial cell
adhesion molecule are linked with aggressive local recurrence of
hepatocellular carcinoma after radiofrequency ablation therapy. Ann
Surg Oncol. 21(Suppl 3): S436–S442. 2014. View Article : Google Scholar : PubMed/NCBI
|