Introduction
Hepatocellular carcinoma (HCC) is one of the most
common malignant cancers worldwide with ~598,000 incidences per
year representing the second leading cause of cancer death with the
5-year survival rate of <5% (1).The primary etiological factor for HCC
is HBV infection (2); however,
inefficient diagnosis of early stage HCC remains a primary causal
factor of the high mortality and poor prognosis (3–6).
Since the identification of α-fetoprotein (AFP) in 1970s, it has
been the only serologic marker that is widely used for the HCC
diagnosis. However, the diagnostic power of AFP has been
continuously questioned and debated. For example, elevated serum
AFP was only observed in ~60–70% of overall HCC patients, while the
proportion was merely 33–65% regarding patients harboring HCCs of
<3 cm in diameter (7,8). Furthermore, the non-specific
elevation of serum AFP was observed in 11–47% of liver cirrhosis
patients (7,9). Although Des-gamma-carboxyprothrombin
(DCP) was once proposed to be a better HCC diagnostic marker,
investigations have reported that it is only positive in 44–47.6%
of the smaller HCCs (10,11).
Because the best currently available diagnostic HCC
markers have significant shortcomings, novel serologic HCC
biomarkers that improve the sensitivity and specificity of HCC
diagnosis, especially in AFP-negative [AFP(−)] individuals, are
greatly needed. In the present study, we define AFP(−) as serum AFP
levels <20 ng/ml.
Several promising HCC biomarkers have been
identified using proteomic strategies. For example, Lee et
al (12) employed
surface-enhanced laser desorption/ionization (SELDI) mass
spectrometry (MS) and two-dimensional gel electrophoresis (2DE)
technologies and found that complement C3a upregulation correlates
with the presence of chronic hepatitis C and hepatitis C virus
(HCV)-related HCC (12). Another
study by Feng et al (13)
used 2DE and matrix assisted laser desorption/ionization-time of
flight tandem mass spectrometry (MALDI-TOF-MS/MS) and identified
heat-shock protein 27 (HSP27) as a potential complementary
biomarker for AFP to improve the diagnosis of AFP(−) (<20 ng/ml)
HCC patients and patients with small HCCs (<5 cm). Even though
gel-based MS provides good visual and physical-chemical
information, it is particularly time and labor intensive, with
relatively low throughput and can have significant intergel
variations (14). Numerous new
post-digestion labeling methods, such as isobaric tags for relative
and absolute quantitation (iTRAQ) (15–17),
have been recommended by the proteomics community to enable deeper
proteome coverage and facilitate biomarker discovery.
In the present study, we performed iTRAQ-based MS to
quantify differentially expressed proteins (DEPs) between plasma
samples from AFP(−) and AFP-positive [AFP(+)] patients. DEPs are
also sometimes referred to as ‘aberrantly expressed proteins’ in
this study when we wish to emphasize their biological and/or
clinical relevance. Our iTRAQ analysis identified 14 aberrantly
expressed proteins specific to the HCC patients. Within this set,
ELISA analysis and immunohistochemistry verified the likely
importance of C-reactive protein (CRP) overexpression in HCC.
Further clinical verification, diagnostic power evaluation and
in vitro experiments were performed to validate the
importance of CRP overexpression in HCC. We propose that CRP is a
potentially useful diagnostic and therapeutic biomarker of AFP(−)
HBV-related HCC.
Materials and methods
Subjects and plasma collection
Two-hundred and eighty-four subjects participated in
this study from January 2013 to March 2014, including 74 AFP
negative hepatocellular carcinoma patients, 60 AFP positive HCC
patients and 70 liver cirrhosis patients, these patients all had
chronic hepatitis B infection. There were 40 chronic hepatitis B
patients and 40 healthy controls. Serum plasma samples were
obtained according to the guidelines given by the HUPO Plasma
Proteome Project (18), and 4 ml
of peripheral blood was collected from each subject. Diagnoses of
chronic hepatitis B, liver cirrhosis and hepatocellular carcinoma
were performed according to the Asian Pacific Association for the
Study of the Liver (APASL), the European Association for the Study
of the Liver (EASL) and the American Association for the Study of
Liver Diseases (AASLD) (19–22).
This study was approved by the Ethics Committee of Chongqing
Medical University. Written informed consent was obtained from all
participants before the treatment. Patient demographics and
clinicopathological data are summarized in Table I.
 | Table IPatient demographic and clinical
characteristics. |
Table I
Patient demographic and clinical
characteristics.
|
Characteristics | Healthy
controls | Chronic hepatitis
B | Liver
cirrhosis | AFP(−)HCC | AFP(+)HCC |
|---|
| Age (years), mean ±
SD | 22±4.5 | 42±13.4 | 44±11.9 | 50±18.9 | 55±20.3 |
| Gender |
| Male | 16 | 22 | 54 | 44 | 32 |
| Female | 24 | 18 | 16 | 30 | 28 |
| ALT (IU/l), mean ±
SD | NA | 453.46±178.94 | 108.8±39.04 | 26.33±4.799 | 47.67±12.45 |
| AFP (ng/ml), mean ±
SD | NA | 111.1±33.89 | 166.5±29.56 | 6.372±0.495 | 765.23±92.28 |
| HBV DNA (log
copies/ml), mean ± SD | NA | 5.67±1.12 | 2.87±0.98 | 3.12±1.02 | 3.33±1.007 |
Abundant protein depletion of plasma
samples and ITRAQ labeling
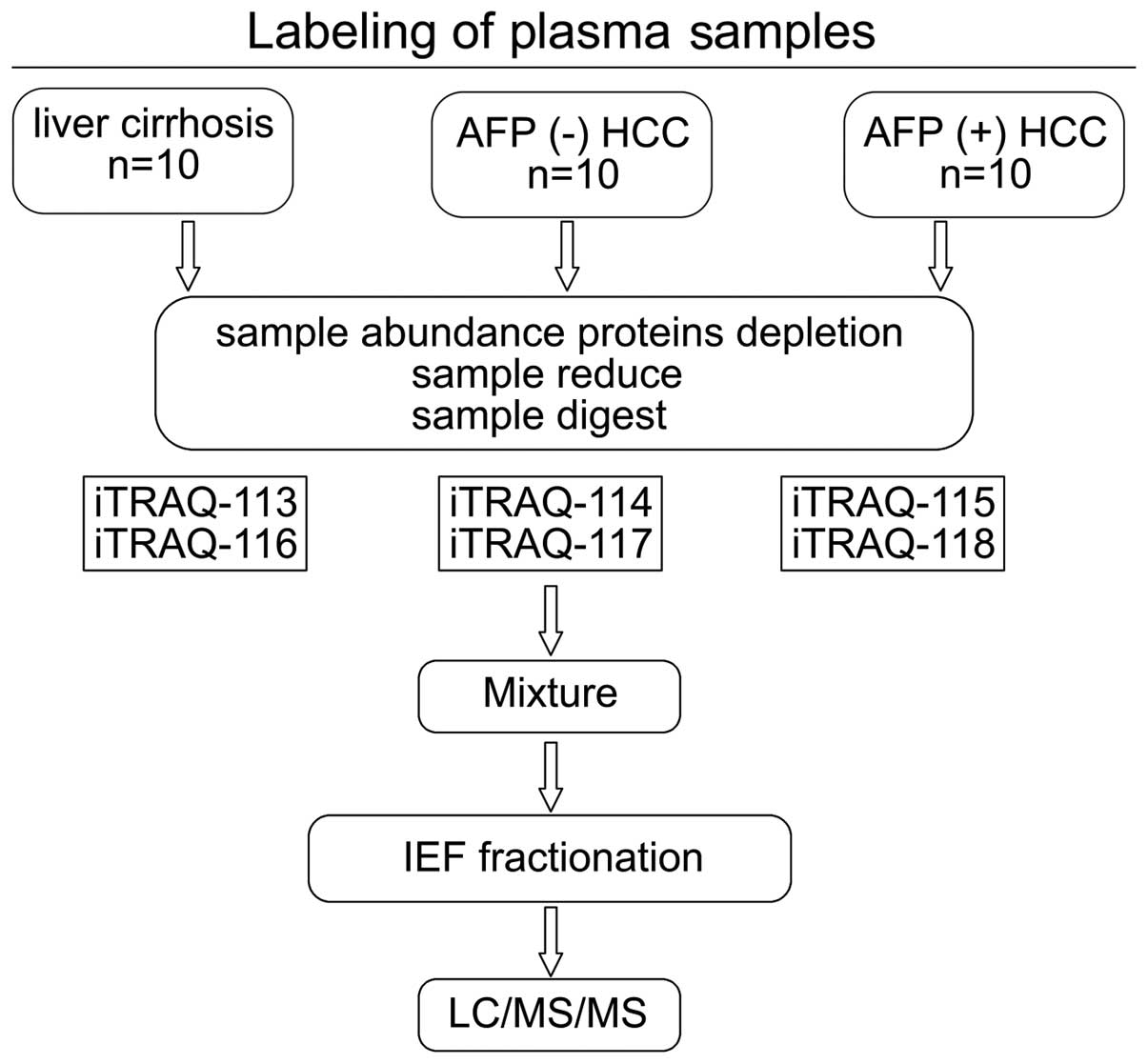
Ten randomly chosen individual samples from each
group were mixed to create three sample pools (Fig. 1). The most abundant proteins were
depleted using an immunodepletion kit (GE Healthcare, Shanghai,
China) as per the manufacturer’s instructions. Immunodepleted
plasma was subjected to protein concentration assays using a 2-D
Quant kit (GE Healthcare). Protein (100 μg) was precipitated from
each pooled group, dissolved in dissolution buffer, denatured,
cysteine blocked, digested with 2 μg of sequencing grade modified
trypsin and labeled using iTRAQ reagents [LC, 113 tag; AFP(−) HCC,
114 tag; and AFP(+) protein, 115 tag] provided by an iTRAQ kit (AB
Sciex Analytical Instrument Trading Co., Shanghai, China) (Fig. 1). For the parallel study, the same
sample set was labeled with the iTRAQ reagents 116, 117 and 118,
respectively (Fig. 1). Peptides
from each sample set were mixed prior to subsequent analysis.
Peptide fractionation
Labeled peptides were fractionated by
immobilized-pH-gradient isoelectric focusing (IPG-IEF), as
previously described (23,24). Briefly, samples were dissolved in a
Pharmalyte (GE Healthcare) and urea solution, rehydrated on a pH
3–10 IPG strip, and then subjected to IEF focusing at 68 kV/h with
an IPGphor system (GE Healthcare). Peptides were then extracted
from the gel using an acetonitrile (ACN) and formic acid solution
(25). The fractions were
lyophilized, and purified with SPE Discovery DSC-18 columns
(Supelco Inc., Bellefonte, PA, USA). The purified peptides were
re-lyophilized and stored at −20°C until use.
MASS spectrometry
Purified peptide fractions were reconstituted in
solvent A [water/ACN (98:2 v/v) with 0.1% formic acid] and
separated using a C18-PepMap column (Thermo Fisher Scientific,
Beijing, China) with a solvent gradient of 2–100% Buffer B (0.1%
formic acid and 98% acetonitrile) in Buffer A at a flow rate of 0.3
μl/min. The peptides were electrosprayed using a nanoelectrospray
ionization source at an ion spray voltage of 2300 eV and analyzed
by a NanoLC-ESI-Triple TOF 5600 system (AB Sciex). The mass
spectrometer was set in the positive ion mode at a mass range of
300–1800 m/z. The two most intensely charged peptides above 20
counts were selected for MS/MS at a dynamic exclusion of 30 sec
(25).
Data were processed by ProteinPilot v2.0 (AB Sciex)
and compared with the International Protein Index (IPI) Human
database v3.77. Cysteine modified by methane thiosulfate (MMTS) was
specified as a fixed modification. Protein identification was based
on a threshold of protein score >1.3. For quantitation, at least
two unique peptides with 95% confidence and a P-value <0.05 were
required.
Bioinformatics
The Gene Ontology was analyzed by PANTHER
(http://www.pantherdb.org/) on biological
processes, protein classes and molecular functions. The signaling
pathway analysis was performed by using the STRING (http://string-db.org/) program.
ELISA
The plasma levels of CRP, SAA, AFP and C9 were
measured in 284 cases using commercial ELISA kits in accordance
with the manufacturer’s instructions. The cut-off value of CRP was
determined using the receiver-operator characteristic curve (ROC)
curve, which was twice the SD above the average of the control
individuals. Human CRP ELISA kit (ab99995) and Complement C9 Human
ELISA kit (ab137972) were purchased from Abcam (Cambridge, UK). The
human SAA ELISA kit (ELH-SAA-001) was purchased from Ray Biotech
(Norcross, GA, USA).
Tissues microarray and
immunohistochemistry (IHC)
IHC evaluation of C9, SAA and CRP was performed with
a commercial tissue microarray (BC03117; Us Biomax Inc., Rockville,
MD, USA) containing 48 unique HCC samples and 22 liver cirrhosis
tissues. Paraffin-embedded liver sections were deparaffinized,
rehydrated and subjected to heat-induced antigen retrieval in 0.01
M sodium citrate buffer for 5 min (26). Next, 3% H2O2
was added to quench the activity of endogenous peroxidase for 5
min. After BSA blocking, the sections were incubated overnight with
primary antibodies for CRP (1:100), SAA (1:100) and C9 (1:100)
(Abcam). The EnVision system with horseradish peroxidase
(DakoCytomation, Glostrup, Denmark) was used for IHC visualization
(26). Gill’s hematoxylin was used
to counterstain slides according to methods previously described
(25,26). Monoclonal antibodies against human
CRP (ab32412), serum amyloid A (SAA, EPR4134), matrix
metalloproteinase 2 (MMP2, EPR1184), matrix metalloproteinase 9
(MMP9, EP1254), signal transducer and activator of transcription 3
(STAT3, E121-21), phosphorylated STAT3 (pY705-STAT3, EP2147Y) and
β-actin (EP1123Y) were purchased from Abcam. Horseradish peroxidase
(HRP)-conjugated secondary antibodies were obtained from Santa Cruz
Biotechnology (Dallas, TX, USA).
Cell lines
The stable HBV-transfected cell line HepG2.2.15, the
human HCC cell line HepG2 (ATCC, Manassas, VA, USA) and the BEL7402
cell line (Cell Bank of the Chinese Academy of Medical Science,
Beijing, China) were cultured in high-glucose DMEM that was
supplemented with 100 μg/ml streptomycin, 0.1% non-essential amino
acids, 100 IU/ml penicillin, 1.0 mM sodium pyruvate, 2 mM glutamine
and 10% FBS at 5% CO2 and 37°C (27).
CRP siRNA transfection, transwell assays
and wound healing
Cell lines were transfected with 100 nm of
CRP-specific siRNA (HSS175221, HSS102299 and HSS102300) or a
negative control plasmid (12935-400) using Lipofectamine 2000
(Invitrogen, Carlsbad, CA, USA). Cell viability was determined with
a trypan blue exclusion assay and only cells that had over 95% live
cells by the trypan blue exclusion assay were used for subsequent
assays.
For wound healing assays, cells were cultured in
6-well plates until they reached 100% confluence. A 200-μl pipet
tip was used to scratch the cell monolayer, followed by washes with
the growth media to remove debris. The resultant gap was monitored
for up to 24 h via a microscope.
Invasion assays were completed by a Cell Invasion
assay kit (Cell Biolabs, Inc., Beijing, China). As determined by
the trypan blue exclusion, ~1×105 viable and
siRNA-transfected cells were seeded onto the upper chamber of a
24-well plate with polycarbonate membrane inserts, and the number
of cells that invaded through the ECM Matrix gel was determined 24
h after seeding by CyQuant GR fluorescent dye (560 nm).
Western blotting
Cells were lysed with RIPA buffer, and a 2-D
Quantification kit (GE Healthcare) was used to determine the
protein concentration. Protein samples were electrophoretically
separated by SDS-PAGE and then transferred onto PVDF membranes.
Membranes were blocked with BSA in Tris-buffered saline solution
with Tween-20 (TBS-T), overnight at 4°C and then incubated with the
primary antibodies (1:500–1:1,000 dilution) for 3 h at room
temperature. Then membranes were incubated with HRP-conjugated
secondary antibodies at a dilution of 1:5,000 after three wahses
with TBST buffer. Finally, membranes were visualized with the
ChemiDoc MP imaging system (Bio-Rad Laboratories, Hercules, CA,
USA).
Supernatant HBV detection and RT-PCR
analysis
Supernatant HBV-DNA was quantified by RT-PCR using a
commercial HBV detection kit (Fosun Diagnostics, Shanghai, China)
on a Roche LightCycler instrument (Roche Molecular Systems,
Alameda, CA, USA). Elecsys HBsAg II and HBeAg quantitative assay
kits were used to detect HBsAg and HBeAg titers, respectively,
using the Roche Cobas e601 electrochemical luminescence analyzer
(Roche Diagnostics GmbH, Mannheim, Germany) (28). To measure the expression levels of
downstream IFN-stimulated genes and type I IFN in transfected
cells, we used gene-specific primers for GAPDH (Hs02758991_g1),
OASL (Hs0 0984390_m1), Mx1 (Hs0 0895608_m1), ISG15 (Hs01921425_s1),
OAS2 (Hs0 0942643_m1), EIF-2α (Hs00230684_m1), IFNβ1
(Hs01077958_s1), OAS1 (Hs00973637_m1), PKR (Hs00169345_m1), IFNα1
(Hs00855471_g1) and OAS3 (Hs00196324_m1) (Life Technologies). The
2−ΔΔCT method (29) was
used to analyze the relative changes in gene expression. All
experiments were performed in triplicate.
Statistical analysis
SPSS software v13.0 (SPSS Inc., Chicago, IL, USA)
was used to perform statistical analysis. Quantitative variables
are presented as the mean and standard deviation (±SD). Comparisons
between groups were analyzed by the Student’s t-test or a
Mann-Whitney U test. Qualitative variables are presented as counts
and percentages, which were analyzed with the χ2 test.
ROC curve analysis of CRP was performed to determine the diagnostic
accuracy of CRP expression levels and 2×2 tables were used to
evaluate sensitivity and specificity. Correlations between CRP and
HBV DNA were determined using a Spearman’s rank correlation
analysis. P<0.05 was considered significant.
Results
MS identification and ITRAQ
quantification of aberrantly expressed proteins
We used iTRAQ-based MS to analyze serum proteins
from the AFP(−) HCC, AFP(+) HCC and LC groups (Fig. 1). We confidently identified and
quantified 510 proteins. The top 30 upregulated and the top 30
downregulated proteins are shown in Table II. We further defined the DEPs
using a ±1.3-fold cut-off in accordance with commonly adopted
iTRAQ-based MS conventions (30,31).
Use of this cut-off is based on the assumption that the estimated
overall data variation from duplicate experiments is ≤30%. Gene
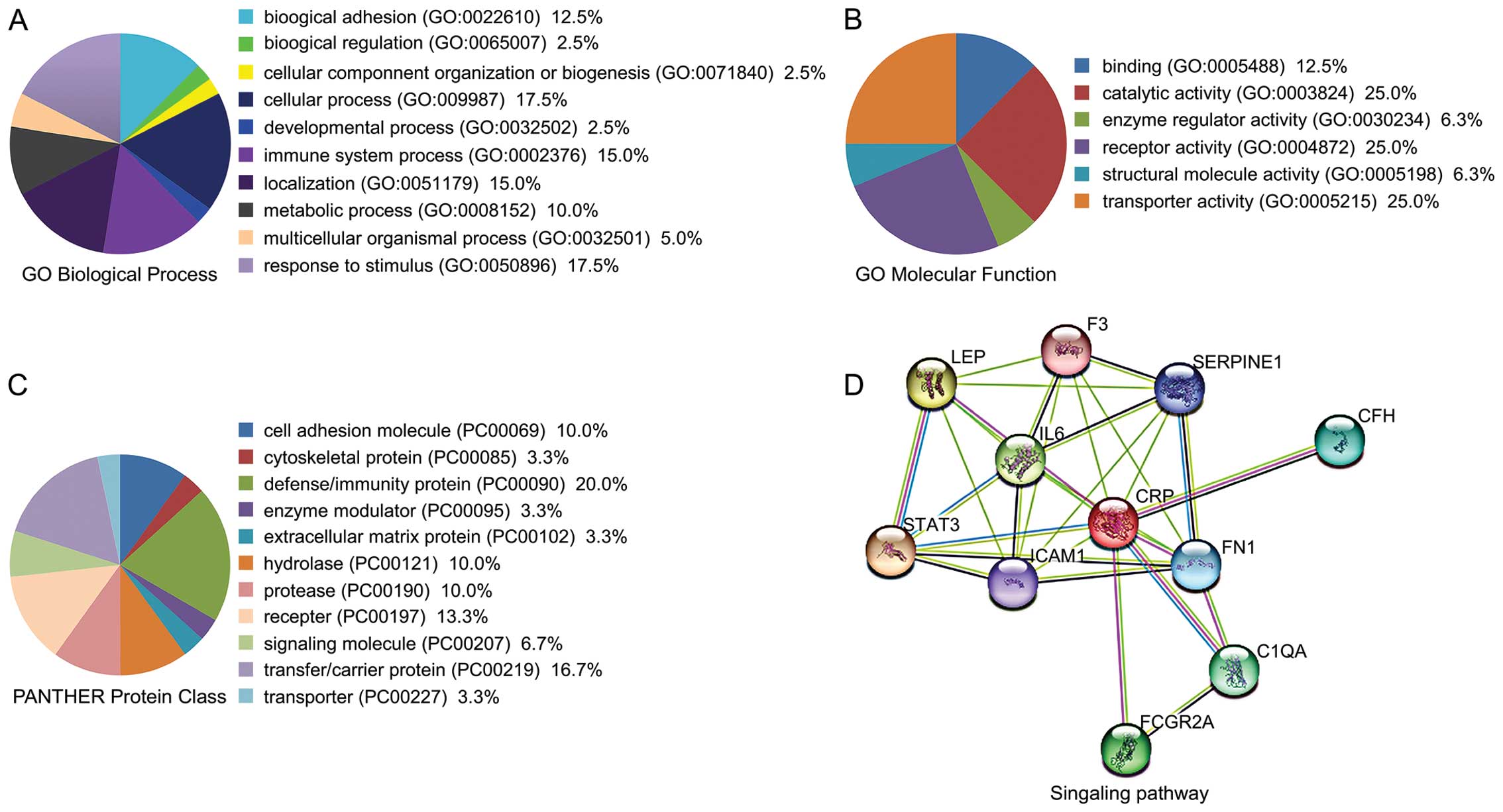
Ontology analysis with PANTHER suggested that the majority of the
DEPs were enzymes or signaling molecules, followed by cell
development regulators and immune-related proteins (Fig. 2A–C). Using STRING analysis, we
identified CRP as the most important node in the DEP network
because it had the greatest connectivity (Fig. 2D).
 | Table IIPartial list of proteins found to be
expressed at different level in liver cirrhosis: AFP(−) HCC and
AFP(+) HCCa. |
Table II
Partial list of proteins found to be
expressed at different level in liver cirrhosis: AFP(−) HCC and
AFP(+) HCCa.
| N | Accession no. | Gene symbol | Pooled liver
cirrhosis: AFP(−) HCC (114:113) | pval 114:113 | Pooled liver
cirrhosis: AFP(+) HCC (115:113) | pval 115:113 | Pooled liver
cirrhosis: AFP(−) HCC (117:116) | pval 117:116 | Pooled liver
cirrhosis: AFP(+) HCC (118:116) | pval 118:116 |
|---|
| Top 30 proteins
upregulated in liver cancer tissues |
| 1 |
IPI:IPI00453473.6 | HIST2H4B | 16.29295921 | 0.001705589 | 9.120108604 | 0.01209232 | 17.53881073 | 0.0012185 | 9.727472305 | 0.00395962 |
| 2 |
IPI:IPI00909530.1 | H3F3A | 11.48153973 | 0.233225003 | 2.91071701 | 0.50077242 | 8.394599915 | 0.2622318 | 6.606935024 | 0.29974699 |
| 3 |
IPI:IPI00963899.3 | HMGCS1 | 8.016780853 | 0.117667101 | 4.786301136 | 0.15475079 | 9.817479134 | 0.1074611 | 1.923092008 | 0.34027699 |
| 4 |
IPI:IPI00007960.4 | POSTN | 7.516229153 | 0.034993611 | 12.94196033 | 0.03113686 | 19.58844948 | 0.0284294 | 17.06081963 | 0.02917336 |
| 5 |
IPI:IPI00022389.1 | CRP | 6.367955208 | 0.000284711 | 7.17794323 | 3.67E-05 | 7.311390877 | 2.11E-05 | 7.447319984 | 0.00180836 |
| 6 |
IPI:IPI00298497.3 | FGB | 6.025596142 | 0.000111177 | 8.709635735 | 6.15E-07 | 3.047894955 | 0.0165862 | 13.30453968 | 1.62E-09 |
| 7 |
IPI:IPI00022246.1 | AZU1 | 5.345643997 | 0.204895303 | 3.191538095 | 0.2823019 | 2.992264986 | 0.2688605 | 3.80189395 | 0.29355779 |
| 8 |
IPI:IPI00745570.2 | SCN9A | 4.965922832 | 0.019038239 | 1.690441012 | 0.06121931 | 1.770109057 | 0.0291526 | 2.703958035 | 0.02286139 |
| 9 |
IPI:IPI00877792.1 | FGG | 4.830587864 | 0.135952994 | 7.79830122 | 0.06993747 | 5.754398823 | 0.108478 | 10.09253025 | 0.04939477 |
| 10 |
IPI:IPI00019581.2 | F12 | 4.613175869 | 0.05299313 | 3.53183198 | 0.12925071 | 1.629295945 | 0.3908164 | 3.191538095 | 0.22002751 |
| 11 |
IPI:IPI00022395.1 | C9 | 4.325138092 | 1.26E-05 | 2.91071701 | 0.01619806 | 3.162277937 | 2.09E-06 | 3.944572926 | 1.49E-05 |
| 12 |
IPI:IPI00646240.3 | HIST2H2BF | 4.246195793 | 0.134902 | 1.706081986 | 0.80735821 | 3.28095293 | 0.4275794 | 3.191538095 | 0.09819763 |
| 13 |
IPI:IPI00980960.1 | CHADL | 4.092607021 | 0.170985803 | 3.28095293 | 0.20105509 | 0.717794299 | 0.5585651 | 1.737800956 | 0.38601881 |
| 14 |
IPI:IPI00745872.2 | ALB | 1.018591046 | 0.005099569 | 0.963828981 | 0.158677 | 1.037528038 | 0.0098839 | 0.981747925 | 0.00147472 |
| 15 |
IPI:IPI00006146.4 | SAA | 3.732501984 | 0.01412376 | 3.076097012 | 0.01969219 | 4.405549049 | 0.0123332 | 2.22843504 | 0.02866358 |
| 16 |
IPI:IPI00025019.3 | PSMB1 | 3.630779982 | 0.186679199 | 3.162277937 | 0.2069978 | 1.614359021 | 0.4323919 | 1.976969957 | 0.32960361 |
| 17 |
IPI:IPI00982101.1 | YWHAZ | 3.499452114 | 0.009089738 | 0.879022479 | 0.36322659 | 1.958845019 | 0.3017248 | 1.224616051 | 0.93211728 |
| 18 |
IPI:IPI00021439.1 | ACTB | 3.40408206 | 0.004408214 | 1.940886021 | 0.03609288 | 2.937649965 | 1.20E-05 | 2.606153011 | 0.00414366 |
| 19 |
IPI:IPI00945846.1 | PRSS1 | 3.34194994 | 0.410066307 | 1.995262027 | 0.71293592 | 2.582259893 | 0.5739074 | 1.923092008 | 0.91428632 |
| 20 |
IPI:IPI00011261.2 | C8G | 3.311311007 | 0.024648501 | 3.250873089 | 0.02026156 | 1.629295945 | 0.085252 | 2.488857031 | 0.02338865 |
| 21 |
IPI:IPI00220642.7 | YWHAG | 3.133285999 | 0.278527111 | 0.061376199 | 0.18025219 | 2.128139019 | 0.450883 | 1.958845019 | 0.49069569 |
| 22 |
IPI:IPI00217945.1 | IL4 | 3.133285999 | 0.209923893 | 10.09253025 | 0.1064933 | 10.76465034 | 0.1039814 | 6.79203701 | 0.12781671 |
| 23 |
IPI:IPI00296608.6 | C7 | 3.104559898 | 0.003662642 | 2.805433989 | 0.02225894 | 1.995262027 | 0.0204516 | 2.630268097 | 0.02887956 |
| 24 |
IPI:IPI00032311.4 | LBP | 2.884032011 | 0.007922285 | 2.630268097 | 0.09581193 | 2.249054909 | 0.0033271 | 3.372873068 | 0.00165596 |
| 25 |
IPI:IPI00019576.1 | F10 | 2.884032011 | 0.29140681 | 2.805433989 | 0.1899274 | 3.133285999 | 0.2494257 | 3.53183198 | 0.09236393 |
| 26 |
IPI:IPI00293925.2 | FCN3 | 2.85758996 | 0.046134971 | 1.380383968 | 0.53549451 | 2.443430901 | 0.1579406 | 2.421029091 | 0.42029911 |
| 27 |
IPI:IPI00021885.1 | FGA | 2.779712915 | 0.000226672 | 5.296635151 | 9.67E-10 | 2.032356977 | 0.0850608 | 4.613175869 | 4.09E-09 |
| 28 |
IPI:IPI00253323.3 | ANKRD57 | 2.679167986 | 0.237978101 | 1.485936046 | 0.4877679 | 2.355048895 | 0.2713818 | 1.445440054 | 0.51068312 |
| 29 |
IPI:IPI00747017.3 | NEK1 | 2.606153011 | 0.245841593 | 3.047894955 | 0.2133164 | 0.920449615 | 0.8856001 | 2.398833036 | 0.26525661 |
| 30 |
IPI:IPI00940615.1 | LAMA2 | 2.511885881 | 0.084028691 | 1.16949904 | 0.56904221 | 0.469894111 | 0.1992138 | 0.981747925 | 0.82043833 |
| Top 30 proteins
downregulated in liver cancer tissues |
| 1 |
IPI:IPI00556155.2 | IGFBP3 | 0.100000001 | 0.059266131 | 0.772680581 | 0.42646229 | 0.319153786 | 0.287104 | 0.57543987 | 0.101595 |
| 2 |
IPI:IPI00020986.2 | LUM | 0.1599558 | 0.000102721 | 0.496592313 | 0.03381877 | 0.334194988 | 0.0043522 | 0.591561615 | 0.01780533 |
| 3 |
IPI:IPI00923627.1 | EML5 | 0.161435902 | 0.184541404 | 2.376840115 | 0.26360521 | 3.34194994 | 0.1925355 | 1.047129035 | 0.73078853 |
| 4 |
IPI:IPI00028064.1 | CTSG | 0.203235701 | 0.193927795 | 1.393157005 | 0.327288 | 1.923092008 | 0.167279 | 1.485936046 | 0.29505801 |
| 5 |
IPI:IPI00304273.2 | APOA4 | 0.222843498 | 2.02E-14 | 0.301995188 | 5.79E-06 | 0.369828194 | 2.64E-14 | 0.35318321 | 4.49E-14 |
| 6 |
IPI:IPI00021855.1 | APOC1 | 0.229086801 | 0.046579931 | 1.056818008 | 0.90821451 | 0.691830993 | 0.3053197 | 1.037528038 | 0.43847549 |
| 7 |
IPI:IPI00021364.1 | CFP | 0.288403213 | 0.000476604 | 0.383707315 | 0.01457084 | 0.487528503 | 0.0025835 | 0.457088202 | 0.00787622 |
| 8 |
IPI:IPI00328113.4 | FBN1 | 0.331131101 | 0.218046799 | 0.01127198 | 0.05582185 | 0.325087309 | 0.214938 | 0.717794299 | 0.56533998 |
| 9 |
IPI:IPI00023673.1 | LGALS3BP | 0.343558013 | 2.35E-05 | 1.330453992 | 0.1234327 | 0.602559626 | 0.0434183 | 1.445440054 | 0.04987133 |
| 10 |
IPI:IPI00026199.2 | GPX3 | 0.363078088 | 0.058937721 | 0.510505021 | 0.64686239 | 0.602559626 | 0.2496072 | 0.369828194 | 0.1118293 |
| 11 |
IPI:IPI00020557.2 | LRP1 | 0.369828194 | 0.115578599 | 1.355188966 | 0.52239323 | 1.08642602 | 0.8344157 | 1.706081986 | 0.3166641 |
| 12 |
IPI:IPI00010779.4 | TPM4 | 0.369828194 | 0.041988332 | 1.127197981 | 0.38104299 | 0.505824685 | 0.7239357 | 0.405508488 | 0.13078029 |
| 13 |
IPI:IPI00022394.2 | C1QC | 0.373250186 | 0.182315201 | 1.106624007 | 0.47615501 | 0.169044107 | 0.4298211 | 1.318256974 | 0.09189624 |
| 14 |
IPI:IPI01009054.1 | GFAP | 0.413047493 | 0.268078297 | 0.630957425 | 0.4229582 | 1.380383968 | 0.5501292 | 0.346736789 | 0.2268386 |
| 15 |
IPI:IPI00433029.1 | IGF1 | 0.452897608 | 0.294940889 | 0.816582382 | 0.70856959 | 0.946237087 | 0.9280483 | 0.679203629 | 0.51287282 |
| 16 |
IPI:IPI00783987.2 | C3 | 0.990831971 | 0.061891649 | 1.018591046 | 5.45E-06 | 1.009253025 | 0.0256407 | 1.047129035 | 0 |
| 17 |
IPI:IPI00032179.3 | SERPINC1 | 0.990831971 | 0.279663414 | 0.301995188 | 0.8061682 | 0.787045777 | 0.3305322 | 0.895364821 | 0.24263071 |
| 18 |
IPI:IPI00018136.1 | VCAM1 | 0.990831971 | 0.941708326 | 1.056818008 | 0.83449608 | 1.009253025 | 0.9862956 | 1.180320978 | 0.24393719 |
| 19 |
IPI:IPI00847381.1 | SEPP1 | 0.990831971 | 0.863334417 | 0.963828981 | 0.98466128 | 0.862978518 | 0.6828699 | 0.847227395 | 0.57136148 |
| 20 |
IPI:IPI00410714.5 | HBA2 | 0.990831971 | 0.870746374 | 0.990831971 | 0.91779089 | 0.824138105 | 0.4009709 | 0.954992592 | 0.76935983 |
| 21 |
IPI:IPI00922213.2 | FN1 | 0.990831971 | 0.992969275 | 0.679203629 | 0.88457477 | 0.564936996 | 0.5626391 | 0.619441092 | 0.7956093 |
| 22 |
IPI:IPI00397834.1 | FERMT3 | 0.990831971 | 0.981856227 | 1.106624007 | 0.70780838 | 0.879022479 | 0.6836015 | 0.963828981 | 0.94302893 |
| 23 |
IPI:IPI00029629.4 | TRIM25 | 0.990831971 | 0.995281518 | 0.855066717 | 0.77631968 | 1.018591046 | 0.957672 | 1.009253025 | 0.96326321 |
| 24 |
IPI:IPI00306844.1 | CRHBP | 0.990831971 | 0.989459097 | 1.887990952 | 0.2508274 | 1.445440054 | 0.373782 | 1.14815402 | 0.64023209 |
| 25 |
IPI:IPI00642126.6 | RNF213 | 0.990831971 | 0.997095108 | 1.770109057 | 0.3765156 | 0.990831971 | 0.996628 | 1.330453992 | 0.59040558 |
| 26 |
IPI:IPI00980673.1 | GOLGA8B | 0.990831971 | 0.996237576 | 1.527565956 | 0.46482059 | 1.432188034 | 0.5220141 | 1.737800956 | 0.389568 |
| 27 |
IPI:IPI00940388.1 | PNPLA6 | 0.990831971 | 0.994333923 | 0.704693079 | 0.5451777 | 1.614359021 | 0.4287863 | 1.028015971 | 0.93621689 |
| 28 |
IPI:IPI00872208.2 | TNRC18 | 0.990831971 | 0.996237576 | 1.527565956 | 0.46482059 | 1.432188034 | 0.5220141 | 1.737800956 | 0.389568 |
| 29 |
IPI:IPI00017696.1 | C1S | 0.981747925 | 0.396650195 | 0.990831971 | 0.93391472 | 0.990831971 | 0.4683169 | 0.981747925 | 0.97989023 |
| 30 |
IPI:IPI00299738.1 | PCOLCE | 0.981747925 | 0.793707728 | 1.028015971 | 0.647003 | 1.037528038 | 0.5697741 | 1.018591046 | 0.52085888 |
Verification of aberrant CRP, SAA and C9
expression
To determine the reliability of the iTRAQ analysis
data, we selected samples from the same sample set we analyzed in
the iTRAQ experiments and employed ELISA assays to test the plasma
levels of several of the most upregulated proteins, including CRP,
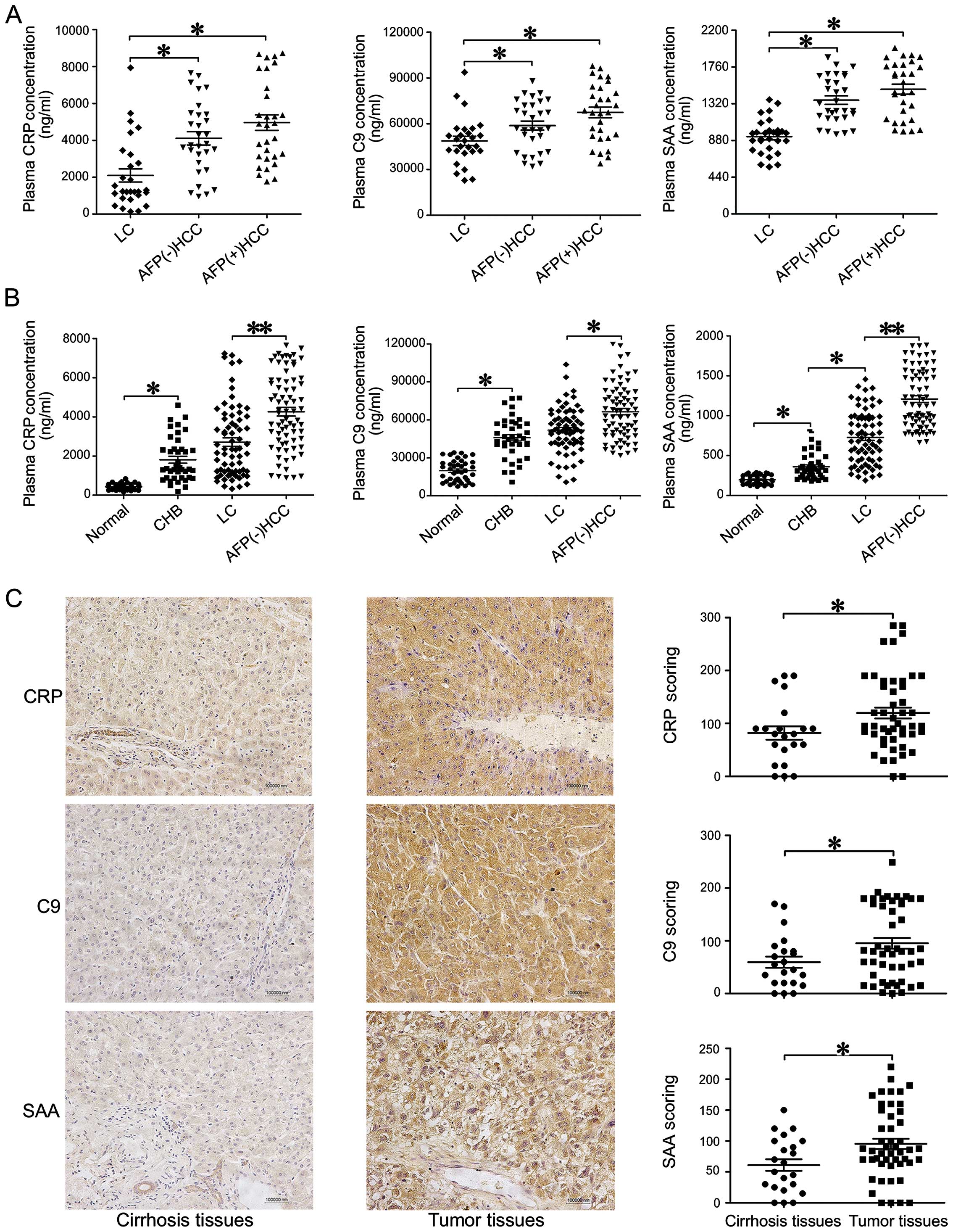
SAA and C9. We examined 90 plasma samples from 30 LC, 30 AFP(−) HCC
and 30 AFP(+) HCC individuals (Fig.
3A). The ELISA measured CRP, SAA and C9 levels were consistent
with the iTRAQ results, as the plasma levels of all three DEPs were
significantly higher in HCC subjects than in LC subjects
(P<0.05, Fig. 3A).
We further tested the serum levels of CRP, C9 and
SAA in all 284 plasma samples by ELISA (Fig. 3B). We found that average CRP
concentrations in the AFP(−) HCC (3932±277 ng/ml) and AFP(+) HCC
(4860±384.3 ng/ml) groups were significantly higher than the CRP
concentrations in the LC (2637±282.4 ng/ml, P<0.001), CHB
(1810±177.9 ng/ml, P<0.0001) and healthy control (424.9±23.95
ng/ml, P<0.0001) groups (Fig.
3B). Similar trends were also observed in the SAA and C9 assays
(Fig. 3B).
IHC analyses of tissue microarrays coincided with
the results of the serum tests, indicating that CRP, SAA and C9
were significantly overexpressed in tumor tissues when compared to
cirrhotic tissues (P<0.05) (Fig.
3C).
Diagnostic power of CRP
Our analysis indicated that CRP was the most
important node in the DEP analysis, and IHC analysis confirmed its
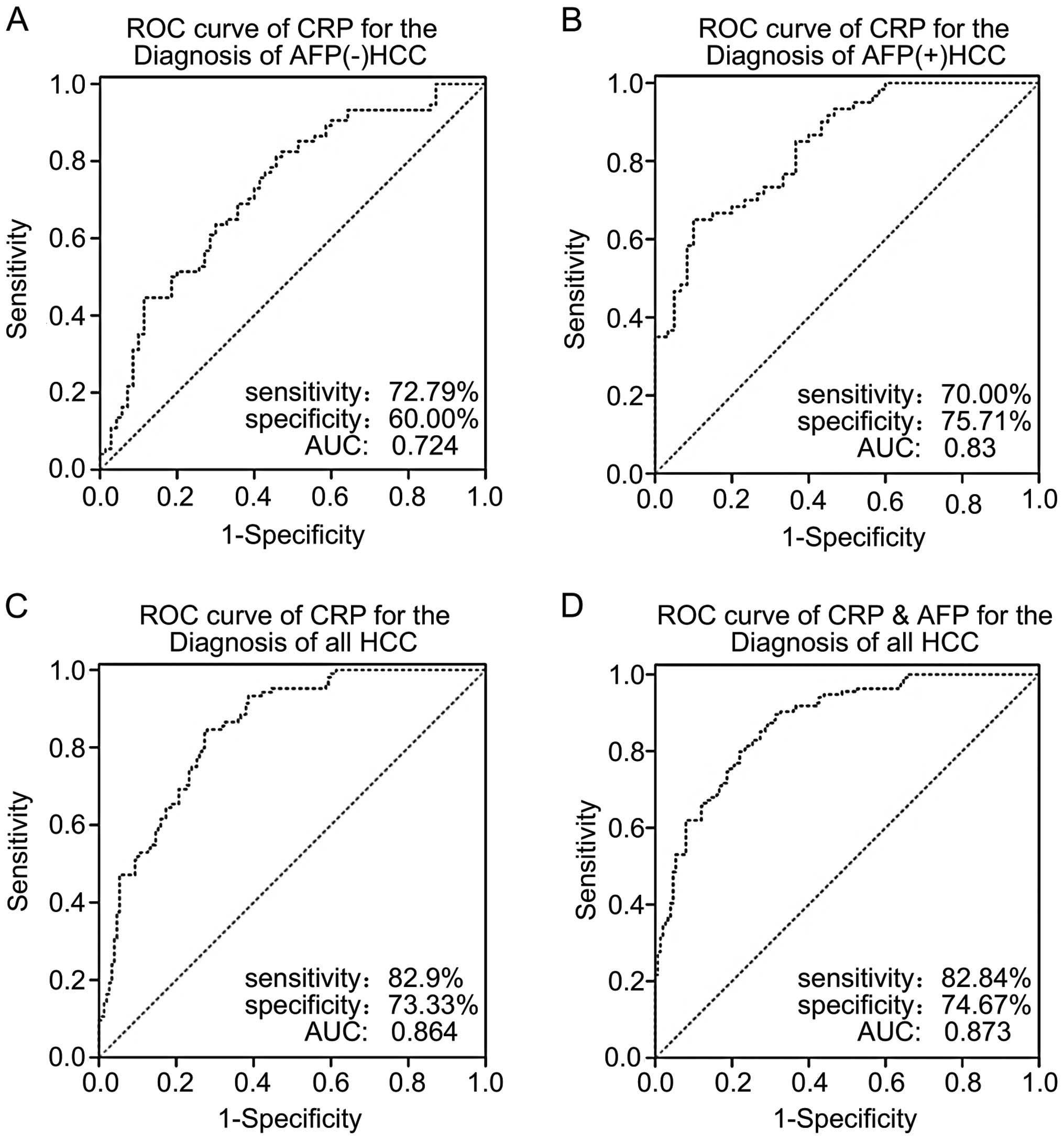
overexpression in clinical samples. Therefore, we performed ROC
analyses to address whether HCC diagnostic tests could be improved
by using CRP expression or combined CRP and AFP expression as
biomarkers for HCC diagnosis. According to our ROC results, the
cut-off value to confirm a positive diagnosis of HCC was 2361.82
ng/ml for CRP. The sensitivity and specificity for AFP(−) HCC
subjects, AFP(+) HCC subjects, all subjects combined were 72.97 and
60%; 70 and 75.71 and 82.09 and 73.33%, respectively (Fig. 4A–C). The positive predictive values
and negative predictive values for all subjects combined, AFP(−)
HCC subjects and AFP(+) HCC subjects were 73.3 and 82.1%; 65.9 and
67.7%; and 71.2 and 74.6%, respectively. The plasma CRP levels of
both the AFP(−) HCC group and all HCC patients combined had
significantly higher predictive accuracy than the plasma CRP levels
of the LC group. The area under ROC curve (AUC) of CRP in all HCC
patients and AFP(−) HCC patients was 0.8642 (95% CI, 0.819–0.902,
P<0.01), and 0.724 (95% CI, 0.643–0.795, P=0.0001), respectively
(Fig. 4A and C). When combining
CRP with AFP, the AUC was 0.873 (95% CI, 0.829–0.910, P<0.01),
which was statistically higher than the AUC of AFP alone
(AUC=0.812, 95% CI, 0.763–0.871, P<0.05) (Fig. 4D). It is important to emphasize
that 65.9% of the patients with AFP(−) serum in the present study
had elevated CRP.
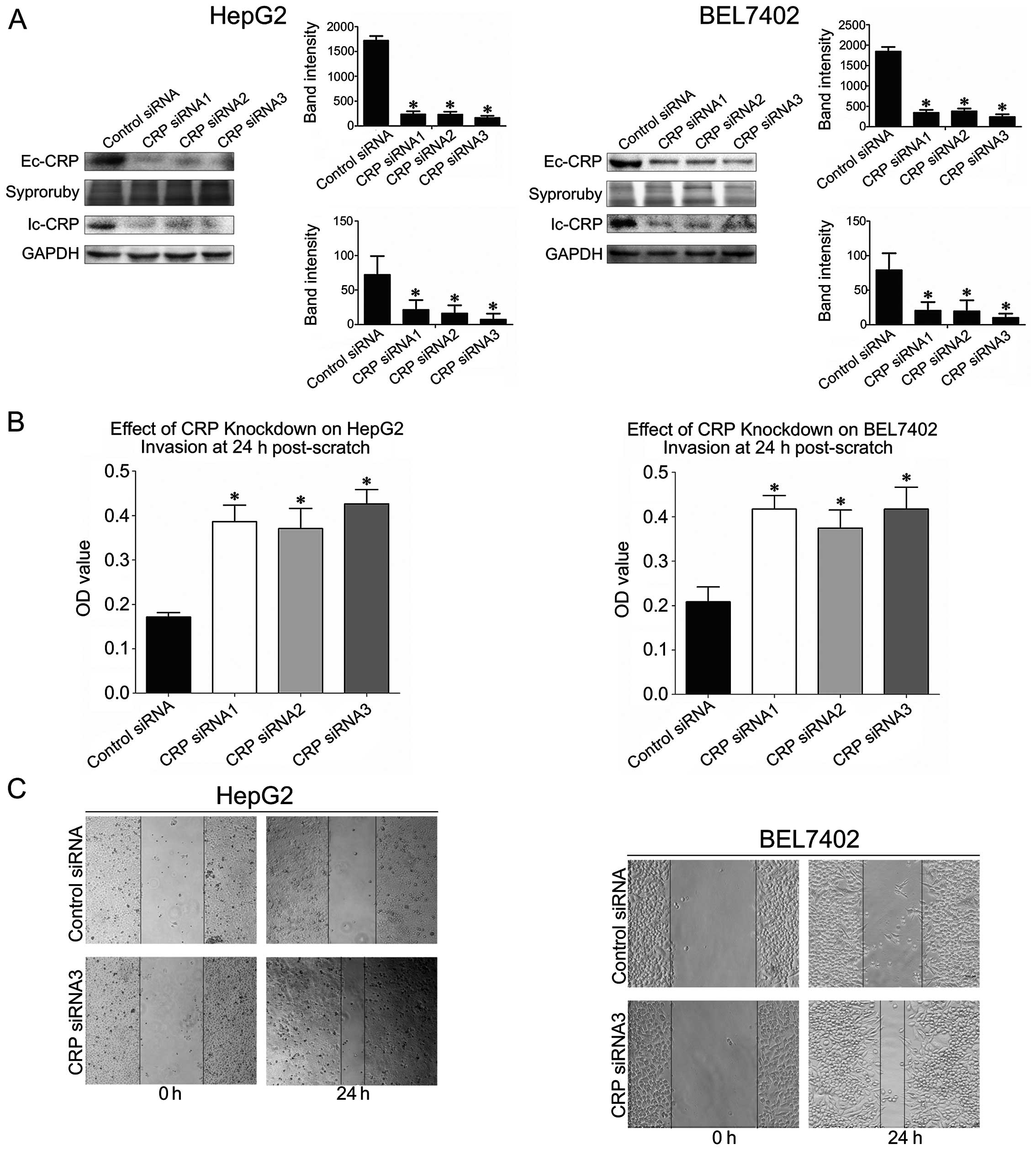
CRP knockdown promotes HCC cell migration
and invasion
To study the role of CRP in tumor cell motility, we
silenced CRP expression in HCC cell lines with CRP-specific siRNA.
According to western blot analysis, we found that each of the
CRP-specific siRNAs significantly downregulated CRP expression in
both HepG2 and BEL7402 cells (Fig.
5A). The assay showed that CRP-specific siRNAs significantly
increased the invasion ability of BEL7402 cells and HepG2 by 47–52
and 40–49%, respectively (P<0.05) (Fig. 5B). Additionally, we found that
CRP-specific siRNA transfection of BEL7402 cells and HepG2 resulted
in a 50–60 and 40–55% increase in wound healing, respectively
(P<0.05) (Fig. 5B).
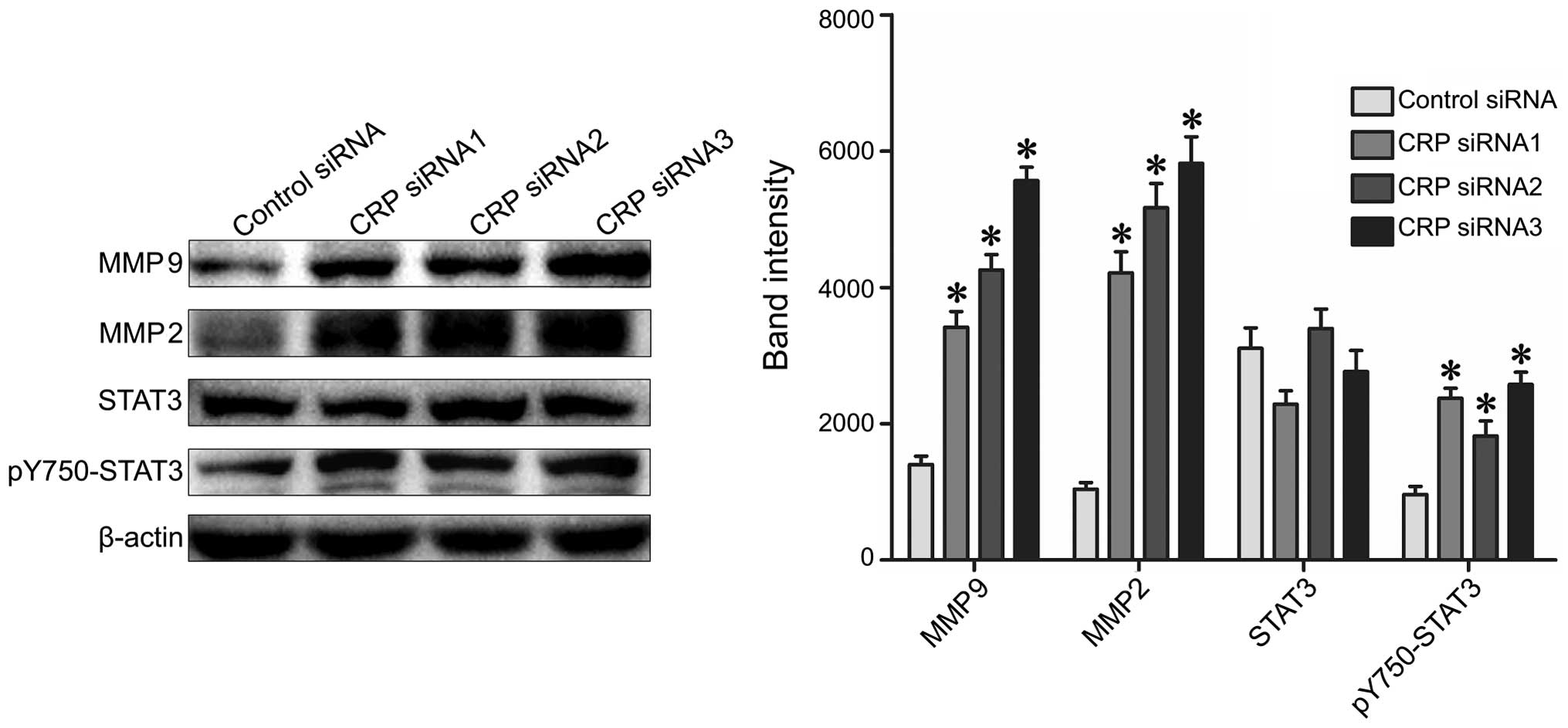
CRP knockdown promotes PY705-STAT3, MMP2
and MMP9 protein expression
To examine the functional consequences of aberrant
CRP expression, we analyzed the expression levels of the STAT3,
pY705-STAT3, MMP2 and MMP9 proteins. These proteins are all known
to participate in the pathogenesis of tumor metastasis (32,33).
We found that CRP-siRNA transfection resulted in significantly
increased expression of pY705-STAT3, MMP2 and MMP9 when compared to
control siRNA in both HepG2 and BEL7402 cells (Fig. 6).
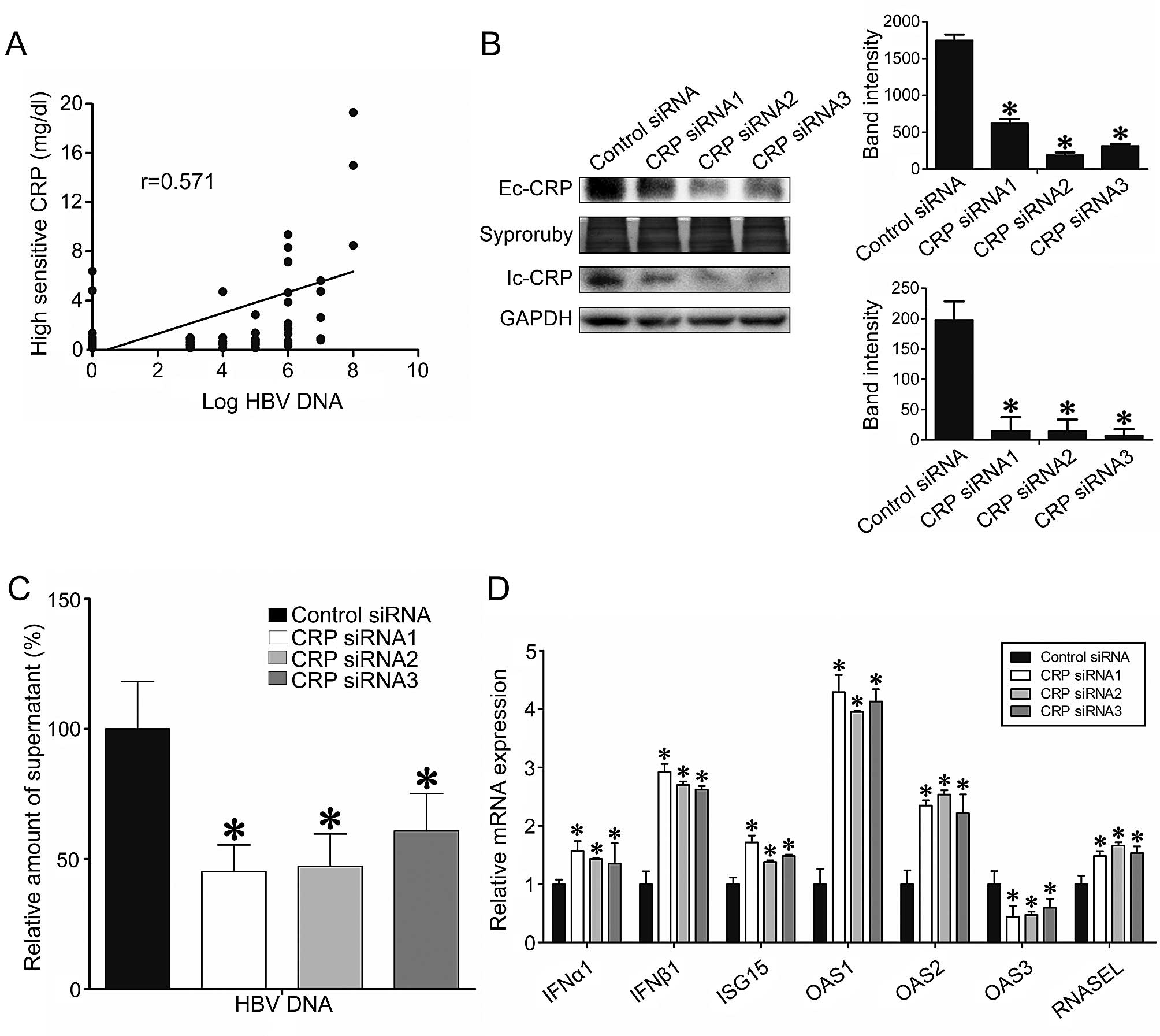
The roles of CRP in HBV replication and
type I IFN-stimulated gene expression
Considering the prevalence of HBV infection in HCC
patients, we investigated whether CRP expression level is
associated with HBV viral load. We observed that CRP plasma levels
were positively correlated with HBV-DNA copy number (Pearson
r=0.571, P<0.05) (Fig. 7A).
Additionally, CRP expression levels were higher in the plasma of
HBV-DNA positive patients than in HBV-DNA negative individuals
(P<0.05) (Fig. 7A). To assess
the effect of CRP expression level on HBV replication, we
transfected HepG2.2.15 cells, which stably produce HBV-DNA, with
CRP specific siRNA. CRP specific siRNA transfection resulted in
successful knockdown of both intracellular and extracellular CRP
(Fig. 7B). We found that the
HBV-DNA titer decreased nearly 2-fold in the CRP-knockdown groups
when compared the control group (Fig.
7C). Furthermore, we determined that CRP knockdown
significantly suppressed the transcription of type I IFNs, which
are known anti-viral cytokines, and resulted in significant
inhibition of genes downstream of IFN, including OAS1,
OAS2, OAS3, RNase L and ISG15
(P<0.05) (Fig. 7D).
Discussion
This functional and clinical proteomics study
demonstrated that CRP is a promising diagnostic biomarker for
AFP(−) and HBV-related HCCs. We observed that CRP has anti-HCC and
anti-HBV activities, and that CRP is an indicator of cancer-related
inflammation and HBV infection in HCC.
CRP is an exquisitely sensitive marker of
inflammation and tissue damage (34). Elevated CRP expression has been
detected in ovarian (35), lung
(36) and colon cancer (37). Based on recent studies, the serum
CRP levels are correlated with the poor prognosis in many
malignancies (37–39). While the molecular mechanism
underlying tumor-related CRP elevation in HCC or other cancers
remains unknown, several possible mechanisms have been proposed.
For instance, cancer growth and tumor-host cell interaction could
increase CRP levels (40).
Additionally, CRP levels might reflect an inflammatory response
activated as a secondary process in reaction to tumor necrosis or
other local tissue damage. Moreover, cancer cells produce cytokines
via autocrine pathways, such as IL-6 and IL-8, which in turn induce
CRP production (41). The
significance of inflammatory signaling through the STAT3 pathway
has been emphasized by numerous studies of HCC and other
malignancies (32,33). This helps to contextualize the
inhibitory role of CRP described by the presenr study as a host
defense mechanism that acts in part by inhibiting STAT3 activation
and the downstream expression of MMP2 and MMP9. Furthermore, we
demonstrated that CRP expression inhibits the migratory and
invasive phenotypes of HCC cells.
We observed that serum CRP was positive in 73.3% of
the HCC patients, and, more specifically, in 65.9% of the AFP(−)
HCC patients. The sensitivity and specificity of CRP expression for
HCC diagnosis in AFP(−) HCC subjects was 72.97 and 60%,
respectively. Additionally, the sensitivity and specificity of
using combined CRP and AFP expression levels for HCC diagnosis
(AUC=0.873) was a significant improvement over using AFP expression
levels alone (AUC=0.812). Therefore, CRP could be a useful
supplementary biomarker to AFP for HCC diagnosis, especially for
AFP(−) HCC. CRP expression has rarely been linked to HBV infection.
Intriguingly, we found a positive correlation between CRP
expression and HBV viral load after analyzing the data of 80
chronic HBV infected patients. We further investigated the
relationship of CRP and HBV with an in vitro model using
HepG2.2.15 cells. The results of these experiments indicated that
CRP serves as a pro-viral protein that promotes HBV replication and
suppresses anti-viral cytokines.
Because they are both acute-phase proteins similar
to CRP, studying the aberrant expression of the SAA and C9 proteins
may help increase our understanding of HCC development (42,43).
SAA is primarily generated by the liver in response to trauma,
infection, inflammation or neoplastic stimuli. It can promote
carcinogenesis by activating the transcriptional factor nuclear
factor kappa-B (NF-κB) (44) and
by inducing the expression of matrix metalloproteinase proteins
(MMPs) (45). Li et al
(46) reported that SAA affects
cell growth and invasion by activating NF-κB and STAT3 signaling in
human (HepG2) and mouse (H22) liver cancer cells.
C9 is the ninth member of the complement components
involved in the formation of the membrane attack complex (MAC)
(42). Preliminary studies
indicate that HCV core proteins attenuate immunity against
infection in part by inhibiting C9 and impairing the membrane
attack complex (47). Previous
proteomics investigations have identified the complement components
(C3, C5 and C9) as major serum proteins overexpressed in variety of
cancers, including familial aggregative HCC (48,49).
However, Ferrin et al (50)
recently reported that C9 was downregulated in HCV-infected HCC
patients, therefore, the role of C9 in liver tumorigenesis needs
further study.
Collectively, we demonstrated a quantitative
proteomic profiles of cirrhosis, AFP (−) HBV-associated HCC and AFP
(+) HBV-associated HCC. We revealed the correlation between the
serum CRP levels and AFP negative HCC patients, and suggest that
CRP might participate in the HBV replication. The lack of smaller
(<3 cm) HCCs, necessitates collection of further tissues and to
study the relation between CRP and the smaller (<3 cm) HCCs, as
our data need further validation with a larger cohort.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (nos. 81171560, 30930082,
81171561, 30972584 and 81372399), the National Science and
Technology Major Project of China (2008ZX10002-006,
2012ZX1002007001, 2011ZX09302005, 2012ZX09303001-001 and
2012ZX10002003).
References
|
1
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Park NH and Chung YH: Molecular mechanisms
of hepatitis B virus-associated hepatocellular carcinoma. Korean J
Hepatol. 13:320–340. 2007.(In Korean). View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen CJ, Yang HI, Su J, Jen CL, You SL, Lu
SN, Huang GT and Iloeje UH; REVEAL-HBV Study Group. Risk of
hepatocellular carcinoma across a biological gradient of serum
hepatitis B virus DNA level. JAMA. 295:65–73. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Taylor-Robinson SD, Foster GR, Arora S,
Hargreaves S and Thomas HC: Increase in primary liver cancer in the
UK, 1979–94. Lancet. 350:1142–1143. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Deuffic S, Poynard T, Buffat L and
Valleron AJ: Trends in primary liver cancer. Lancet. 351:214–215.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
El-Serag HB and Mason AC: Rising incidence
of hepatocellular carcinoma in the United States. N Engl J Med.
340:745–750. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Taketa K: Alpha-fetoprotein: Reevaluation
in hepatology. Hepatology. 12:1420–1432. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen DS, Sung JL, Sheu JC, Lai MY, How SW,
Hsu HC, Lee CS and Wei TC: Serum alpha-fetoprotein in the early
stage of human hepatocellular carcinoma. Gastroenterology.
86:1404–1409. 1984.PubMed/NCBI
|
|
9
|
Lok AS and Lai CL: alpha-Fetoprotein
monitoring in Chinese patients with chronic hepatitis B virus
infection: Role in the early detection of hepatocellular carcinoma.
Hepatology. 9:110–115. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tsai SL, Huang GT, Yang PM, Sheu JC, Sung
JL and Chen DS: Plasma des-gamma-carboxyprothrombin in the early
stage of hepatocellular carcinoma. Hepatology. 11:481–488. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Soga K, Watanabe T, Aikawa K, Toshima M,
Shibasaki K and Aoyagi Y: Serum des-gamma-carboxyprothrombin level
by a modified enzyme immunoassay method in hepatocellular
carcinoma: Clinical significance in small hepatocellular carcinoma.
Hepatogastroenterology. 45:1737–1741. 1998.PubMed/NCBI
|
|
12
|
Lee IN, Chen CH, Sheu JC, Lee HS, Huang
GT, Chen DS, Yu CY, Wen CL, Lu FJ and Chow LP: Identification of
complement C3a as a candidate biomarker in human chronic hepatitis
C and HCV-related hepatocellular carcinoma using a proteomics
approach. Proteomics. 6:2865–2873. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Feng JT, Liu YK, Song HY, Dai Z, Qin LX,
Almofti MR, Fang CY, Lu HJ, Yang PY and Tang ZY: Heat-shock protein
27: A potential biomarker for hepatocellular carcinoma identified
by serum proteome analysis. Proteomics. 5:4581–4588. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Görg A, Obermaier C, Boguth G, Harder A,
Scheibe B, Wildgruber R and Weiss W: The current state of
two-dimensional electrophoresis with immobilized pH gradients.
Electrophoresis. 21:1037–1053. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gygi SP, Rist B, Gerber SA, Turecek F,
Gelb MH and Aebersold R: Quantitative analysis of complex protein
mixtures using isotope-coded affinity tags. Nat Biotechnol.
17:994–999. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mirgorodskaya OA, Kozmin YP, Titov MI,
Körner R, Sönksen CP and Roepstorff P: Quantitation of peptides and
proteins by matrix-assisted laser desorption/ionization mass
spectrometry using 18O-labeled internal standards. Rapid
Commun Mass Spectrom. 14:1226–1232. 2000. View Article : Google Scholar
|
|
17
|
Ross PL, Huang YN, Marchese JN, Williamson
B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, et
al: Multiplexed protein quantitation in Saccharomyces cerevisiae
using amine-reactive isobaric tagging reagents. Mol Cell
Proteomics. 3:1154–1169. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rai AJ, Gelfand CA, Haywood BC, Warunek
DJ, Yi J, Schuchard MD, Mehigh RJ, Cockrill SL, Scott GB, Tammen H,
et al: HUPO Plasma Proteome Project specimen collection and
handling: Towards the standardization of parameters for plasma
proteome samples. Proteomics. 5:3262–3277. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lok AS and McMahon BJ: Chronic hepatitis
B: Update 2009. Hepatology. 50:661–662. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liaw YF, Leung N, Kao JH, Piratvisuth T,
Gane E, Han KH, Guan R, Lau GK and Locarnini S; Chronic Hepatitis B
Guideline Working Party of the Asian-Pacific Association for the
Study of the Liver. Asian-Pacific consensus statement on the
management of chronic hepatitis B: A 2012 update. Hepatol Int.
6:531–561. 2012. View Article : Google Scholar
|
|
21
|
European Association for the Study of the
Liver. EASL clinical practice guidelines on the management of
ascites, spontaneous bacterial peritonitis, and hepatorenal
syndrome in cirrhosis. J Hepatol. 53:397–417. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bruix J and Sherman M; American
Association for the Study of Liver Diseases. Management of
hepatocellular carcinoma: An update. Hepatology. 53:1020–1022.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang Y, Lim SK, Choong LY, Lee H, Chen Y,
Chong PK, Ashktorab H, Wang TT, Salto-Tellez M, Yeoh KG, et al:
Cathepsin S mediates gastric cancer cell migration and invasion via
a putative network of metastasis-associated proteins. J Proteome
Res. 9:4767–4778. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tong SW, Yang YX, Hu HD, An X, Ye F, Hu P,
Ren H, Li SL and Zhang DZ: Proteomic investigation of
5-fluorouracil resistance in a human hepatocellular carcinoma cell
line. J Cell Biochem. 113:1671–1680. 2012.
|
|
25
|
Yang Y, Toy W, Choong LY, Hou P, Ashktorab
H, Smoot DT, Yeoh KG and Lim YP: Discovery of SLC3A2 cell membrane
protein as a potential gastric cancer biomarker: Implications in
molecular imaging. J Proteome Res. 11:5736–5747. 2012.PubMed/NCBI
|
|
26
|
Ho J, Kong JW, Choong LY, Loh MC, Toy W,
Chong PK, Wong CH, Wong CY, Shah N and Lim YP: Novel breast cancer
metastasis-associated proteins. J Proteome Res. 8:583–594. 2009.
View Article : Google Scholar
|
|
27
|
Lepiller Q, Abbas W, Kumar A, Tripathy MK
and Herbein G: HCMV activates the IL-6-JAK-STAT3 axis in HepG2
cells and primary human hepatocytes. PLoS One. 8:e595912013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tong SW, Yang YX, Hu HD, An X, Ye F, Ren
H, Li SL and Zhang DZ: HSPB1 is an intracellular antiviral factor
against hepatitis B virus. J Cell Biochem. 114:162–173. 2013.
View Article : Google Scholar
|
|
29
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gan CS, Chong PK, Pham TK and Wright PC:
Technical, experimental, and biological variations in isobaric tags
for relative and absolute quantitation (iTRAQ). J Proteome Res.
6:821–827. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhou C, Simpson KL, Lancashire LJ, Walker
MJ, Dawson MJ, Unwin RD, Rembielak A, Price P, West C, Dive C, et
al: Statistical considerations of optimal study design for human
plasma proteomics and biomarker discovery. J Proteome Res.
11:2103–2113. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xie TX, Wei D, Liu M, Gao AC, Ali-Osman F,
Sawaya R and Huang S: Stat3 activation regulates the expression of
matrix metalloproteinase-2 and tumor invasion and metastasis.
Oncogene. 23:3550–3560. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yu H, Pardoll D and Jove R: STATs in
cancer inflammation and immunity: A leading role for STAT3. Nat Rev
Cancer. 9:798–809. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yang J, Wezeman M, Zhang X, Lin P, Wang M,
Qian J, Wan B, Kwak LW, Yu L and Yi Q: Human C-reactive protein
binds activating Fcgamma receptors and protects myeloma tumor cells
from apoptosis. Cancer Cell. 12:252–265. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hefler LA, Concin N, Hofstetter G, Marth
C, Mustea A, Sehouli J, Zeillinger R, Leipold H, Lass H, Grimm C,
et al: Serum C-reactive protein as independent prognostic variable
in patients with ovarian cancer. Clin Cancer Res. 14:710–714. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xu M, Zhu M, Du Y, Yan B, Wang Q, Wang C
and Zhao J: Serum C-reactive protein and risk of lung cancer: A
case-control study. Med Oncol. 30:3192013. View Article : Google Scholar
|
|
37
|
Nozoe T, Matsumata T, Kitamura M and
Sugimachi K: Significance of preoperative elevation of serum
C-reactive protein as an indicator for prognosis in colorectal
cancer. Am J Surg. 176:335–338. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Fujikawa K, Matsui Y, Oka H, Fukuzawa S
and Takeuchi H: Serum C-reactive protein level and the impact of
cytoreductive surgery in patients with metastatic renal cell
carcinoma. J Urol. 162:1934–1937. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Nozoe T, Saeki H and Sugimachi K:
Significance of preoperative elevation of serum C-reactive protein
as an indicator of prognosis in esophageal carcinoma. Am J Surg.
182:197–201. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Mantovani A, Allavena P, Sica A and
Balkwill F: Cancer-related inflammation. Nature. 454:436–444. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Heikkilä K, Ebrahim S and Lawlor DA: A
systematic review of the association between circulating
concentrations of C reactive protein and cancer. J Epidemiol
Community Health. 61:824–833. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Tschopp J, Podack ER and Müller-Eberhard
HJ: Ultrastructure of the membrane attack complex of complement:
Detection of the tetramolecular C9-polymerizing complex C5b-8. Proc
Natl Acad Sci USA. 79:7474–7478. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Urieli-Shoval S, Linke RP and Matzner Y:
Expression and function of serum amyloid A, a major acute-phase
protein, in normal and disease states. Curr Opin Hematol. 7:64–69.
2000. View Article : Google Scholar
|
|
44
|
Betts JC, Cheshire JK, Akira S, Kishimoto
T and Woo P: The role of NF-kappa B and NF-IL6 transactivating
factors in the synergistic activation of human serum amyloid A gene
expression by interleukin-1 and interleukin-6. J Biol Chem.
268:25624–25631. 1993.PubMed/NCBI
|
|
45
|
Migita K, Kawabe Y, Tominaga M, Origuchi
T, Aoyagi T and Eguchi K: Serum amyloid A protein induces
production of matrix metalloproteinases by human synovial
fibroblasts. Lab Invest. 78:535–539. 1998.PubMed/NCBI
|
|
46
|
Li Y, Cai L, Wang H, Wu P, Gu W, Chen Y,
Hao H, Tang K, Yi P, Liu M, et al: Pleiotropic regulation of
macrophage polarization and tumorigenesis by formyl peptide
receptor-2. Oncogene. 30:3887–3899. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kim H, Meyer K, Di Bisceglie AM and Ray R:
Hepatitis C virus suppresses C9 complement synthesis and impairs
membrane attack complex function. J Virol. 87:5858–5867. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Chong PK, Lee H, Loh MC, Choong LY, Lin Q,
So JB, Lim KH, Soo RA, Yong WP, Chan SP, et al: Upregulation of
plasma C9 protein in gastric cancer patients. Proteomics.
10:3210–3221. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Narayanasamy A, Ahn JM, Sung HJ, Kong DH,
Ha KS, Lee SY and Cho JY: Fucosylated glycoproteomic approach to
identify a complement component 9 associated with squamous cell
lung cancer (SQLC). J Proteomics. 74:2948–2958. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ferrín G, Ranchal I, Llamoza C,
Rodríguez-Perálvarez ML, Romero-Ruiz A, Aguilar-Melero P,
López-Cillero P, Briceño J, Muntané J, Montero-Álvarez JL, et al:
Identification of candidate biomarkers for hepatocellular carcinoma
in plasma of HCV-infected cirrhotic patients by 2-D DIGE. Liver
Int. 34:438–446. 2013. View Article : Google Scholar : PubMed/NCBI
|