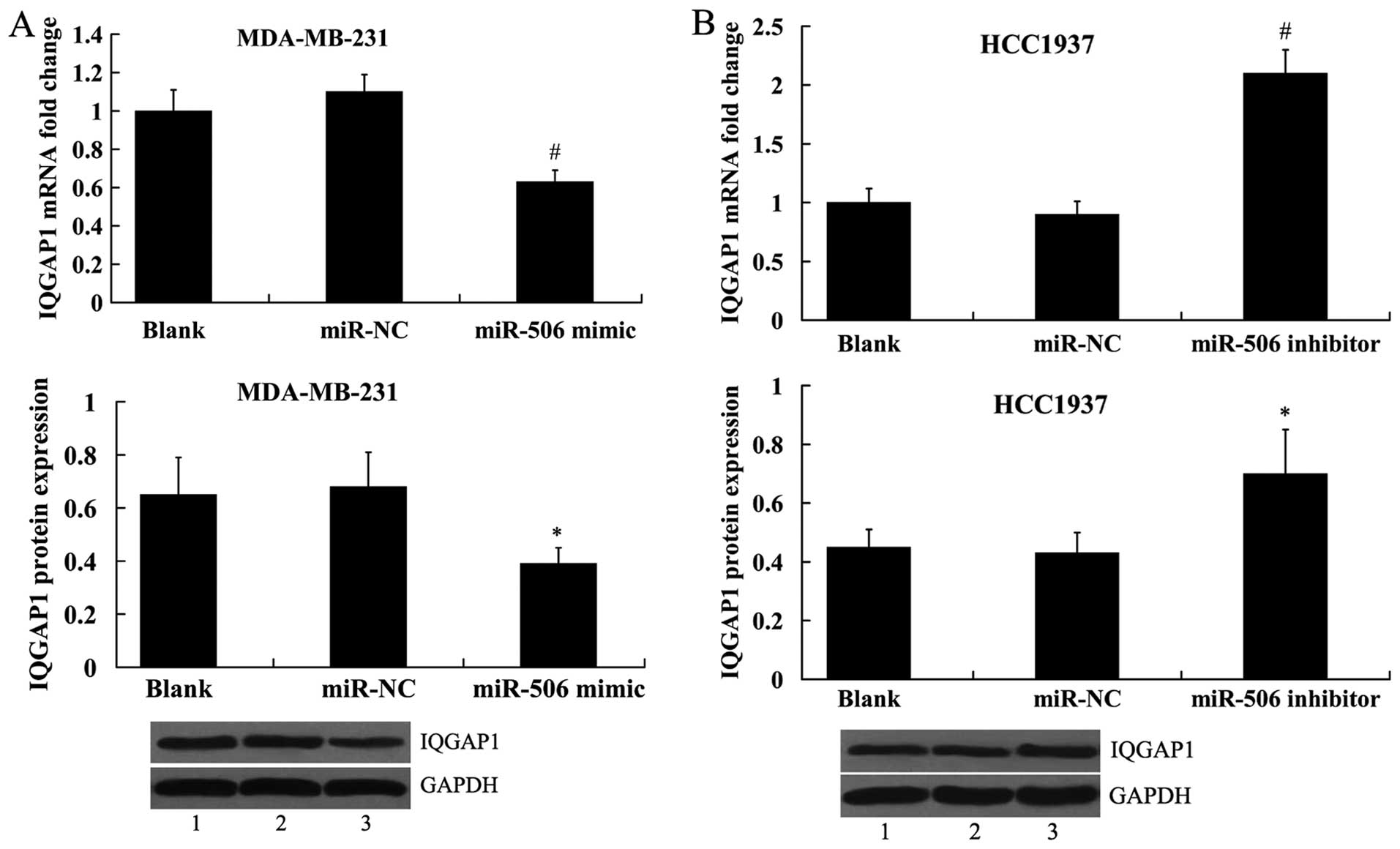
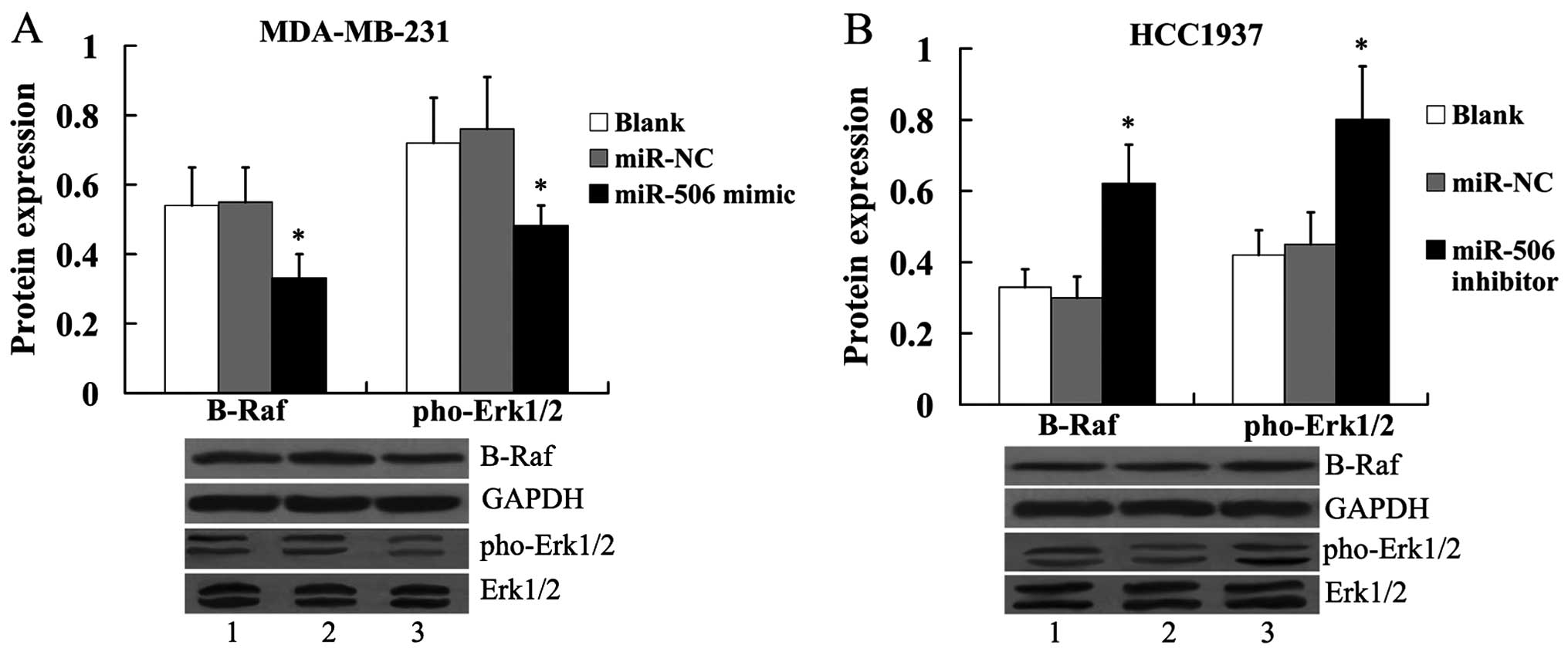
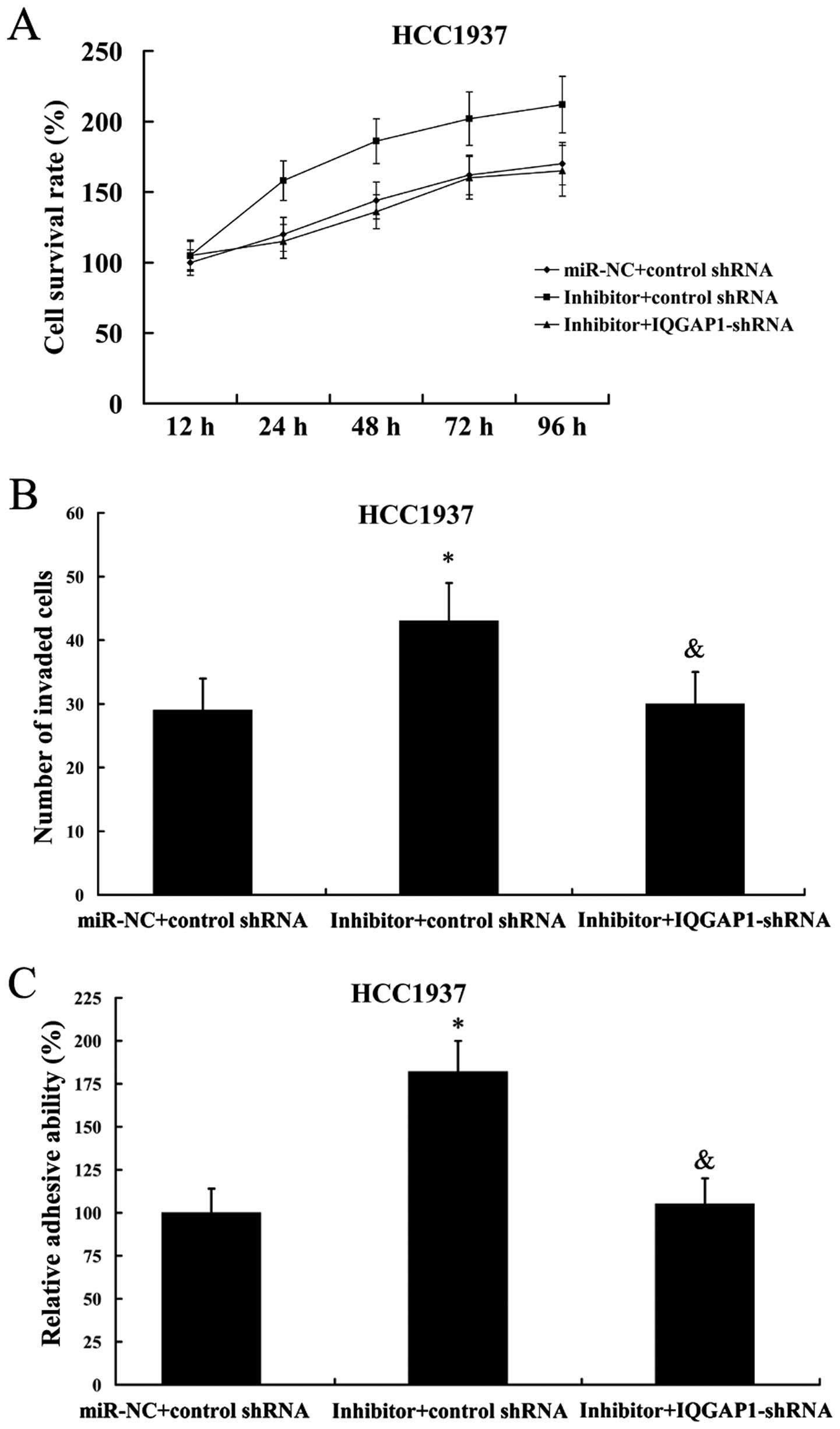
|
1
|
Badar F, Faruqui ZS, Ashraf A and Uddin N:
Third world issues in breast cancer detection. J Pak Med Assoc.
57:137–140. 2007.PubMed/NCBI
|
|
2
|
Jamal S, Mamoon N, Moghal S, Mushtaq S and
Luqman M: Carcinoma breast: A histopathological audit. J Coll
Physicians Surg Pak. 16:117–119. 2006.PubMed/NCBI
|
|
3
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chaffer CL and Weinberg RA: A perspective
on cancer cell metastasis. Science. 331:1559–1564. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nelson KM and Weiss GJ: MicroRNAs and
cancer: Past, present, and potential future. Mol Cancer Ther.
7:3655–3660. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu G, Sun Y, Ji P, Li X, Cogdell D, Yang
D, Parker Kerrigan BC, Shmulevich I, Chen K, Sood AK, et al:
MiR-506 suppresses proliferation and induces senescence by directly
targeting the CDK4/6-FOXM1 axis in ovarian cancer. J Pathol.
233:308–318. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Arora H, Qureshi R and Park WY: miR-506
regulates epithelial mesenchymal transition in breast cancer cell
lines. PLoS One. 8:e642732013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wen SY, Lin Y, Yu YQ, Cao SJ, Zhang R,
Yang XM, Li J, Zhang YL, Wang YH, Ma MZ, et al: miR-506 acts as a
tumor suppressor by directly targeting the hedgehog pathway
transcription factor Gli3 in human cervical cancer. Oncogene.
34:717–725. 2015. View Article : Google Scholar
|
|
10
|
Streicher KL, Zhu W, Lehmann KP,
Georgantas RW, Morehouse CA, Brohawn P, Carrasco RA, Xiao Z, Tice
DA, Higgs BW, et al: A novel oncogenic role for the miRNA-506-514
cluster in initiating melanocyte transformation and promoting
melanoma growth. Oncogene. 31:1558–1570. 2012. View Article : Google Scholar
|
|
11
|
Buffa FM, Camps C, Winchester L, Snell CE,
Gee HE, Sheldon H, Taylor M, Harris AL and Ragoussis J:
microRNA-associated progression pathways and potential therapeutic
targets identified by integrated mRNA and microRNA expression
profiling in breast cancer. Cancer Res. 71:5635–5645. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhao Y, Liu H, Li Y, Wu J, Greenlee AR,
Yang C and Jiang Y: The role of miR-506 in transformed 16HBE cells
induced by anti-benzo[a]pyrene-trans-7,8-dihydrodiol-9,10-epoxide.
Toxicol Lett. 205:320–326. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang Y, Cui M, Sun BD, Liu FB, Zhang XD
and Ye LH: MiR-506 suppresses proliferation of hepatoma cells
through targeting YAP mRNA 3′UTR. Acta Pharmacol Sin. 35:1207–1214.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Weissbach L, Settleman J, Kalady MF,
Snijders AJ, Murthy AE, Yan YX and Bernards A: Identification of a
human rasGAP-related protein containing calmodulin-binding motifs.
J Biol Chem. 269:20517–20521. 1994.PubMed/NCBI
|
|
15
|
Hart MJ, Callow MG, Souza B and Polakis P:
IQGAP1, a calmodulin-binding protein with a rasGAP-related domain,
is a potential effector for cdc42Hs. EMBO J. 15:2997–3005.
1996.PubMed/NCBI
|
|
16
|
Noritake J, Watanabe T, Sato K, Wang S and
Kaibuchi K: IQGAP1: A key regulator of adhesion and migration. J
Cell Sci. 118:2085–2092. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
White CD, Brown MD and Sacks DB: IQGAPs in
cancer: A family of scaffold proteins underlying tumorigenesis.
FEBS Lett. 583:1817–1824. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Johnson M, Sharma M and Henderson BR:
IQGAP1 regulation and roles in cancer. Cell Signal. 21:1471–1478.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lu SH, Jiang XJ, Xiao GL, Liu DY and Yuan
XR: miR-124a restoration inhibits glioma cell proliferation and
invasion by suppressing IQGAP1 and β-catenin. Oncol Rep.
32:2104–2110. 2014.PubMed/NCBI
|
|
20
|
Chen F, Zhu HH, Zhou LF, Wu SS, Wang J and
Chen Z: IQGAP1 is overexpressed in hepatocellular carcinoma and
promotes cell proliferation by Akt activation. Exp Mol Med.
42:477–483. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nakamura H, Fujita K, Nakagawa H, Kishi F,
Takeuchi A, Aute I and Kato H: Expression pattern of the scaffold
protein IQGAP1 in lung cancer. Oncol Rep. 13:427–431.
2005.PubMed/NCBI
|
|
22
|
Wang XX, Li XZ, Zhai LQ, Liu ZR, Chen XJ
and Pei Y: Overexpression of IQGAP1 in human pancreatic cancer.
Hepatobiliary Pancreat Dis Int. 12:540–545. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Erdemir HH, Li Z and Sacks DB: IQGAP1
binds to estrogen receptor-α and modulates its function. J Biol
Chem. 289:9100–9112. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jadeski L, Mataraza JM, Jeong HW, Li Z and
Sacks DB: IQGAP1 stimulates proliferation and enhances
tumorigenesis of human breast epithelial cells. J Biol Chem.
283:1008–1017. 2008. View Article : Google Scholar
|
|
25
|
Brown MD and Sacks DB: IQGAP1 in cellular
signaling: Bridging the GAP. Trends Cell Biol. 16:242–249. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ren JG, Li Z and Sacks DB: IQGAP1
modulates activation of B-Raf. Proc Natl Acad Sci USA.
104:10465–10469. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Schaeffer HJ and Weber MJ:
Mitogen-activated protein kinases: Specific messages from
ubiquitous messengers. Mol Cell Biol. 19:2435–2444. 1999.PubMed/NCBI
|
|
28
|
Roy M, Li Z and Sacks DB: IQGAP1 is a
scaffold for mitogen-activated protein kinase signaling. Mol Cell
Biol. 25:7940–7952. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Roy M, Li Z and Sacks DB: IQGAP1 binds
ERK2 and modulates its activity. J Biol Chem. 279:17329–17337.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
McCubrey JA, Steelman LS, Chappell WH,
Abrams SL, Wong EW, Chang F, Lehmann B, Terrian DM, Milella M,
Tafuri A, et al: Roles of the Raf/MEK/ERK pathway in cell growth,
malignant transformation and drug resistance. Biochim Biophys Acta.
1773:1263–1284. 2007. View Article : Google Scholar
|