Introduction
Hepatocellular carcinoma (HCC) is one of the most
common malignant cancers and the third most fatal form of neoplasia
worldwide. It is mostly associated with multiple factors including
HBV/HCV virus infection (1,2),
exposure to hepatocarcinogens such as aflatoxin B1, and exposure to
alcohol (3,4). Previous studies have demonstrated
multiple regulatory pathways involved in HCC. However, the exact
mechanism for the pathogenesis of HCC is not yet clearly
understood.
KLF4 gene is located on chromosome 9 and has 5
exons. Multiple transcripts of KLF4 and their protein isoforms (for
example, KLF4α, KLF4β, KLF4γ and KLF4δ) have been reported
(5). However, little attention has
been paid to the putative non-coding transcript variant, KLF4-003
(Ensembl: ENST00000493306) possibly due to the fact that it has no
protein product.
Recent genomic and transcriptomic projects have
unraveled an astounding large number of non-coding RNAs (ncRNAs) in
the human genome (6,7). A class of small ncRNAs has been
identified, some of which have been linked to neoplastic
transformation (8–11). Another class of ncRNAs, long ncRNAs
(lncRNAs) were tentatively defined as ncRNAs with >200
nucleotides in length and featured with diversity of their
sequences and complexity of the mechanisms involved. Accumulating
evidence supports the possibility that lncRNA acts as genetic
regulators or riboregulators (12). Altered lncRNA levels were found to
be responsible for the aberrant expression of gene products that
might be related to cancer biology. For example, metastasis
associated lung adenocarcinoma transcript 1, a human lncRNA was
found upregulated in metastasizing non-small cell lung carcinomas
and is evolutionarily conserved among mammals (13). Another lncRNA, the paternally
imprinted gene H19 (14), can
function as a tumor suppressor in some tumor types and may play a
significant role in tumorigenesis in other types (15–17).
It is notable that Hepcarcin, also an lncRNA transcript, was found
highly upregulated in human HCCs as well as other five
non-hepatocellular human carcinomas and may serve as an informative
marker for HCCs. Although a few aberrantly expressed protein-coding
genes and several lncRNAs have been identified in HCC, novel
molecular markers with early-diagnostic or risk-assessment value
are still urgently needed. It is of paramount importance to
elucidate the relationships between clinical traits and molecular
changes in HCC for developing new strategies that can be used in
HCC diagnosis, treatment and prognosis.
Extensive studies have been carried out on KLF4 in
the field of stem cells and oncology, including a recent study
showing that high cytoplasmic expression of KLF4 was associated
with better disease-specific survival and was an independently
favorable prognostic factor in HCC (18). However, little is known with regard
to its non-coding transcript variants. Here, we report the
identification of the splice variant KLF4-003 and elucidate the
characteristics of this variant, including its differential
expression and epigenetic regulation in HCC, association with HCC
traits and the potential diagnostic significance as a biomarker for
HCC.
Materials and methods
Cellculture
All cell lines used in this study were purchased
from the American Type Culture Collection. WRL68 (a human fetal
liver cell line) and two human hepatocellular carcinoma cell lines
including HepG2 and Huh7 were cultured in Dulbecco's modified
Eagle's medium (DMEM) (Thermo Fisher Scientific, Inc., Waltham, MA
USA) supplemented with 10% (v/v) fetal bovine serum, 100 U/ml
penicillin and 100 μg/ml streptomycin, while Hep3B (a human
hepatocellular carcinoma cell line) was cultured in Roswell Park
Memorial Institute-1640 medium (Thermo Fisher Scientific, Inc.)
with the same supplements. The other cells from different tissues
including HeLa (from cervical cancer), HONE1 (from nasopharyngeal
cancer), OB (from osteoblast), Saos2 (from osteosarcoma) and BMSC
(bone marrow stromal cells) were also cultured in DMEM medium at
37°C, 5% CO2 humidified incubator.
Patient samples
Cancerous HCC tissues and adjacent non-tumor tissue
(≥1 cm away from the tumor edge) were collected from patients who
underwent surgical resection at the Prince of Wales Hospital, Hong
Kong, China. Written consents was obtained from each patient prior
to tissue harvesting and the study protocol was approved by the
ethics committee of the Chinese University of Hong Kong. The
tissues were immediately snap-frozen in liquid nitrogen and stored
at −80°C pending analysis. Further details of the patients are
given in Table I.
 | Table IDemographic and clinical features of
HCC patients. |
Table I
Demographic and clinical features of
HCC patients.
| | KLF4-003
expression | |
|---|
| |
| |
|---|
| Pathological
parameter | Total | Reduced | Non-reduced | P-value |
|---|
| Gender |
| Male | 42 | 36 | 6 | 1.000 |
| Female | 12 | 10 | 2 | |
| Age |
| <60 | 34 | 29 | 5 | 1.000 |
| ≥60 | 20 | 17 | 3 | |
| HBsAg |
| Positive | 45 | 39 | 6 | 0.607 |
| Negative | 9 | 7 | 2 | |
| AJCC staging |
| Stage I | 38 | 31 | 7 | 0.342 |
| Stage II | 6 | 5 | 1 | |
| Stage III | 10 | 10 | 0 | |
| Cirrhosis |
| Yes | 24 | 22 | 2 | 0.230 |
| No | 30 | 24 | 6 | |
| Recurrencea |
| Yes | 14 | 4 | 10 | 0.045 |
| No | 17 | 11 | 6 | |
RNA extraction, reverse transcription
PCR, DNA sequencing for KLF4-003
Total RNAs from different cell lines or HCC tissues
were extracted by using TRIzol reagent (Thermo Fisher Scientific,
Inc.) following the manufacturer's instructions. Nuclear and
cytoplasmic RNAs were extracted by using the NE-PER Nuclear and
Cytoplasmic Extraction reagents (Thermo Fisher Scientific, Inc.)
followed by TRIzol reagent extraction. cDNA was synthesized by 1 μg
RNA with QuantiTect Rev Transcription kit (Qiagen, GmbH. Hilden,
Germany) according to the protocol provided by the manufacturer.
The reverse transcription reaction mixture was incubated at 42°C
for 15 min for reverse transcription followed by denaturation at
95°C for 3 min. KLF4-003 in each cell line was identified with PCR
amplification followed by DNA sequencing (service provided by BGI).
The PCR reaction contains 1.25 U of AmpliTaq Gold DNA polymerase
(Applied Biosystems, Foster City, CA, USA), 1X PCR buffer, 200 μM
dNTP mixture, and 200 nM of each primer in a final volume of 50 μl.
The forward primer sequence is 5′-TCC CGG CTT CCA TCC CCA CCC-3′.
The reverse primer is 5′-GGT CCT TTT CCG GGG CCA CGA TC-3′. PCR
were performed by DNA denaturing at 94°C for 3 min, followed by 35
cycles of 94°C for 30 sec, 60°C for 30 sec, 72°C for 1 min with a
final extension of 72°C for 7 min. For all primers, the same PCR
condition was used.
Bioinformatics analysis
The exon-intron patterns of KLF4-003 transcript were
constructed by Ensembl browser tool (http://asia.ensembl.org/index.html). The CpG island
was revealed with Genome Browser tool of UCSC (University of
California, Santa Cruz) (http://genome.ucsc.edu/cgiBin/hgGateway).
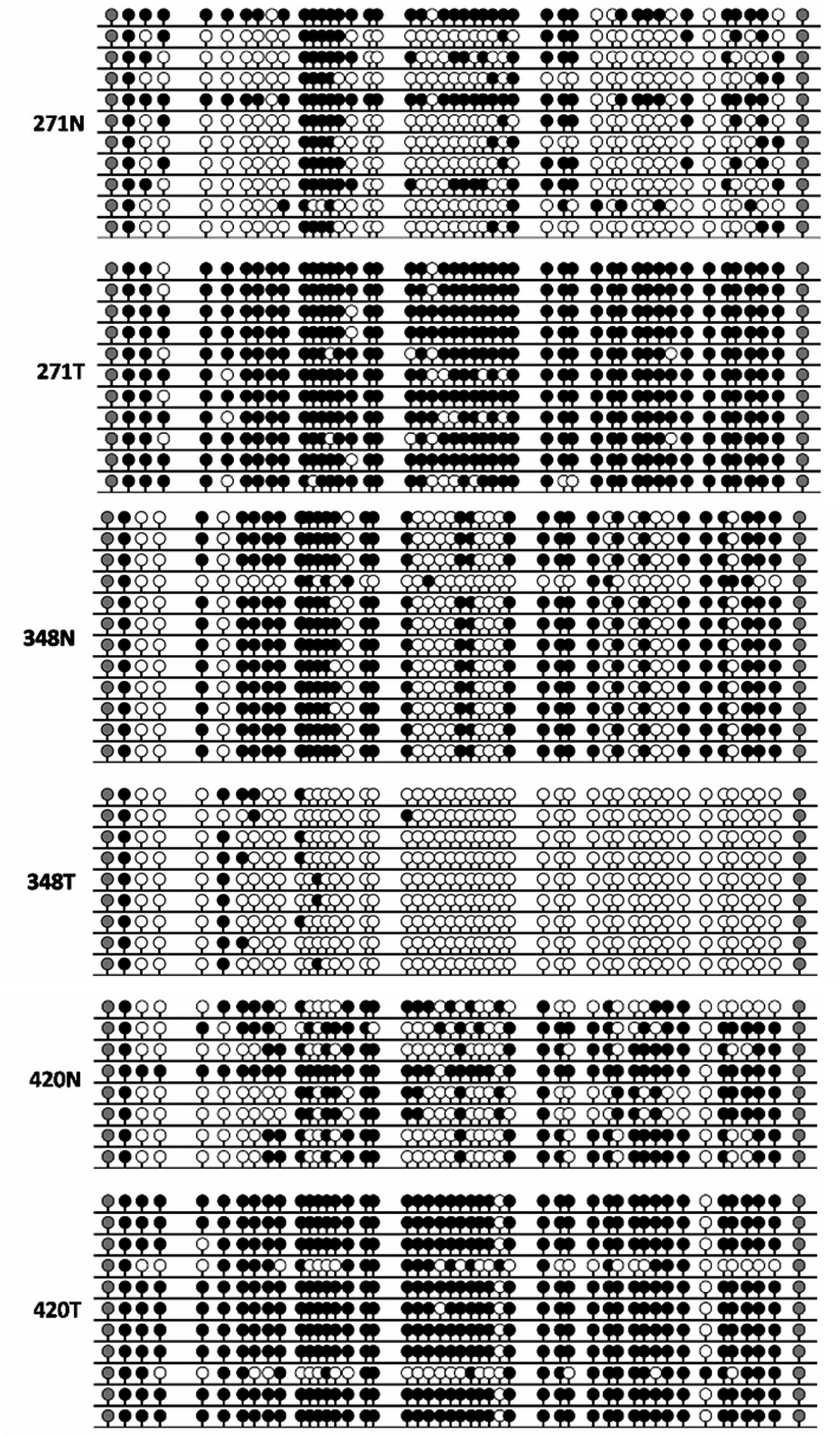
Bisulfite sequencing
Genomic DNA was extracted from cells or frozen
tissues using QIAamp DNA Mini kit (Qiagen, GmbH) following the
manufacturer's instructions. Sodium bisulphate treatment was
performed with the EZ DNA Methylation-Gold kit (Zymo Research,
Freiburg, Germany). Twenty nanograms of treated DNA were used as a
template for PCR amplification using ZymoTaq DNA polymerase (Zymo
Research) and primers targeting a fragment of non-promoter CpG
island proximal to the retained intron of KLF4-003. Forward primer,
5′-GGT TTT TAG TTT ACG TTG TAT AGT GTT GG-3′ and reverse primer,
5′-CCG TAA CGC CAA CCA AAC AAC T-3′ were used for PCR
amplification. The PCR products were cloned into pGEM-T Easy vector
(Promega, Madison, WI, USA) with the manufacturer's standard
protocol for sequencing to determine the methylation status of the
CpG sites within KLF4-003 gene. In total, 73 colonies were
sequenced.
5-aza-2′-deoxycytidine (5-aza-dC)
treatment
Cells were seeded at a density of 1×105
cells per well in 6-well plate one day before treatment. Dimethyl
sulfoxide dissolved 5-aza-dC (Sigma, St. Louis, MO, USA), a
demethylating agent, was added to the cells at different
concentration, 0, 2 or 5 μM. The 5-aza-dC and medium were refreshed
every day. The cells were harvested for total RNA extraction 4 days
post-treatment. The expression level of KLF4-003 was determined by
real-time (RT) PCR.
Cloning of KLF4-003
To construct the pCMV-Myc-KLF4-003 vector, PCR was
performed by gene specific primers containing BglII site and
XhoI site, respectively. The forward primer sequence is
5′-GGC AGA TCT TGG CTG TCA GCG ACG CGC T-3′. The reversed primer is
5′-GGC CTC GAG TTA AAA ATG CCT CTT CAT GTG T-3′. PCR products were
purified using Wizard™ SV Gel and PCR Clean-up system (Promega)
following the manufacturer's instructions. Purified PCR products
and pCMV-Myc vector (Clontech, Mountain View, CA, USA) were
submitted for BglII and XhoI (New England Biolabs,
Ipswich, MA, USA) double digestion. Digestion products were
resolved in 1% agarose gel, followed by purification of the DNA
products and ligation with DNA insert using T4 ligase (New England
Biolabs). The entire mixture of ligation products were transformed
into E. coli competent cells (DH5α) followed by recovery in
800 μl 2% LB (Luria Bertani) (USB Corp., Cleveland, OH, USA)
medium. E. coli clones carrying the desired inserts were
selected by ampicillin resistance and further cultured for plasmid
extraction using the Mini Plus™ plasmid DNA Extraction system
(Viogene, Sunnyvale, CA, USA). The sequence of cloned
KLF4-003 in pCMV-Myc vector was determined and confirmed by
automated DNA sequencing (service provided by BGI).
Transient transfection of KLF4-003 in
human liver cell line
Hep3B cells were seeded in 6-well plate at an
initial density of 2×105 cells per well. When the cells
were 70% confluent, plasmid, pCMV-Myc, pCMV-KLF4-003 were
transfected into the cells using Lipofectamine™ 2000 (Thermo Fisher
Scientific, Inc.) according to the manufacturer's procedures,
respectively. Cells were incubated at 37°C in 5% CO2 for
24–72 h prior to RT-PCR and western blot assay.
RT-PCR
RT-PCR was performed using 2X Power SYBR Green PCR
Master Mix (Thermo Fisher Scientific, Inc.) with ABI 7500 Fast
RT-PCR system (Thermo Fisher Scientific, Inc.). The PCR cycling
conditions included an initial denaturation of 95°C for 10 min
followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min.
KLF4-003 was amplified by forward primer 5′-TCG GGA CAC ACG GGA TGA
T-3′ paired with reverse primer 5′-GCC CGC GTA ATC ACA AGT GT-3′.
While forward primer 5′-CAT TAC CAA GAG CTC ATG CC-3′ paired with
reverse primer 5′-GCC CGC GTA ATC ACA AGT GT-3′ were used for KLF4
amplification. The house-keeping gene β-actin was measured for
normalization using forward primer 5′-GCC CCG CGA GCA CAG AGC-3′
paired with reverse primer 5′-TGC CGG AGC CGT TGT CGA-3′.
Comparative CT method (2−ΔΔCT) was used to
calculate the relative level of the KLF4 and KLF4-003 mRNA.
Western blot analysis
Total proteins were extracted from Hep3B cells with
RIPA lysis and extraction buffer (Thermo Fisher Scientific, Inc.)
following the manufacturer's protocol. The target proteins were
separated on sodium dodecyl sulfate polyacrylamide gel
electrophoresis and transferred onto PVDF membrane. Specific
antibodies including anti-Myc and anti-KLF4 (Santa Cruz
Biotechnology, USA) were used for protein detection. The signals of
target proteins were visualized on X-ray film after treatment with
Western Lightning Chemiluminescence Reagent Plus (Perkin-Elmer Life
Science, MA, USA).
Statistical analysis
Statistical significance between groups was analyzed
by Student's t-test. Correlation analysis was used to examine the
association between the expression of KLF4-003 and clinical
parameters of HCC patients. Receiver operating characteristic (ROC)
curve was plotted by SPSS (version 16.0) to evaluate the diagnostic
value for differentiating between HCC cancer and benign diseases.
The other figures were created with GraphPad Prism5 (GraphPad, San
Diego, CA, USA). P-value of ≤0.05 was considered statistically
significant.
Results
Sequence analysis and expression profile
of KLF4-003
According to the database of Ensembl, KLF4-003 is a
non-coding transcript of KLF4 gene (ENSG00000136826). Compared with
the vulgate transcript of KLF4, an intron retention was observed in
KLF4-003 (Fig. 1A). This retained
intron is 102 nucleotides in length and located near a non-promoter
CpG island in KLF4-003 gene (Fig.
1B). By RT-PCR and DNA sequencing, KLF4-003 transcript was
identified in a series of cell lines including WRL68, HONE1, OB,
BMSC, HeLa and Soas2 cells (Fig.
1C).
KLF4-003 is downregulated in liver cancer
cell lines and HCC tissues
To determine the expression levels of KLF4-003,
RT-PCR was performed in a number of liver cancer cell lines using
β-actin as a housekeeping control. Compared with WRL68 normal liver
cells, the expression of KLF4-003 mRNA was significantly
downregulated in the three examined HCC cell lines including Huh7,
HepG2 and Hep3B (P<0.001, Fig.
2A).
We then examined the expression levels of KLF4-003
in the HCC tissues and their adjacent normal counterparts of HCC
patients using RT-PCR. Among 54 pairs of clinical specimens,
KLF4-003 were significantly downregulated (P<0.001) by
>2-fold in 46 pairs (78%) of HCC tissues when compared with
their normal counterparts (Fig.
2B). The reduced expression of KLF4-003 was found significantly
associated with HCC recurrence (Table
I, P=0.045) in the follow-up of 31 HCC patients. However, no
correlation was detected between KLF4-003 expression level and
other clinical parameters including sex, age, tumor size,
differentiation status, The American Joint Committee on Cancer
(AJCC) staging and HBV infection status (data not shown).
Hypermethylation of non-promoter CpG
island of KLF4-003 in HCC tissues
Epigenetic regulation has been closely related to
altered gene expression. One feature of KLF4-003 is that it retains
an intron of KLF4. Interestingly, a non-promoter CpG island
proximal to this unique intron was observed. To investigate whether
this CpG island is responsible for the decreased expression of
KLF4-003 in HCC, we first determine the methylation status of this
CpG island in HCC samples by bisulfite sequencing. Significant
differences of methylation status were detected between three pairs
of investigated HCC specimens and their paracancerous tissues
(Fig. 3).
Demethylation rescues the expression of
KLF4-003 in Hep3B cells
To investigate whether the reduced expression of
KLF4-003 was caused by hypermethylation of the proximal
non-promoter CpG island proximal to the retained intron, we
determined the methylation status of this CpG island in Hep3B
cells. As shown in Fig. 4A,
hypermethylation was observed in the selected region. Then we
treated the Hep3B cells with two concentrations of 5-aza-dC for 4
days. The expression of KLF4-003 was examined by RT-PCR. We found
that KLF4-003 was significantly increased in Hep3B cells treated
with 2 and 5 μM of 5-aza-dC in a dose-dependent manner compared
with untreated cells (Fig. 4B).
This finding suggested that hypermethylation of the selected
non-promoter CpG island might be responsible for the reduced
expression of KLF4-003 in HCC.
Observation of the value of using
KLF4-003 as a diagnostic marker
To evaluate the diagnostic significance of KLF4-003
in HCC, the differences of KLF4-003 expression between HCC tissue
and matched paracancerous tissues were compared based on the cutoff
value (0.778) from the ROC curve. The area under ROC curve (AUC)
reached 0.803 (95% CI=0.719–0.886, P<0.001, Fig. 5A). The sensitivity was 0.889 and
specificity was 0.389. For comparison, ROC curve for KLF4 was also
created (Fig. 5B). The AUC of KLF4
ROC curve was 0.616 (95% CI=0.468–0.764, P=0.136). It was obvious
that KLF4-003 had a better diagnostic value than KLF4.
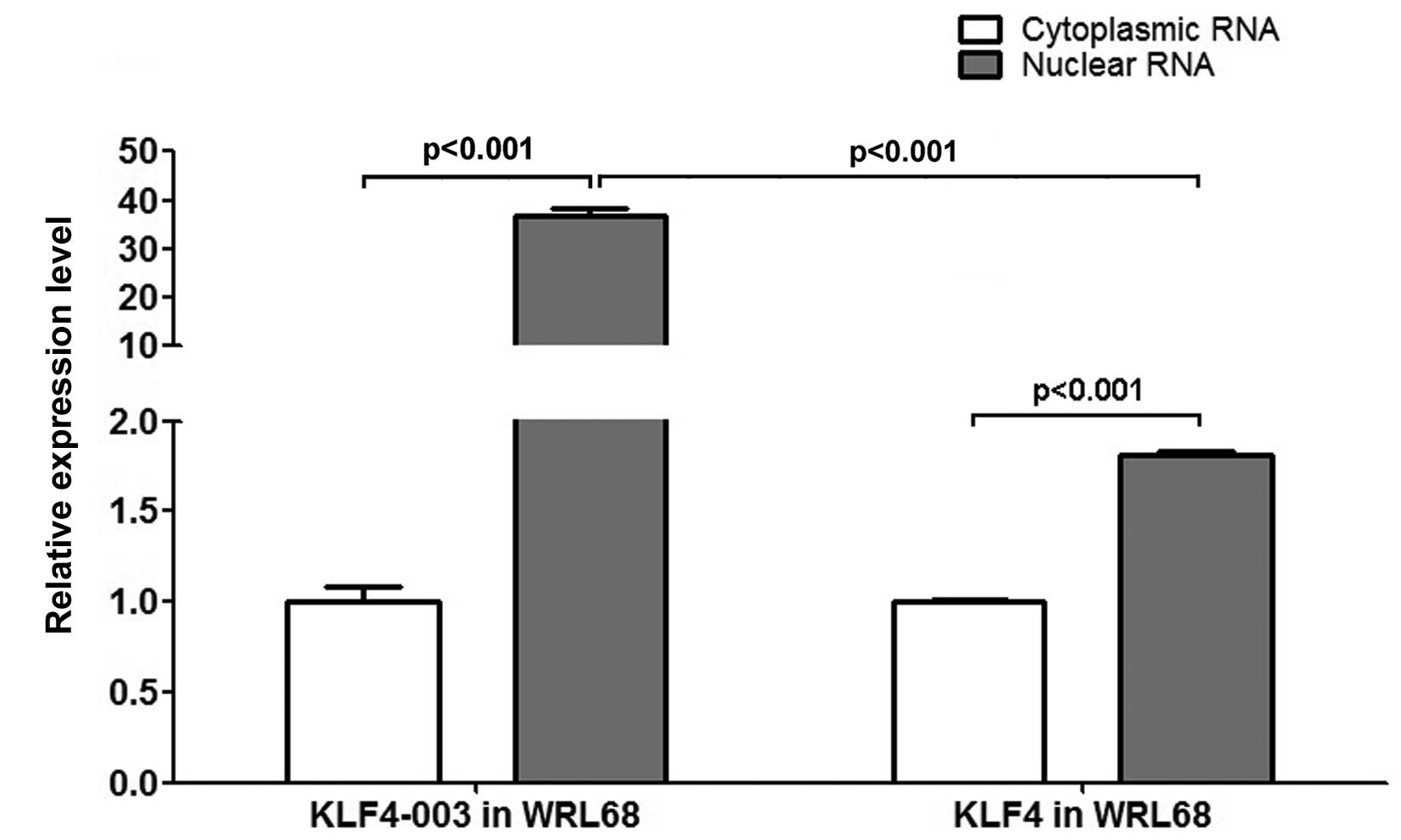
KLF4-003 tends to remain in the
nucleus
To investigate the cellular distribution of KLF4-003
RNA, RT-PCR was performed in WRL68 cells. In contrast to KLF4, the
nuclear RNA level of KLF4-003 was >36-fold higher than the
cytoplasmic RNA level (Fig. 6).
Such observation was consistent with the property of the
lncRNA.
KLF4-003 protein is endogenously
undetectable in liver cells
Since KLF4-003 open reading frame (ORF) encodes a
putative protein that is 34 amino acids longer than KLF4 protein,
which enable KLF4-003 to be distinguished from KLF4 by western blot
assay. To determine whether KLF4-003 was expressed in liver cells,
the KLF4-003 ORF was cloned into pCMV-Myc vector and transiently
transfected into Hep3B cells. We confirmed that the fusion protein
KLF4-003 with Myc tag was detected, whereas, the endogenous
KLF4-003 protein was undetectable (Fig. 7).
Discussion
Tumor markers have drawn increased attention as they
could provide potential targets for diagnosis and therapeutic
intervention. Recent studies have found the expression profiles of
miRNAs were significantly altered in human neoplasms (9,11),
which could be used in tumor diagnosis to distinguish tumors from
normal tissues. Despite the huge number of lncRNAs identified so
far, definitely characterized lncRNAs only accounted for <1%
(19). Recent studies showed many
lncRNAs were deregulated in various solid tumors and a number of
lncRNAs could regulate cancer metastasis by directly targeting
chromatin modification complexes, indicating that lncRNAs may play
an important role in tumorigenesis and cancer development (20,21).
HCC is one of the most common malignant carcinomas in the world. It
is of great importance to elucidate the functional role of lncRNA
in HCC, which may significantly contribute to the better
understanding, diagnosis and treatment of HCC conditions. An
increasing number of evidence has suggested that deregulated
expression of microRNAs (miRNAs) have considerable potential in
predicting the prognosis of HCC patients (22,23).
In the present study, we identified a putative lncRNA, KLF4-003 in
human HCC samples that clearly distinguishes HCC from their
corresponding normal tissues. We demonstrated that HCC patients
with lower KLF4-003 expression had a significantly increased risk
of recurrence. ROC analysis indicated that the lncRNA KLF4-003
could serve as a potential tumor marker for HCC. Such observation
further underlined the potential importance of the lncRNA in the
molecular cell biology of neoplasia.
By comparative analyses, ncRNA promoters have been
found more conserved than those of protein-coding genes (24). Many studies have linked the
promoter-associated DNA methylation with transcriptional silencing
of associated genes (25–28). Notably, non-promoter genomic DNA
methylation has also been found within both intronic and exonic
regions of numerous genes and could also elicit repressive effects
on gene expression (29). By
bisulfite sequencing, significant differences were detected between
the methylation status of KLF4-003 in HCC specimens and their
adjacent normal controls, which indicated that hypermethylation
might be responsible for the downregulation of lncRNA KLF4-003. To
verify the silencing effect of hypermethylation on KLF4-003,
demethylation was performed in Hep3B cells. We found that the
expression of KLF4-003 was dramatically rescued upon demethylation
treatment in Hep3B cells. This observation supported the notion
that non-promoter genomic DNA methylation could also result in gene
silencing.
Much effort has been made to investigate the
mechanisms of the relationship between DNA methylation and
associated gene repression. At least four possible mechanisms have
become apparent thus far. Firstly, the association of DNA-binding
factors with their cognate DNA recognition sequences might be
inhibited by the modification of cytosine bases (30). Secondly, methyl CpG binding
proteins could recognize the methylated DNA, target chromatin
remodeling co-repressor complexes and consequently mediate the
silencing of gene expression (31). Thirdly, DNA methyltransferase
enzymes themselves might be involved in setting up the silenced
state besides their catalytic activities (32). Finally, transcriptional elongation
could be affected by DNA methylation as the capacity of RNA
polymerase II (Pol II) to transcribe through the methylated regions
could be dampened by the DNA methylation (32). Although each of the above
mechanisms may possibly elicit repressive effect on the expression
of KLF4-003 in HCC, the exact role of this non-promoter CpG
hypermethylation on KLF4-003 expression remains elusive.
It is interesting that KLF4-003 has an ORF although
no endogenous protein product could be obtained. However, the
majority of KLF4-003 was located in the nucleus. It has been shown
that nuclear-localized lncRNAs are only transiently expressed and
more likely unstable (33). The
rapid degradation of nuclear-localized KLF4-003 may not permit
sufficient KLF4-003 transcripts to complete the transportation from
the nucleus to the cytoplasm for subsequent translation. It is
notable that KLF4-003 fusion protein could be detected in Hep3B
cells when highly overexpressed. Since the overexpressed KLF4-003
transcript did not contain 5′- and 3′-UTR, it is possible that the
5′- or 3′-UTR of endogenous KLF4-003 transcript contained protein
binding sites which halted its transportation from the nucleus to
the cytoplasm and thereby decreased the chance of this transcript
to be translated in the cytoplasm. However, the exact mechanism for
the predominant nuclear localization of the KLF4-003 transcript
remains unclear.
In conclusion, we identified the non-protein-coding
transcript KLF4-003, which was downregulated in most of the
examined HCC specimens compared with their paracancerous tissues.
This is possibly due to the differential hypermethylation of the
non-promoter CpG islands in the KLF4-003 gene. The reduced
expression of KLF4-003 is significantly associated with the HCC
recurrence and might serve as a potential diagnostic marker with
high sensitivity for human HCCs.
Acknowledgements
The research team was supported by the Scheme B
Funding on the Centre for Microbial Genomics and Proteomics from
the Chinese University of Hong Kong, China.
Abbreviations:
|
KLF4
|
Krüppel-like factor 4
|
|
HCC
|
human hepato-cellular carcinoma
|
|
RT-PCR
|
real-time polymerase chain
reaction
|
|
ROC
|
receiver operating characteristic
|
|
AUC
|
area under ROC curve
|
|
ncRNAs
|
non-coding RNAs
|
|
lncRNAs
|
long non-coding RNAs
|
|
DMEM
|
Dulbecco's modified Eagle's medium
|
|
ORF
|
open reading frame
|
|
AJCC
|
American Joint Committee on Cancer
|
References
|
1
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: Epidemiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Blum HE: Hepatocellular carcinoma: Therapy
and prevention. World J Gastroenterol. 11:7391–7400. 2005.
|
|
3
|
Soini Y, Chia SC, Bennett WP, Groopman JD,
Wang JS, DeBenedetti VM, Cawley H, Welsh JA, Hansen C, Bergasa NV,
et al: An aflatoxin-associated mutational hotspot at codon 249 in
the p53 tumor suppressor gene occurs in hepatocellular carcinomas
from Mexico. Carcinogenesis. 17:1007–1012. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Donato F, Tagger A, Gelatti U, Parrinello
G, Boffetta P, Albertini A, Decarli A, Trevisi P, Ribero ML,
Martelli C, et al: Alcohol and hepatocellular carcinoma: The effect
of lifetime intake and hepatitis virus infections in men and women.
Am J Epidemiol. 155:323–331. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wei D, Wang L, Kanai M, Jia Z, Le X, Li Q,
Wang H and Xie K: KLF4α up-regulation promotes cell cycle
progression and reduces survival time of patients with pancreatic
cancer. Gastroenterology. 139:2135–2145. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Scherer SW, Cheung J, MacDonald JR,
Osborne LR, Nakabayashi K, Herbrick JA, Carson AR, Parker-Katiraee
L, Skaug J, Khaja R, et al: Human chromosome 7: DNA sequence and
biology. Science. 300:767–772. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ota T, Suzuki Y, Nishikawa T, Otsuki T,
Sugiyama T, Irie R, Wakamatsu A, Hayashi K, Sato H, Nagai K, et al:
Complete sequencing and characterization of 21,243 full-length
human cDNAs. Nat Genet. 36:40–45. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Meltzer PS: Cancer genomics: Small RNAs
with big impacts. Nature. 435:745–746. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Erdmann VA, Barciszewska MZ, Szymanski M,
Hochberg A, de Groot N and Barciszewski J: The non-coding RNAs as
riboregulators. Nucleic Acids Res. 29:189–193. 2001. View Article : Google Scholar :
|
|
13
|
Ji P, Diederichs S, Wang W, Böing S,
Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et
al: MALAT-1, a novel noncoding RNA, and thymosin beta4 predict
metastasis and survival in early-stage non-small cell lung cancer.
Oncogene. 22:8031–8041. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
O'Neill MJ: The influence of non-coding
RNAs on allele-specific gene expression in mammals. Hum Mol Genet.
14(Suppl 1): R113–R120. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Steenman MJ, Rainier S, Dobry CJ, Grundy
P, Horon IL and Feinberg AP: Loss of imprinting of IGF2 is linked
to reduced expression and abnormal methylation of H19 in Wilms'
tumour. Nat Genet. 7:433–439. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lottin S, Adriaenssens E, Dupressoir T,
Berteaux N, Montpellier C, Coll J, Dugimont T and Curgy JJ:
Overexpression of an ectopic H19 gene enhances the tumorigenic
properties of breast cancer cells. Carcinogenesis. 23:1885–1895.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Manoharan H, Babcock K, Willi J and Pitot
HC: Biallelic expression of the H19 gene during spontaneous
hepatocarcinogenesis in the albumin SV40 T antigen transgenic rat.
Mol Carcinog. 38:40–47. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hsu HT, Wu PR, Chen CJ, Hsu LS, Yeh CM,
Hsing MT, Chiang YS, Lai MT and Yeh KT: High cytoplasmic expression
of Krüppel-like factor 4 is an independent prognostic factor of
better survival in hepatocellular carcinoma. Int J Mol Sci.
15:9894–9906. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Calin GA, Liu CG, Ferracin M, Hyslop T,
Spizzo R, Sevignani C, Fabbri M, Cimmino A, Lee EJ, Wojcik SE, et
al: Ultraconserved regions encoding ncRNAs are altered in human
leukemias and carcinomas. Cancer Cell. 12:215–229. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tsai MC, Spitale RC and Chang HY: Long
intergenic noncoding RNAs: New links in cancer progression. Cancer
Res. 71:3–7. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hou J, Lin L, Zhou W, Wang Z, Ding G, Dong
Q, Qin L, Wu X, Zheng Y, Yang Y, et al: Identification of miRNomes
in human liver and hepatocellular carcinoma reveals miR-199a/b-3p
as therapeutic target for hepatocellular carcinoma. Cancer Cell.
19:232–243. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ji J, Shi J, Budhu A, Yu Z, Forgues M,
Roessler S, Ambs S, Chen Y, Meltzer PS, Croce CM, et al: MicroRNA
expression, survival, and response to interferon in liver cancer. N
Engl J Med. 361:1437–1447. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Carninci P, Kasukawa T, Katayama S, Gough
J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al:
RIKEN Genome Exploration Research Group and Genome Science Group
(Genome Network Project Core Group): The transcriptional landscape
of the mammalian genome. Science. 309:1559–1563. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Boyes J and Bird A: DNA methylation
inhibits transcription indirectly via a methyl-CpG binding protein.
Cell. 64:1123–1134. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Compere SJ and Palmiter RD: DNA
methylation controls the inducibility of the mouse
metallothionein-I gene lymphoid cells. Cell. 25:233–240. 1981.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kass SU, Landsberger N and Wolffe AP: DNA
methylation directs a time-dependent repression of transcription
initiation. Curr Biol. 7:157–165. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Siegfried Z, Eden S, Mendelsohn M, Feng X,
Tsuberi BZ and Cedar H: DNA methylation represses transcription in
vivo. Nat Genet. 22:203–206. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hsieh CL: Stability of patch methylation
and its impact in regions of transcriptional initiation and
elongation. Mol Cell Biol. 17:5897–5904. 1997.PubMed/NCBI
|
|
30
|
Watt F and Molloy PL: Cytosine methylation
prevents binding to DNA of a HeLa cell transcription factor
required for optimal expression of the adenovirus major late
promoter. Genes Dev. 2:1136–1143. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sarraf SA and Stancheva I: Methyl-CpG
binding protein MBD1 couples histone H3 methylation at lysine 9 by
SETDB1 to DNA replication and chromatin assembly. Mol Cell.
15:595–605. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Klose RJ and Bird AP: Genomic DNA
methylation: The mark and its mediators. Trends Biochem Sci.
31:89–97. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Clark MB, Johnston RL, Inostroza-Ponta M,
Fox AH, Fortini E, Moscato P, Dinger ME and Mattick JS: Genome-wide
analysis of long noncoding RNA stability. Genome Res. 22:885–898.
2012. View Article : Google Scholar : PubMed/NCBI
|