Introduction
Gliomas are the most common and aggressive types of
tumors in central nervous system. It has been proved that dozens of
molecules and signal pathways are involved in the genetic
alterations and chromosome aberrations during tumorigenesis. The
abnormal abilities of invasion, proliferation, migration and
apoptosis of glioma cells are the main obstacles to better
prognosis (1–3). Despite the achievements in
conventional treatments, the median survival period of patients
suffering from glioblastoma multiforme (GBM), a type of grade IV
glioma, remains at 12 months over the past few decades (4). Hence, discovering new therapeutic
targets and developing mechanism-based approaches is an urgent
need.
MicroRNAs (miRNA or miR) are a group of endogenous
non-coding small RNAs which are expressed in the vast majority of
eukaryotes (5). They cause gene
silencing by targeting specifically 3′-untranslated regions (3′UTR)
of mRNAs, leading to the degradation or repression of mRNA
translation (6,7). Some miRNA precursors can form two
stably expressing mature miRNAs, which are distinguished by adding
‘−3p' or ‘−5p' to the end of the name according to their procession
positions. Emerging evidence has strongly indicated that this
post-transcriptional regulation by miRNAs has major implications in
many physiological and pathological processes, and is associated
with the initiation and progression of human cancer, including
glioma (8–11). Some miRNAs are proved to be crucial
oncogenes or tumor suppressors of glioma (12–14).
miR-21, miR-10b, miR-93 and miR-155 are upregulated
in gliomas and have been shown to be significant contributors in
tumor growth (13,15–17);
while miR-7, miR-124, miR-128 and miR-137 are downregulated with
tumor suppressor activities (16,18–20).
Diverse expression profiles between non-neoplasm and glioma tissues
can partly elucidate the functions of miRNAs in tumorigenesis and
therefore can be utilized to predict novel therapeutic targets. We
noticed that miR-95 was reported to have different expression
profiles between glioma tissues and non-neoplasm brain tissues by
sequencing analysis (21). The
results of functional research on miR-95 seem to be contradictory,
as miR-95 promotes the growth of colorectal, pancreatic, prostate
and breast cancer (22–24), while has anticancer activity in
hepatic, brain and neck cancer (25,26).
However, miR-95 has not been previously reported in gliomas. So
here we recruited miR-95-3p as a candidate to explore its
biological function in glioma.
In this study, we first compared the expression
level of miR-95-3p in glioma and non-neoplasm brain tissues by
qRT-PCR, and evaluated its influence on the proliferation,
invasion, cell cycle and apoptosis of glioma in vitro. We
discovered the reverse relationship between CELF2 and miR-95-3p
expression both in cell lines and human samples, and utilized
luciferase reporter assay to confirm that CELF2 was a novel target
of miR-95-3p. Rescue experiment showed that downregulation of CELF2
could largely abolish the effect induced by repression of
miR-95-3p. Taken together, we conclude that miR-95-3p affects
proliferation, apoptosis and invasion of glioma cells by targeting
CELF2, and we identified miR-95-3p as a putative therapeutic target
and CELF2 as a potential tumor suppressor.
Materials and methods
Clinical sample collection
Human glioma samples and non-neoplasm brain samples
were collected from the Neurosurgery Department of the Second
Affiliated Hospital of Hebei Medical University in 2014. Fifteen
females and 35 males were included, with ages ranging from 30 to
60. Thirty-five tumor samples, including 4 grade I, 5 grade II
glioma, 17 grade III and 9 grade IV tissues (GBM), were obtained by
surgical resection, and 5 non-neoplasma samples as control were
obtained by intracranial decompression for severe traumatic brain
injury. Diagnoses were established histologically by two
pathologists according to the WHO classification guidelines. All
samples were stored at −80°C in liquid nitrogen immediately after
resection. Written informed consent was obtained from all patients
and the study was approved by the ethics committee of The Second
Affiliated Hospital of Hebei Medical University.
Cell culture and transfection
Human glioma cell lines U87 and U251 were obtained
from the American Type Culture Collection (Manassas, VA, USA) and
were maintained in α-MEM (Gibco BRL, Grand Island, NY, USA)
supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin
and 100 μg/ml streptomycin at 37°C with 5% CO2. The
miR-95-3p ASO, control ASO, CELF2 siRNA and control siRNA were
synthesized and purified by GenePharma, Shanghai, China. The RNA
sequences mentioned above were as follows: miR-95-3p ASO:
5′-TGCTCAATAAATACCCGTTGAA-3′; Ctrl ASO:
5′-TTATCGCCATGTCCAATGAGGCT-3′; CUGBP2 siRNA:
5′-GCAAACCUUACUGAUCCUA-3′; Ctrl siRNA:
5′-UAAGGCUAUGAAGAGAUAC-3′.
The control RNAs contain random sequences and are
predicted not to have any interactions in cells. Cell transfection
was performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA,
USA) according to the manufacturer's instructions. To transfect RNA
oligonucleotides, 10 pmol of miRNA antisense oligonucleotides and
ctrl ASO were used. For rescue experiments, cells were
contransfected with 10 pmol of miRNA ASO and siRNA or control siRNA
in a 96-well plate.
RNA extraction and real-time quantitative
RT-PCR
Total cell RNA was extracted using TRIzol reagent
(Invitrogen) according to the manufacturer's instructions. miRNA
level was measured by qRT-PCR. All the primers of miRNAs for TaqMan
miRNA assays were purchased from Genewiz (Beijing, China). Human U6
small nuclear RNA was used as an internal normalized reference. The
forward and reverse primers of miR-95-3p and U6 are as follows:
miR-95-3p F/R, 5′-TGCGGTTCAACGGGTATTTATTG-3′/5′-CCAGTG
CAGGGTCCGAGGT-3′; U6 F/R, 5′-TGCGGGTGCTCGCTT
CGGCAGC-3′/5′-CCAGTGCAGGGTCCGAGGT-3′. RNA (5 μg) was mixed with 1
μl of MMLV reverse transcriptase (Promega, Madison, WI, USA) to
synthesize complementary DNA according to the manufacturer's
instructions. The expression of miR-95-3p in human samples was
quantified using SYBR® Premix Ex Taq™ (Takara, Tokyo,
Japan) in the ABI PRISM® 7500 real-time PCR system
(Applied Biosystems, Foster City, CA, USA). Real-time PCR was
performed according to the manufacturer's instructions. Melting
curves were used to evaluate non-specific amplification. All
experiments were performed using biological triplicates and
experimental duplicates. The relative expression levels were
evaluated via the 2−ΔΔCt method.
Western blotting
U87 or U251 cells were processed with RIPA lysis
buffer (Saierbio, Tianjing, China) for 48 h before collecting
protein samples. Total protein (32 μl) and 8 μl of loading buffer
(5X) was boiled for 3 min and cooled down for 3 min on ice.
Heat-denatured protein samples were resolved by 10%
SDS-polyacrylamide gel (SDS-PAGE) electrophoresis under 100 V and
transferred to hybond-nitrocellulose membranes (Millipore, USA)
under 350 mA at 4°C. The membrane was blocked in Blotto solution
for 2 h, and then incubated for 12 h at 4°C with primary rabbit
anti-human monoclonal CELF2 antibody (1:1,000, Abcam, Cambridge,
UK) overnight for combination. The GAPDH expression level was
measured using rabbit anti-human monoclonal GAPDH antibody
(1:3,000, Saier, Tianjing, China) as a loading control. Then the
membrand was incubated for 2 h with HRP marked goat anti-rabbit
second antibody (1:5,000, Saier, Tianjing, China). The bound
antibody was detected with the use of Western
Lightning®-ECL, Enhanced Chemiluminescence Substrate
(Perkin-Elmer, NEL100001EA). The band density of CELF2 was
quantified after normalization with the density of GAPDH.
MTT assay
U87 (1×104) or U251 cells were seeded
into each well of a 96-well plate one day before transfection.
Following a 48-h incubation after transfection, 20 μl of MTT
reagent (5 mg/ml) was added into each well and the plate was
incubated for another 4 h. The supernatant was removed, and 150 μl
of DMSO was added into each well to dissolve the precipitate. The
absorbance value was recorded at 570 nm using a microplate reader
(BioTek, Winooski, VT, USA). All experiments were performed using
biological triplicates and experimental duplicates, and the data
are presented as mean ± SD.
Flow cytometry
The apoptosis analysis was performed with Annexin
V/7-AAD apoptosis detection kit (KeyGen, Nanjing, China) according
to the manufacturer's instructions. Briefly, 5×105 cells
were planted into a 6-well plate and treated with 0.1% trypsin and
0.02% EDTA, and were washed twice with cold PBS before collection.
The cells were mixed with 100 μl of 1X binding buffer and
resuspended. Annexin V (10 μl) was added and incubated for 30 min.
Then 380 μl of 1X binding buffer and 10 μl of 7-AAD was added
sequently and fully mixed with the cells. The fluorescence values
of Annexin V and 7-AAD were measured by flow cytometry (Beckman
Coulter, CA, USA).
For cell cycle assay, 5×105 cells were
planted into a 6-well plate, washed by 1X PBS once and treated with
0.1% trypsin and 0.02% EDTA for digestion. The cells were suspended
in 1 ml culture medium and centrifuged under 12,000 × g for 5 min.
The supernatant was discarded and 1 ml 1X PBS was added into the
cells then centrifuged for 5 min. The cells were stained with PI
for 30 min and then investigate by flow cytometry. All the
procedures were conducted according to the manufacturer's
instructions.
Cell invasion assay
Cell invasion ability was determined by Corning
transwell insert chambers (Corning, USA) and BD BioCoat Matrigel
(BD Biosciences, Bedford, MA, USA). U87 or U251 cells were
harvested and trypsinized in serum-free medium after transfection.
Then the cell suspension (200 μl, 1×105 cells) was added
into the chamber and incubated 24 h at 37°C. Cells that invaded the
Matrigel and passed through the filter were fixed with 20% methanol
for 30 min, stained with crystal violet for 15 min, and then imaged
under ×100 magnification Nikon 90I light microscope (Nikon, Japan)
and counted in 6 randomized fields.
Vector construction
For overexpression of miR-95-3p, the human
pre-miR-95 sequence was amplified and cloned into pcDNA3.1-hisA
constructs (Invitrogen) to generate pcDNA3.1-miR-95 expression
vector. The primers were miR-95-KL-F: CCGGAATTCGAAGGTAGGATTGTGA CAC
and miR-95-KL-R: CGCGGATCCGGAGGGATGGAT GAATGAC.
For luciferase reporter assay, the 3′UTR of the
human CELF2 gene containing the putative binding site was PCR
amplified using the following primers: wt-CELF2-Fw: AAA CTAGCGG
CCGCTAGTCTAGGAATGGGCTTTTTTTCACCGTTGAA CCCTAGACCTGT; wt-CELF2-Rv:
CTAGACAGGTCTAGG GTTCAACGGTGAAAAAAAGCCCATTCCTAGACTAGC GGCCGCTAGTTT.
3′UTR of CELF2 gene containing the mutant binding site used the
following primers: mut-CELF2-Fw: AAACTAGCGGCCGCTAGTCTAGGAATGGGCTTT
TTTTCAGGCAACTACCCTAGACCTGT; mut-CELF2-Rv:
CTAGACAGGTCTAGGGTAGTTGCCTGAAAAAAAGCC CATTCCTAGACTAGCGGCCGCTAGTTT.
Fragments in CELF2 3′UTR containing the binding site (wt-CELF2) or
mutated site (mut-CELF2) were, respectively, double-digested with
XbaI/EcoRI and cloned downstream of firefly
luciferase coding region sites of pmirGLO plasmid (Promega,
Madison, WI, USA).
Luciferase reporter assay
The binding site was replaced with its complementary
sequence to generate mutation. U251 cells were plated in a 96-well
plate and then co-transfected with 0.25 μg of wt-CELF2 or mut-CELF2
expression vector and 10 pmol of miR-95-3p ASO or control ASO using
Lipofectamine 2000 reagent. Cells were collected 48 h after
transfection and analyzed using the Dual-Luciferase Reporter Assay
System (Promega, Madison, WI, USA). Luciferase activity was
detected by F200/M200 microplate fluorescence reader (Tecan
Infinite). The co-transfected Renilla luciferase control vector was
used as an internal control to correct the differences in both
transfection and the harvesting efficiency. All the regents and
instruments were performed according to the manufacturer's
instructions. Transfection experiments were performed in duplicates
and repeated at least thrice in independent experiments.
Statistical analysis
All tests and vector graphics were done using PRISM
version 5.0 (GraphPad Software Inc., San Diego, CA, USA)
statistical software. The experiments were carried out for three
times independently. Data are mean ± standard deviation except when
indicated otherwise. Spearman's rank correlation test was used for
association analysis between miR-95-3p and CELF2 level data.
Student's t-test was used to analyze the statistical significance
between two groups. One-way ANOVA was used to compare multiple
groups. P<0.05 was considered to be significant (shown in
figures as: *P<0.05;
**P<0.01;***P<0.001).
Results
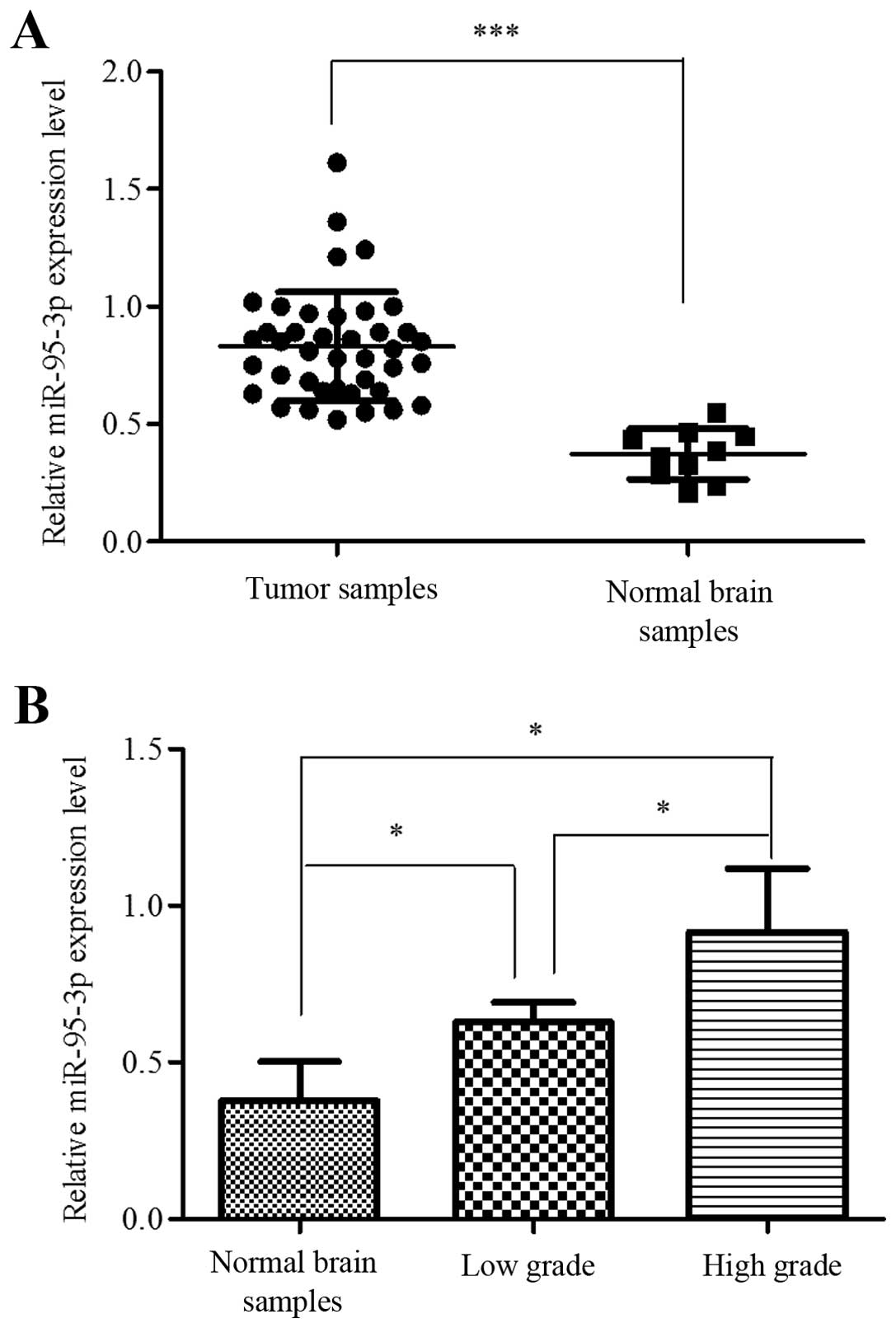
miR-95-3p is significantly upregulated in
human glioma tissues
To determine the role of miR-95-3p in tumorigenesis
of human glioma, we compared the expression level of miR-95-3p in
glioma samples and non-neoplasma brain samples. In this study, 35
tumor samples were used as experimental group and 5 non-neoplasma
samples as control. Case number, age, gender, WHO grade and
pathological report of each patient were recorded. The qRT-PCR
results showed that miR-95-3p was significantly overexpressed in
glioma tissues compared to control group (Fig. 1A). We also analyzed the correlation
between expression level and pathological grade. One-way ANOVA test
showed that the differences between non-neoplasm tissues, low grade
glioma (grade I and II) and high grade glioma (grade III and IV)
were significant, while the differences between grade I and II or
between grade III and IV were not (Fig. 1B).
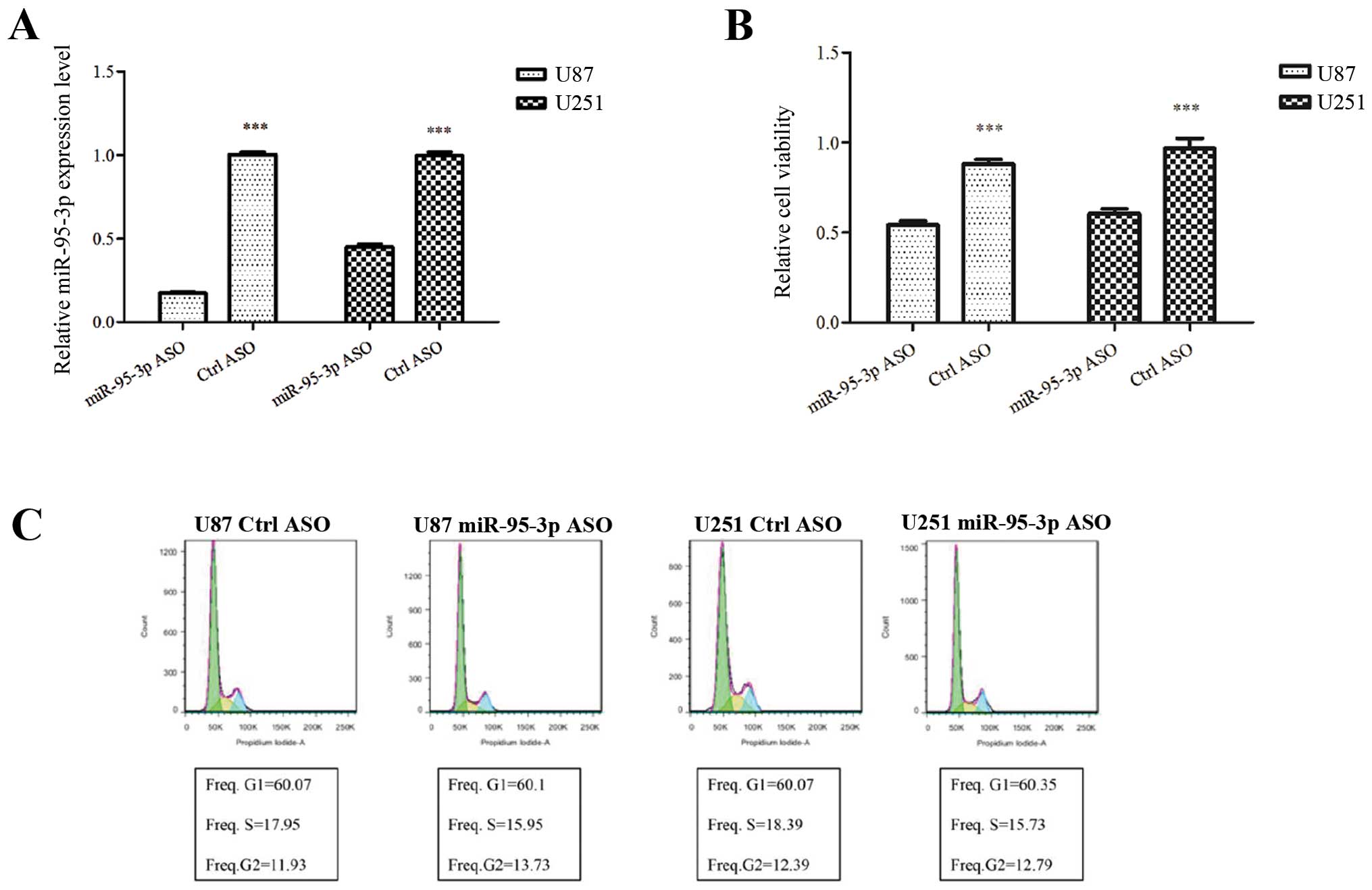
Downregulation of miR-95-3p manifests
tumor-suppressive functions in glioma cell lines
To understand the function of miR-95-3p in glioma
cells, expression of miR-95-3p was downregulated in U87 and U251
cell lines for a loss-of-function behavior. Antisense
oligonucleotide (ASO) was introduced into the cells to repress
miR-95-3p expression. Control cells were transfected with false
ASO. Forty-eight hours later, miR-95-3p expression levels were
examined by qRT-PCR. As shown in Fig.
2A, miR-95-3p was downregulated in U87 and U251 cells after
transfection of ASO. Then we investigated the nature of the cells
with different miR-95-3p levels.
MTT assay was performed to investigate the effect of
miR-95-3p on cell proliferation. We found that transfection of U87
cells with miR-95-3p ASO significantly decreased the relative
absorbance of MTT, which was consistant with that of U251 cells
(Fig. 2B). The data indicated an
anti-proliferation effect of downregulation of miR-95-3p on glioma
cells. Then, we conducted flow cytometry to analyze the effect of
miR-95-3p on the cell cycle and apoptosis. Our experiment failed to
prove any difference in the cell cycle. The proportion of U87 or
U251 cells in G1, S and G2 phase stayed almost unchanged after
transfection with miR-95-3p ASO, compared to control group
(Fig. 2C). As to apoptosis, U87 or
U251 cells transfected with miR-95-3p ASO had a significant
increase in number stained by Annexin V and/or 7-AAD, compared to
cells with false ASO (Fig. 2D).
The data showed that downregulation of miR-95-3p induces cell
apoptosis. Finally, cell invasiveness was measured by transwell
assay in order to evaluate metastasis potential. The cells were
imaged and counted after 24 h of incubation in the transwell
chamber. Our experiments showed less U87 or U251 cells transfected
with miR-95-3p ASO invaded through the membrane than those with
false ASO, which indicated downregulation of miR-95-3p decreased
cell invasiveness (Fig. 2E). Taken
together, the results manifested that downregulation of miR-95-3p
has therapeutic effects on glioma proliferation, invasion and
apoptosis.
CELF2 is a bona fide target of
miR-95-3p
To delineate the mechanism by which miR-95-3p
regulates the biological behavior of glioma cells, we searched the
online program Targetscan for the target. Among all the protein
candidates predicted, we finally recruited CELF2 (NM_001025077) as
a putative target and identified one potential binding site for
miR-95-3p (Fig. 3A and B). To
confirm that CELF2 is a bona fide target of miR-95-3p,
luciferase reporter assays were performed. Fig. 3C shows the mode pattern of pmirGLO
vector. We demonstrated the fact that miR-95-3p ASO could
significantly increase luciferase activity, which could be
abrogated by the mutation of the binding site (Fig. 3D).
We explored the expression profiles of CELF2 and
miR-95-3p in 7 human samples. Each piece of sample used in this
experiment for western blot assay came from the same tissue with
that for qRT-PTR to make their results comparable (Fig. 4A and B). The results supported a
negative correlation between CELF2 and miR-95-3p expression level
in human tissues (Fig. 4C). We
modified the level of miR-95-3p in U251 cells by tansfecting with
ASO or expression vector, to investigate the effect of miR-95-3p on
CELF2 in vitro. Compared to control cells, CELF2 was
downregulated in cells overexpressing miR-95-3p while upregulated
in cells low-expressing miR-95-3p (Fig. 4D). Based on the above, we proved
that miR-95-3p suppresses CELF2 gene expression by regulating 3′UTR
at the post-transcription level.
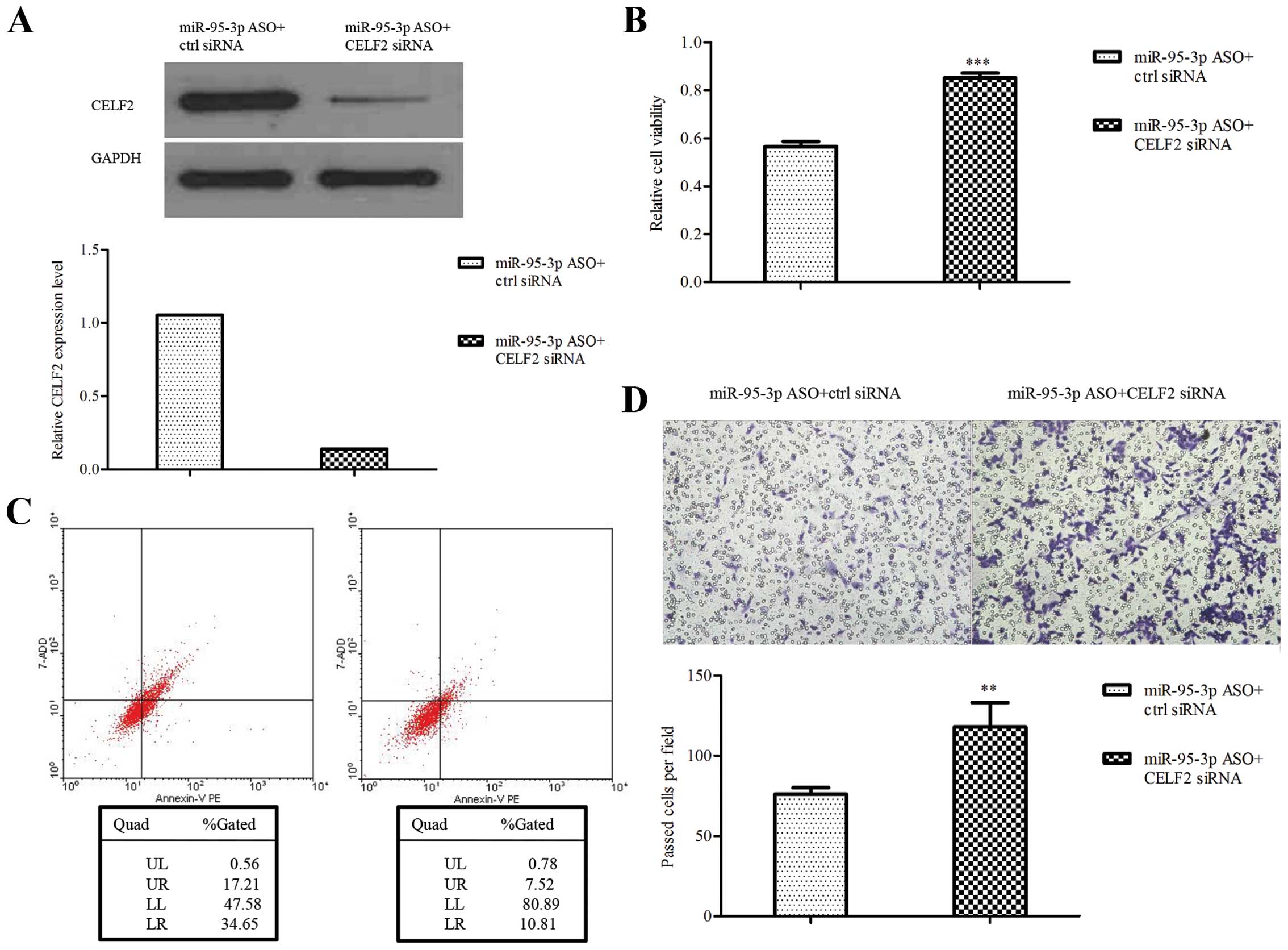
CELF2 rescues the effect of miR-95-3p on
glioma cells
To determine whether miR-95-3p regulates glioma cell
biological behavior by targeting CELF2, CELF2 siRNA was used to
perform rescue experiments. The downregulation of CELF2 by siRNA
was validated by western blotting (Fig. 5A). The results showed that U251
cells transfected with miR-95-3p ASO plus CELF2 siRNA were
increased in cell proliferation and invasion while decreased in
apoptosis, compared to control (Fig.
5B–D). The data further suggest that CELF2 is a functional
downstream target of miR-95-3p in regulating glioma biological
behavior.
Discussion
Given the fact that cancer cells develop
extraordinary characteristics which make them refractory to
therapies, it is crucial to understand the underlying molecular
biological mechanism. Evidence has shown miRNAs are implicated in
many pathological and physiological processes, and we focused on
miRNAs to explore their interaction with gliomas. The functions of
miR-95-3p has been studied previously. Huang et al (22) found miR-95 could promote
proliferation in human colorectal carcinoma cells. It has also been
pointed out that miR-95 overexpression increased tumor growth and
resistance to radiation treatment in prostate cancer cells
(23). Our preliminary test showed
that the expression difference of miR-95 was significant between
glioma and non-neoplasm brain samples. So we picked out miR-95-3p,
one of miR-95 mature sequences, for study. We analyzed our data and
tried to identify correlations of miR-95-3p expression level with
pathological grade and found miR-95-3p level increased with WHO
grade.
As miR-95-3p has hardly been investigated in glioma
previously, we planned functional experiments on multiple aspects
of tumorigenesis, including proliferation, cell cycle, apoptosis
and invasion. In order to set up the experimental group, miR-95-3p
ASO was transfected into glioma cells to reduce the miR-95-3p
level. In MTT assay, we observed downregulation of miR-95-3p
repressed glioma cells proliferation. MTT results reflected the
change of mitochondria viability, or the amount of living cells,
regardless of the cause. To explain how the living cells decreased,
we further analyzed the cell cycle and apoptosis by flow cytometry.
Our results manifested no significant difference in cell cycle
between experimental and control group, which indicated miR-95-3p
does not affect glioma cell growth rate. Apoptosis analysis showed
a significantly increased number of cell death in the experimental
group. So we inferred that miR-95-3p suppresses glioma cell
proliferation by inducing apoptosis, rather than by slowing down
cell mitosis. As invasiveness is an important factor in tumor
metastasis, we also performed transwell test to investigate the
function of miR-95-3p on this aspect. Less cells transfected with
miR-95-3p invaded through the membrane than control, as we
expected.
In the following phase of the study, we aimed to
understand the mechanism by which miR-95-3p mediates glioma
behavior. We searched the online programs TargetScan, mirWalk and
PicTar to identify the target of miR-95-3p and focused on CELF2.
CELF (CUGBP- and ETR-3-like family) is a group of RNA-binding
proteins encoding various functions in post-transcriptional level,
including alternative splicing, RNA editing and mRNA translation
(27). These proteins contain 3
RNA recognition motifs (RRMs), which are 80–90 amino acid domains
with conserve sequences for proteins to bind RNAs (28). Two of the RRMs are located near the
N-terminus and separated from the third one by an intervening
bridge segment (29). CELF2 (also
known as CUGBP2, NAPOR2, ETR3), one of the CELF family members, is
expressed ubiquitously, albeit at higher level in mouse cells
(30). In view of the fact that
CELF2 was found involved in neuroblastoma cell apoptosis (31) and is encoded by a gene located on
chromosome 10, which shows monosomy in ≤60% and partial loss in 80%
of high grade gliomas (32,33),
we hypothesized CELF2 may be implicated in the development of
glioma as well. We employed luciferase reporter assay to verify our
theory. The results indicated that miR-95-3p ASO significantly
increased luciferase activity, which could be abrogated by mutation
on the predicted binding site. In the experiments of western
blotting and qRT-PCR, our data implied the levels of CELF2 and
miR-95-3p were negatively correlated both in cell lines and human
samples. The conclusion that CELF2 is a direct target of miR-95-3p
was drawn.
To determine whether miR-95-3p regulates glioma
cells biological behavior by CELF2, rescue experiments were
performed. From the results we found that the effects of miR-95-3p
ASO on glioma cells were rescued by decreased expression of CELF2.
Thus, the miR-95-3p-CELF2 regulation mechanism on glioma was
established. Furthermore, CELF2 was also reported to promote cell
apoptosis by mediating Mcl-1, and downregulation of CELF2 by
prostaglandin E2 represses mitotic catastrophe induced by radiation
in colon cancer cells (34,35).
Subramaniam et al (36)
also found that CELF2 plays a role in mitotic catastrophe of
pancreatic cancer cells. It can be inferred that CELF2 may play an
important role in the mitotic process, although miR-95-3p does not,
and there may be other pathways related to CELF2 regulating
glioma.
In conclusion, this study demonstrates miR-95-3p is
upregulated in glioma samples and downregulation of miR-95-3p
inhibits proliferation, invasion and promotes apoptosis of glioma
cells by targeting CELF2. Thus, we identified miR-95-3p as a
putative therapeutic target and CELF2 as a potential tumor
suppressor.
Acknowledgements
We thank Mr. Shuang Chen and Yong Pu (Saierbio,
Tianjing, China) for their technical advice.
References
|
1
|
Giese A, Bjerkvig R, Berens ME and
Westphal M: Cost of migration: Invasion of malignant gliomas and
implications for treatment. J Clin Oncol. 21:1624–1636. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lefranc F, Brotchi J and Kiss R: Possible
future issues in the treatment of glioblastomas: Special emphasis
on cell migration and the resistance of migrating glioblastoma
cells to apoptosis. J Clin Oncol. 23:2411–2422. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Singh SK, Clarke ID, Hide T and Dirks PB:
Cancer stem cells in nervous system tumors. Oncogene. 23:7267–7273.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Furnari FB, Fenton T, Bachoo RM, Mukasa A,
Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, et al:
Malignant astrocytic glioma: Genetics, biology, and paths to
treatment. Genes Dev. 21:2683–2710. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Eulalio A, Huntzinger E and Izaurralde E:
Getting to the root of miRNA-mediated gene silencing. Cell.
132:9–14. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Carthew RW: Gene regulation by microRNAs.
Curr Opin Genet Dev. 16:203–208. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang B, Pan X, Cobb GP and Anderson TA:
microRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007. View Article : Google Scholar
|
|
9
|
Lujambio A and Lowe SW: The microcosmos of
cancer. Nature. 482:347–355. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bagnoli M, De Cecco L, Granata A,
Nicoletti R, Marchesi E, Alberti P, Valeri B, Libra M, Barbareschi
M, Raspagliesi F, et al: Identification of a chrXq27.3 microRNA
cluster associated with early relapse in advanced stage ovarian
cancer patients. Oncotarget. 2:1265–1278. 2011.
|
|
11
|
Iorio MV and Croce CM: microRNA
involvement in human cancer. Carcinogenesis. 33:1126–1133. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Godlewski J, Nowicki MO, Bronisz A, Nuovo
G, Palatini J, De Lay M, Van Brocklyn J, Ostrowski MC, Chiocca EA
and Lawler SE: MicroRNA-451 regulates LKB1/AMPK signaling and
allows adaptation to metabolic stress in glioma cells. Mol Cell.
37:620–632. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gabriely G, Yi M, Narayan RS, Niers JM,
Wurdinger T, Imitola J, Ligon KL, Kesari S, Esau C, Stephens RM, et
al: Human glioma growth is controlled by microRNA-10b. Cancer Res.
71:3563–3572. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li Y, Guessous F, Zhang Y, Dipierro C,
Kefas B, Johnson E, Marcinkiewicz L, Jiang J, Yang Y, Schmittgen
TD, et al: MicroRNA-34a inhibits glioblastoma growth by targeting
multiple oncogenes. Cancer Res. 69:7569–7576. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chan JA, Krichevsky AM and Kosik KS:
MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells.
Cancer Res. 65:6029–6033. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ciafrè SA, Galardi S, Mangiola A, Ferracin
M, Liu CG, Sabatino G, Negrini M, Maira G, Croce CM and Farace MG:
Extensive modulation of a set of microRNAs in primary glioblastoma.
Biochem Biophys Res Commun. 334:1351–1358. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhou J, Wang W, Gao Z, Peng X, Chen X,
Chen W, Xu W, Xu H, Lin MC and Jiang S: MicroRNA-155 promotes
glioma cell proliferation via the regulation of MXI1. PLoS One.
8:e830552013. View Article : Google Scholar :
|
|
18
|
Godlewski J, Nowicki MO, Bronisz A,
Williams S, Otsuki A, Nuovo G, Raychaudhury A, Newton HB, Chiocca
EA and Lawler S: Targeting of the Bmi-1 oncogene/stem cell renewal
factor by microRNA-128 inhibits glioma proliferation and
self-renewal. Cancer Res. 68:9125–9130. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Silber J, Lim DA, Petritsch C, Persson AI,
Maunakea AK, Yu M, Vandenberg SR, Ginzinger DG, James CD, Costello
JF, et al: miR-124 and miR-137 inhibit proliferation of
glioblastoma multiforme cells and induce differentiation of brain
tumor stem cells. BMC Med. 6:142008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bier A, Giladi N, Kronfeld N, Lee HK,
Cazacu S, Finniss S, Xiang C, Poisson L, deCarvalho AC, Slavin S,
et al: MicroRNA-137 is downregulated in glioblastoma and inhibits
the stemness of glioma stem cells by targeting RTVP-1. Oncotarget.
4:665–676. 2013.PubMed/NCBI
|
|
21
|
Skalsky RL and Cullen BR: Reduced
expression of brain-enriched microRNAs in glioblastomas permits
targeted regulation of a cell death gene. PLoS One. 6:e242482011.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Huang Z, Huang S, Wang Q, Liang L, Ni S,
Wang L, Sheng W, He X and Du X: MicroRNA-95 promotes cell
proliferation and targets sorting Nexin 1 in human colorectal
carcinoma. Cancer Res. 71:2582–2589. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huang X, Taeb S, Jahangiri S, Emmenegger
U, Tran E, Bruce J, Mesci A, Korpela E, Vesprini D, Wong CS, et al:
miRNA-95 mediates radioresistance in tumors by targeting the
sphingolipid phosphatase SGPP1. Cancer Res. 73:6972–6986. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang Y, Li M, Wang H, Fisher WE, Lin PH,
Yao Q and Chen C: Profiling of 95 microRNAs in pancreatic cancer
cell lines and surgical specimens by real-time PCR analysis. World
J Surg. 33:698–709. 2009. View Article : Google Scholar
|
|
25
|
Xiao Z, Ching Chow S, Han Li C, Chun Tang
S, Tsui SK, Lin Z and Chen Y: Role of microRNA-95 in the anticancer
activity of Brucein D in hepatocellular carcinoma. Eur J Pharmacol.
728:141–150. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nurul-Syakima AM, Yoke-Kqueen C, Sabariah
AR, Shiran MS, Singh A and Learn-Han L: Differential microRNA
expression and identification of putative miRNA targets and
pathways in head and neck cancers. Int J Mol Med. 28:327–336.
2011.PubMed/NCBI
|
|
27
|
Barreau C, Paillard L, Méreau A and
Osborne HB: Mammalian CELF/Bruno-like RNA-binding proteins:
Molecular characteristics and biological functions. Biochimie.
88:515–525. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Keene JD: RNA recognition by autoantigens
and autoantibodies. Mol Biol Rep. 23:173–181. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ladd AN and Cooper TA: Multiple domains
control the subcellular localization and activity of ETR-3, a
regulator of nuclear and cytoplasmic RNA processing events. J Cell
Sci. 117:3519–3529. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang GS, Kearney DL, De Biasi M, Taffet G
and Cooper TA: Elevation of RNA-binding protein CUGBP1 is an early
event in an inducible heart-specific mouse model of myotonic
dystrophy. J Clin Invest. 117:2802–2811. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Choi DK, Ito T, Mitsui Y and Sakaki Y:
Fluorescent differential display analysis of gene expression in
apoptotic neuroblastoma cells. Gene. 223:21–31. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pershouse MA, Stubblefield E, Hadi A,
Killary AM, Yung WK and Steck PA: Analysis of the functional role
of chromosome 10 loss in human glioblastomas. Cancer Res.
53:5043–5050. 1993.PubMed/NCBI
|
|
33
|
Ransom DT, Ritland SR, Moertel CA, Dahl
RJ, O'Fallon JR, Scheithauer BW, Kimmel DW, Kelly PJ, Olopade OI,
Diaz MO, et al: Correlation of cytogenetic analysis and loss of
heterozygosity studies in human diffuse astrocytomas and mixed
oligo-astrocytomas. Genes Chromosomes Cancer. 5:357–374. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Subramaniam D, Natarajan G, Ramalingam S,
Ramachandran I, May R, Queimado L, Houchen CW and Anant S:
Translation inhibition during cell cycle arrest and apoptosis:
Mcl-1 is a novel target for RNA binding protein CUGBP2. Am J
Physiol Gastrointest Liver Physiol. 294:G1025–G1032. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Natarajan G, Ramalingam S, Ramachandran I,
May R, Queimado L, Houchen CW and Anant S: CUGBP2 down-regulation
by prostaglandin E2 protects colon cancer cells from
radiation-induced mitotic catastrophe. Am J Physiol Gastrointest
Liver Physiol. 294:G1235–G1244. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Subramaniam D, Ramalingam S, Linehan DC,
Dieckgraefe BK, Postier RG, Houchen CW, Jensen RA and Anant S: RNA
binding protein CUGBP2/CELF2 mediates curcumin-induced mitotic
catastrophe of pancreatic cancer cells. PLoS One. 6:e169582011.
View Article : Google Scholar : PubMed/NCBI
|