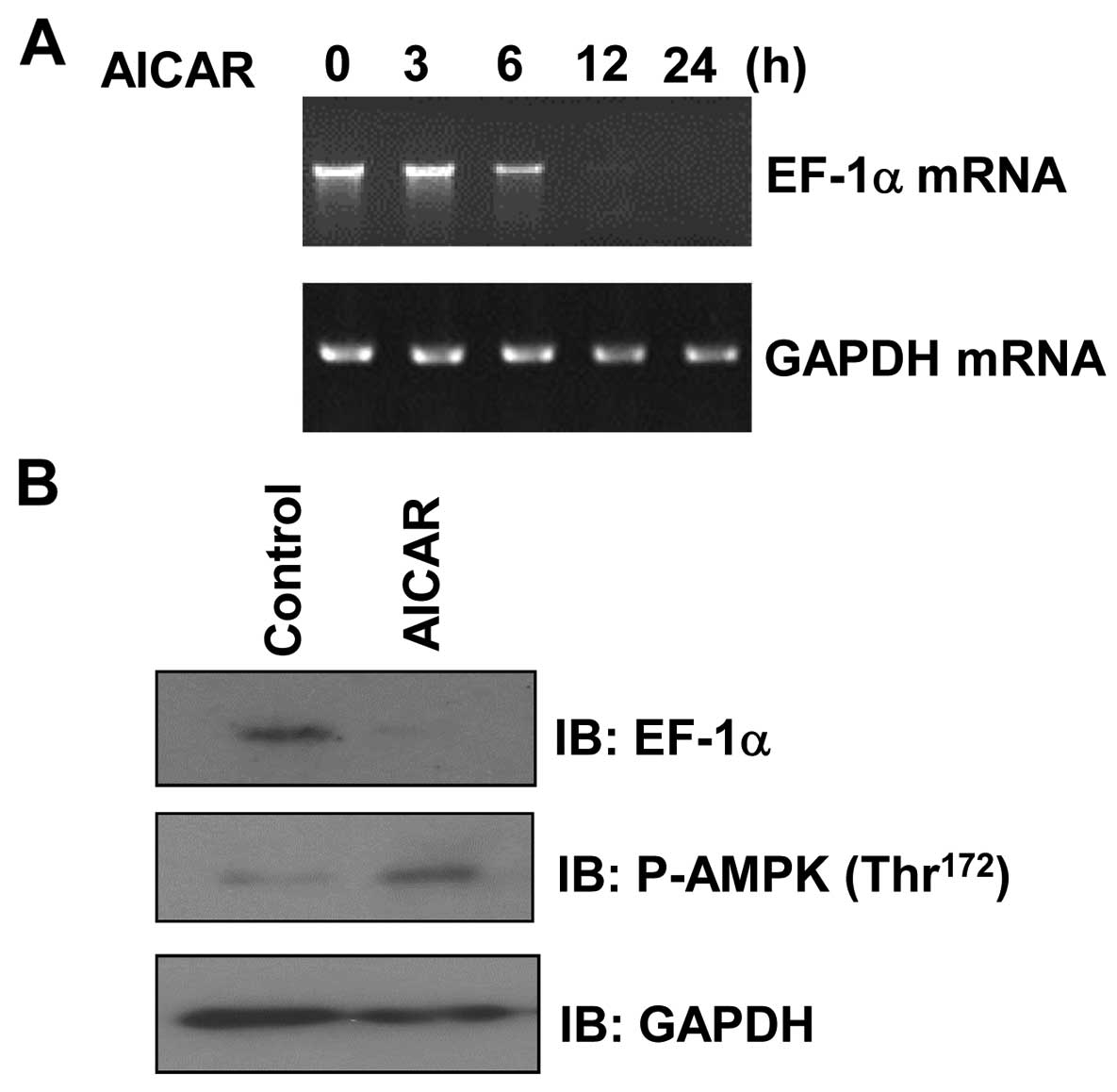
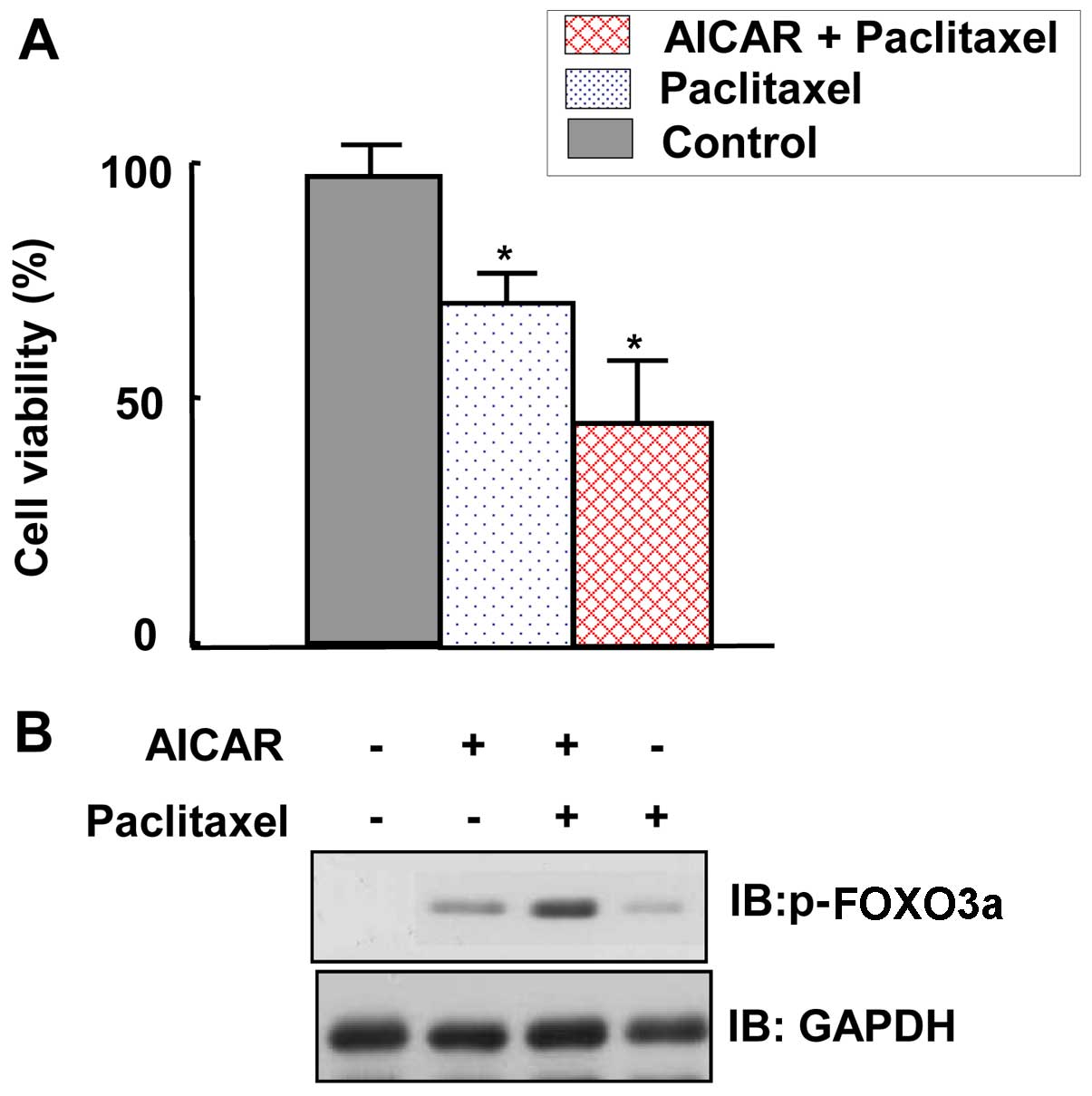
|
1
|
Wani MC, Taylor HL, Wall ME, Coggon P and
McPhail AT: Plant antitumor agents. VI. The isolation and structure
of taxol, a novel antileukemic and antitumor agent from Taxus
brevifolia. J Am Chem Soc. 93:2325–2327. 1971. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Schiff PB, Fant J and Horwitz SB:
Promotion of microtubule assembly in vitro by taxol. Nature.
277:665–667. 1979. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Horwitz SB, Lothstein L, Manfredi JJ,
Mellado W, Parness J, Roy SN, Schiff PB, Sorbara L and Zeheb R:
Taxol: Mechanisms of action and resistance. Ann N Y Acad Sci. 466(1
Dynamic Aspec): 733–744. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Holmes FA: Combination chemotherapy with
Taxol (paclitaxel) in metastatic breast cancer. Ann Oncol. 5(Suppl
6): S23–S27. 1994.PubMed/NCBI
|
|
5
|
Beer TM, El-Geneidi M and Eilers KM:
Docetaxel (taxotere) in the treatment of prostate cancer. Expert
Rev Anticancer Ther. 3:261–268. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Markman M: Taxol: An important new drug in
the management of epithelial ovarian cancer. Yale J Biol Med.
64:583–590. 1991.PubMed/NCBI
|
|
7
|
Ettinger DS: Overview of paclitaxel
(Taxol) in advanced lung cancer. Semin Oncol. 20(Suppl 3): 46–49.
1993.PubMed/NCBI
|
|
8
|
Horwitz SB: Taxol (paclitaxel): Mechanisms
of action. Ann Oncol. 5(Suppl 6): S3–S6. 1994.PubMed/NCBI
|
|
9
|
Hardie DG, Carling D and Carlson M: The
AMP-activated/SNF1 protein kinase subfamily: Metabolic sensors of
the eukaryotic cell? Annu Rev Biochem. 67:821–855. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hardie DG: Minireview: the AMP-activated
protein kinase cascade: the key sensor of cellular energy status.
Endocrinology. 144:5179–5183. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hardie DG and Sakamoto K: AMPK: A key
sensor of fuel and energy status in skeletal muscle. Physiology
(Bethesda). 21:48–60. 2006. View Article : Google Scholar
|
|
12
|
Garcia-Gil M, Pesi R, Perna S, Allegrini
S, Giannecchini M, Camici M and Tozzi MG:
5′-aminoimidazole-4-carboxamide riboside induces apoptosis in human
neuroblastoma cells. Neuroscience. 117:811–820. 2003. View Article : Google Scholar
|
|
13
|
Kefas BA, Cai Y, Kerckhofs K, Ling Z,
Martens G, Heimberg H, Pipeleers D and Van de Casteele M:
Metformin-induced stimulation of AMP-activated protein kinase in
beta-cells impairs their glucose responsiveness and can lead to
apoptosis. Biochem Pharmacol. 68:409–416. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Meisse D, Van de Casteele M, Beauloye C,
Hainault I, Kefas BA, Rider MH, Foufelle F and Hue L: Sustained
activation of AMP-activated protein kinase induces c-Jun N-terminal
kinase activation and apoptosis in liver cells. FEBS Lett.
526:38–42. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Saitoh M, Nagai K, Nakagawa K, Yamamura T,
Yamamoto S and Nishizaki T: Adenosine induces apoptosis in the
human gastric cancer cells via an intrinsic pathway relevant to
activation of AMP-activated protein kinase. Biochem Pharmacol.
67:2005–2011. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Talukder AH, Jorgensen HF, Mandal M,
Mishra SK, Vadlamudi RK, Clark BF, Mendelsohn J and Kumar R:
Regulation of elongation factor-1alpha expression by growth factors
and anti-receptor blocking antibodies. J Biol Chem. 276:5636–5642.
2001. View Article : Google Scholar
|
|
17
|
Bourne HR, Sanders DA and McCormick F: The
GTPase superfamily: Conserved structure and molecular mechanism.
Nature. 349:117–127. 1991. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
de Vos AM, Tong L, Milburn MV, Matias PM,
Jancarik J, Noguchi S, Nishimura S, Miura K, Ohtsuka E and Kim SH:
Three-dimensional structure of an oncogene protein: Catalytic
domain of human c-H-ras p21. Science. 239:888–893. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jurnak F: Structure of the GDP domain of
EF-Tu and location of the amino acids homologous to ras oncogene
proteins. Science. 230:32–36. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
la Cour TF, Nyborg J, Thirup S and Clark
BF: Structural details of the binding of guanosine diphosphate to
elongation factor Tu from E. coli as studied by X-ray
crystallography. EMBO J. 4:2385–2388. 1985.PubMed/NCBI
|
|
21
|
Pai EF, Krengel U, Petsko GA, Goody RS,
Kabsch W and Wittinghofer A: Refined crystal structure of the
triphosphate conformation of H-ras p21 at 1.35 A resolution:
Implications for the mechanism of GTP hydrolysis. EMBO J.
9:2351–2359. 1990.PubMed/NCBI
|
|
22
|
Valencia A, Chardin P, Wittinghofer A and
Sander C: The ras protein family: Evolutionary tree and role of
conserved amino acids. Biochemistry. 30:4637–4648. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ann DK, Moutsatsos IK, Nakamura T, Lin HH,
Mao PL, Lee MJ, Chin S, Liem RK and Wang E: Isolation and
characterization of the rat chromosomal gene for a polypeptide
(pS1) antigenically related to statin. J Biol Chem.
266:10429–10437. 1991.PubMed/NCBI
|
|
24
|
Lee S, Francoeur AM, Liu S and Wang E:
Tissue-specific expression in mammalian brain, heart, and muscle of
S1, a member of the elongation factor-1 alpha gene family. J Biol
Chem. 267:24064–24068. 1992.PubMed/NCBI
|
|
25
|
Lee S, LeBlanc A, Duttaroy A and Wang E:
Terminal differentiation-dependent alteration in the expression of
translation elongation factor-1 alpha and its sister gene, S1, in
neurons. Exp Cell Res. 219:589–597. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sanders J, Brandsma M, Janssen GM, Dijk J
and Möller W: Immunofluorescence studies of human fibroblasts
demonstrate the presence of the complex of elongation factor-1 beta
gamma delta in the endoplasmic reticulum. J Cell Sci.
109:1113–1117. 1996.PubMed/NCBI
|
|
27
|
Taniguchi S, Miyamoto S, Sadano H and
Kobayashi H: Rat elongation factor 1 alpha: Sequence of cDNA from a
highly metastatic fos-transferred cell line. Nucleic Acids Res.
19:69491991. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Paik JH, Kollipara R, Chu G, Ji H, Xiao Y,
Ding Z, Miao L, Tothova Z, Horner JW, Carrasco DR, et al: FoxOs are
lineage-restricted redundant tumor suppressors and regulate
endothelial cell homeostasis. Cell. 128:309–323. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Nakae J, Biggs WH III, Kitamura T, Cavenee
WK, Wright CV, Arden KC and Accili D: Regulation of insulin action
and pancreatic beta-cell function by mutated alleles of the gene
encoding forkhead transcription factor Foxo1. Nat Genet.
32:245–253. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ogg S, Paradis S, Gottlieb S, Patterson
GI, Lee L, Tissenbaum HA and Ruvkun G: The Fork head transcription
factor DAF-16 transduces insulin-like metabolic and longevity
signals in C. elegans. Nature. 389:994–999. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lin K, Dorman JB, Rodan A and Kenyon C:
daf-16: An HNF-3/forkhead family member that can function to double
the life-span of Caenorhabditis elegans. Science. 278:1319–1322.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hwangbo DS, Gershman B, Tu MP, Palmer M
and Tatar M: Drosophila dFOXO controls lifespan and regulates
insulin signalling in brain and fat body. Nature. 429:562–566.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Giannakou ME, Goss M, Jünger MA, Hafen E,
Leevers SJ and Partridge L: Long-lived Drosophila with
overexpressed dFOXO in adult fat body. Science. 305:3612004.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Furuyama T, Yamashita H, Kitayama K,
Higami Y, Shimokawa I and Mori N: Effects of aging and caloric
restriction on the gene expression of Foxo1, 3, and 4 (FKHR,
FKHRL1, and AFX) in the rat skeletal muscles. Microsc Res Tech.
59:331–334. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xuan Z and Zhang MQ: From worm to human:
Bioinformatics approaches to identify FOXO target genes. Mech
Ageing Dev. 126:209–215. 2005. View Article : Google Scholar
|
|
36
|
Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo
P, Hu LS, Anderson MJ, Arden KC, Blenis J and Greenberg ME: Akt
promotes cell survival by phosphorylating and inhibiting a Forkhead
transcription factor. Cell. 96:857–868. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Dijkers PF, Medema RH, Lammers JW,
Koenderman L and Coffer PJ: Expression of the pro-apoptotic Bcl-2
family member Bim is regulated by the forkhead transcription factor
FKHR-L1. Curr Biol. 10:1201–1204. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Medema RH, Kops GJ, Bos JL and Burgering
BM: AFX-like Forkhead transcription factors mediate cell-cycle
regulation by Ras and PKB through p27kip1. Nature. 404:782–787.
2000. View
Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kops GJ, Dansen TB, Polderman PE, Saarloos
I, Wirtz KW, Coffer PJ, Huang TT, Bos JL, Medema RH and Burgering
BM: Forkhead transcription factor FOXO3a protects quiescent cells
from oxidative stress. Nature. 419:316–321. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Nemoto S, Fergusson MM and Finkel T:
Nutrient availability regulates SIRT1 through a forkhead-dependent
pathway. Science. 306:2105–2108. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tran H, Brunet A, Grenier JM, Datta SR,
Fornace AJ Jr, DiStefano PS, Chiang LW and Greenberg ME: DNA repair
pathway stimulated by the forkhead transcription factor FOXO3a
through the Gadd45 protein. Science. 296:530–534. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lee SS, Kennedy S, Tolonen AC and Ruvkun
G: DAF-16 target genes that control C. elegans life-span and
metabolism. Science. 300:644–647. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Murphy CT, McCarroll SA, Bargmann CI,
Fraser A, Kamath RS, Ahringer J, Li H and Kenyon C: Genes that act
downstream of DAF-16 to influence the lifespan of Caenorhabditis
elegans. Nature. 424:277–283. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Mammucari C, Milan G, Romanello V, Masiero
E, Rudolf R, Del Piccolo P, Burden SJ, Di Lisi R, Sandri C, Zhao J,
et al: FoxO3 controls autophagy in skeletal muscle in vivo. Cell
Metab. 6:458–471. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhao J, Brault JJ, Schild A, Cao P, Sandri
M, Schiaffino S, Lecker SH and Goldberg AL: FoxO3 coordinately
activates protein degradation by the autophagic/lysosomal and
proteasomal pathways in atrophying muscle cells. Cell Metab.
6:472–483. 2007. View Article : Google Scholar : PubMed/NCBI
|