Introduction
PCa is the most commonly diagnosed invasive cancer
and second only to lung cancer as a cause of cancer death in males
in the United States and other Western countries (1). The growth of PCa cells depends on the
presence of androgens, a group of steroid hormones. Most prostate
cancers are androgen-dependent and respond to the anti-androgens or
androgen-deprivation therapy. Unfortunately, patients who have
undergone androgen ablation therapy frequently develop a
hormone-refractory state, for which there is no curative therapy
(2).
AR signaling plays a crucial role in the development
of androgen-independent prostate cancer. AR, a member of the
steroid receptor subfamily of nuclear receptors, is a
ligand-activated transcription factor that regulates the expression
of target genes to mediate the biological function of androgens
(3). AR is composed of three major
domains: an NH2-terminal transcriptional activation domain (NTD), a
central DNA-binding domain (DBD) and a COOH-terminal ligand-binding
domain (LBD) (4). After binding to
androgen, AR formats the receptor homodimerization and translocates
to the nucleus. By recruiting to androgen response elements located
within AR-target genes, it can regulate the expression of AR-target
genes.
Once activated by a ligand, AR interacts with
various cofactors that facilitate transcription according to the
general transcriptional machinery (5). Deregulation of AR activity by
positive and negative cofactors is likely to play key a key role in
prostate cancer progression (6).
It has been reported that aberrant regulation of coregulator
expression or activity allows weak androgens to activate AR target
genes responsible for cell growth, eventually leading to the
development of hormone-refractory prostate cancers (7). An increasing number of AR-binding
proteins have been recognized to date, however, details remain
largely elusive. Therefore, finding novel and more effective
cofactors of AR signaling is of great interest in the present
research.
The LIM-only (LMO) proteins are a family of nuclear
transcription coregulators, which are characterized by the
exclusive presence of two tandem LIM domains and no other
functional domains (8). The LIM
domain is an ~55-residue, highly conserved cysteine-rich
zinc-binding motif (9,10). To date, four LMO proteins
(LMO1–LMO4) have been identified. These proteins are required for
many developmental processes, and are implicated in the initiation
and progression of several cancers, including T cell leukemia,
breast cancer and neuroblastoma (11). LMO1 was initially found as T cell
oncogene through its association with chromosomal translocations
occurring in T-cell acute lymphoblastic leukemia (12). Although LMO1 has been implicated in
cancer, most previous studies have only focused on defining its
role in T-cell acute lymphoblastic leukemia and neuroblastoma
pathogenesis.
This is the first report on the investigation into
the role of LMO1 as a potential AR-interacting protein and its
function in the regulation of AR transcriptional activity in PCa
cells. It has been found that LMO1 expression is significantly
increased with a higher level of aggressiveness in human PCa.
Together, our data suggest that LMO1, as a novel coactivator of AR,
can form a complex with AR and activate AR transcriptional activity
in the progression of human prostate cancer.
Materials and methods
Plasmid construction
AR, ARE-luc and PSA-luc were presented by Dr Yue
Zhao (China Medical University). GST-tagged, GFP-tagged,
Myc/His-tagged and Flag-tagged LMO1 (full-length and deletions)
were constructed by PCR amplification and subcloned into
pGEX-5X-1/2 (GE Healthcare, Piscataway, NJ, USA), pEGFP-C1
(Clontech Laboratories, Mountain View, CA, USA), pcDNA3.1Myc/HisA
(Invitrogen, San Diego, CA, USA) and p3xFlag CMV (Sigma-Aldrich,
St. Louis, MO, USA) vectors, respectively. All primer sequences for
generating these constructs are provided in Table I.
 | Table IThe sequences of primers used in
generating various constructs. |
Table I
The sequences of primers used in
generating various constructs.
| Primers name | Primer
sequences |
|---|
| LMO1 | F:
GACACGGAGGGATCCAGATGATGGTGCTGGAC | R:
CCAGGCGCGAATTCTTACTGAACTTGGGATTC |
| LMO1 (1–83) | F:
GACACGGAGGGATCCAGATGATGGTGCTGGAC | R:
GTTCCCTGGAATTCCAAAGAGCCTC |
| LMO1 (24–83) | F:
GAAGCGGATCCCCTGTGCGGGCTGTAAC | R:
GTTCCCTGGAATTCCAAAGAGCCTC |
| LMO1 (83–156) | F:
CTACCGGATCCTCTTTGGCACCACAG | R:
CCAGGCGCGAATTCTTACTGAACTTGGGATTC |
| LMO1 (88–147) | F:
GCACCAGGATCCACTGTGCTGCTTGC | R:
AGGTGCGAATTCCCTGCCCTTCCTC |
| LMO1 (24–147) | F:
GAAGCGGATCCCCTGTGCGGGCTGTAAC | R:
AGGTGCGAATTCCCTGCCCTTCCTC |
| FLAG-LMO1 | F:
CGGAGGGAATTCGATGATGGTGCTGGAC | R:
GGCAGGGGATCCTTACTGAACTTGGGATTC |
| AR | F:
CCAAGGATCCGGATGGAAGTGCAGTTAGGGC | R:
TCCAATCTCGAGCTGGGTGTGGAAATAGATG |
| AR (1–559) | F:
CCAAGGATCCGGATGGAAGTGCAGTTAGGGC | R:
CTCCACAGAATTCGCAGGTCTTCTGGGG |
| AR (505–676) | F:
ATGTGTGGATCCCTGGCGGCATGGTG | R:
CAATGGCGAATTCGACATTCAGAAAGATG |
| AR (633–919) | F:
CCGGAGGATCCAGAAACTTGGTAATC | R:
CTCCACAGAATTCGCAGGTCTTCTGGGG |
Cell culture and transfections
HEK-293, CV1 and PC-3 cell lines were cultured in
Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal
bovine serum (FBS). The prostate CWR22RV1 cells, were cultured in
RPMI-1640 medium containing 10% FBS. Charcoal-stripped serum (CSS)
was purchased from HyClone (Thermo Fisher Scientific, Waltham, MA,
USA). Lipofectamine 2000 (Invitrogen) was used for transfection.
Cells at ~60% confluence were transfected for 6 h and incubated
with phenol red-free medium containing 10% CSS for 16 h. Cell
extracts were prepared following another 16-h treatment with 10 nM
dihydrotestosterone (DHT) (Sigma-Aldrich). LMO1-siRNA and control
siRNA were purchased from Shanghai GeneChem Co. (Shanghai,
China).
GST pull-down assay
In vitro GST pull-down has been previously
described in detail (13). In
vitro transcription and translation of LMO1 proteins were
performed by using the TNT-coupled transcription and translation
system (Promega Biotech Co., Madison, WI, USA). Equal amounts of
GST, GST-fusion proteins immobilized by GST Sepharose Beads
(Amersham Biosciences) were incubated with in vitro
translated protein at 4°C for 2 h. Protein bound to beads was
eluted by 2X SDS-loading buffers and analyzed using western blot
analyses.
Immunoprecipitation and western blot
analysis
The cells were washed with cold PBS twice before
being lysed in IP lysis buffer supplemented with PMSF and protease
inhibitor cocktail (Roche, Basel, Switzerland). Cell lysates were
collected, washed and incubated at 4°C. Then, protein A agarose
slurry (GE Healthcare) preloaded with antibodies or normal IgG was
added to an equal amount of cell extracts and incubated overnight
at 4°C. Washed precipitated proteins were analyzed by western blot
analysis.
Denatured protein were analyzed by SDS-PAGE and
transferred to a PVDF membrane (Millipore, Billerica, MA, USA).
Samples were incubated and detected with indicated antibodies.
Anti-AR, anti-PSA and anti-P21 antibodies were from Thermo Fisher
Scientific and anti-LMO1 antibody was from Abcam (Cambridge, MA,
USA). Anti-myc antibody was purchased from Santa Cruz Biotechnology
(Santa Cruz, CA, USA), anti-Flag antibody was from Shanghai
Genomics, Inc., (Shanghai, China) and anti-GAPDH antibody was from
Shanghai KangChen (Shanghai, China).
Immunofluorescence staining
Cells were fixed in 4% paraformaldehyde and
permeabilized with 0.2% Triton X. Then they were blocked with
normal goat serum, after which they were incubated with the primary
antibody, followed by incubation with Alexa Flour 546 secondary
antibody (Molecular Probes, Eugene, OR, USA). Nuclei were
visualized by TO-PRO-3 (Molecular Probes) and the stained cells
were observed using a Leica/Olympus laser confocal scanning
microscope.
Tissue samples and
immunohistochemistry
Tissue specimens were collected from 97 patients
with PCa who underwent surgery in Shengjing Hospital and First
Hospital of China Medical University. All human tissues were
collected and used according to protocols approved by the Ethics
Committee of China Medical University. The histological grade of
cancers was based on the criteria set by the World Health
Organization.
Immunohistochemisty was previously specified
(14), and the intensity values of
immunohistochemical results were determined by HSCORE (histological
score) (15). The HSCORE was
calculated using two indices of proportion (Pί) and intensity
(ί). The proportion (Pί) represented the percentage of positive
cells. The intensity (ί) was classified as 0 (no staining), or 1+
(light brown staining), or 2+ (brown staining) or 3+ (heavy brown
staining). HSCORE was obtained for each slide by using the
following algorithm: HSCORE = ∑Pίxί. Where ί is 0, 1, 2, 3 and
Pί varies from 0.0 to 1.0, HSCOREs ranged from a minimum of zero
in cases with no staining to a maximum of 3.0 in cases in which all
the cells stained with maximal intensity. The HSCORE was determined
by two independent observers.
Luciferase assay
Luciferase assay has been previously described
(16). CV1, PC-3 and CWR22RV1
cells were transfected with indicated reporter and
Renilla-encoding plasmids with or without DHT. Then, the
supernatants from cells were measured for luciferase activities
using the Dual-luciferase reporter assay system (Promega) and a
Berthold Luminometer LB9570 (EG & G. Berthold, Freiburg,
Germany). Measured luciferase values were normalized to internal
Renilla control. Each experiment was repeated in
triplicate.
Real-time PCR
Real-time PCR has been described (17). Total RNA was isolated from CWR22RV1
cells using TRIzol reagent (Invitrogen). cDNA was synthesized by
reverse transcription using an RT reaction kit (Takara) according
to the manufacturer's instruction. Real-time PCR was performed
using LightCycler 480 (Roche) and SYBR® Premix ExTaq
(Takara) as a DNA-specific fluorescent dye. The housekeeping gene
β-actin was used as an endogenous control. PCR was performed in
triplicate for each reaction. The sequences of PCR primers were as
follows: P21 forward, 5′-TGTCCGTCAG AACCCATGC-3′ and P21 reverse,
5′-AAAGTCGAAGTTCCA TCGCTC-3′; PSA forward, 5′-CACCTGCTCGGGTGATTC
TG-3′ and PSA reverse, 5′-CCACTTCCGGTAATGCACCA-3′.
Statistical analysis
All statistical calculations were performed using
the SPSS (16.0) statistical software (SPSS, Inc., Chicago, IL,
USA). Immunohistochemistry data were analyzed by one-way ANOVA. For
luciferase assay and real-time PCR, two-sided Student's t-test was
used to determine the significant difference. Data are presented as
means ± standard errors (SE). Statistical significance was accepted
at P<0.05.
Results
Identification of LMO1 as an interacting
protein of AR
To investigate the potential physical and functional
interactions between LMO1 and AR, as a first step, we performed
in vitro GST pull-down assays to demonstrate the ability of
LMO1 and AR to interact directly. An interaction between AR and
GST-LMO1 was observed (Fig. 1A),
suggesting that LMO1 is an AR-binding protein.
To define the interacting domains within AR that
were required for the observed interaction, we used three deletion
constructs of AR (NTD, DBD and LBD) in GST pull-down assays. In
vitro-translated LMO1 was bound to LBD domain rather than NTD
or DBD domain (Fig. 1B). The
results indicate that AR interacts with LMO1 primarily via its
LBD.
We then identified the AR-binding domain in LMO1
using a series of LMO1 deletion constructs. GST pull-down assays
showed that deletion of either of the LMO1 LIM domains would not
abolish the ability of the LMO1 binding to the AR (Fig. 1C), which suggests that both of the
LIM domains of LMO1 are required for the cooperation with AR.
Subsequently, immunoprecipitation assays were used
to show the interaction of endogenous LMO1 and AR in the
AR-positive PCa cell line CWR22RV1. Cell lysates were subjected to
immunoprecipitate with AR antibody and then underwent
immunoblotting analysis with antibodies to LMO1 and AR. Endogenous
LMO1 was immunoprecipitated by the AR antibody, but not by the
control IgG antibody, confirming the physiological association
between endogenous LMO1 and AR (Fig.
1D). Furthermore, the same experiments carried out in a
reciprocal manner revealed that endogenous AR was also
immunoprecipitated by the LMO1 antibody (Fig. 1E). In addition, the interaction was
significantly enhanced in the presence of DHT. Taken together, our
findings suggest that LMO1 can interact with AR both in
vitro and in vivo.
LMO1 cotranslocates into the nucleus with
AR in an androgen-dependent manner
To observe the intracellular localization of LMO1
and AR, we cotransfected HEK293 cells with GFP-LMO1 and
pcDNA3.1-AR, followed by treatment with DHT. LMO1 was primarily
distributed in both cytoplasmic and nuclear compartments, and then
became concentrated in the nucleus after DHT treatment (Fig. 2, first column) from
immunofluorescence analysis. As we know, AR translocated into the
nucleus upon DHT exposure (Fig. 2,
second column). Interestingly, merged images revealed that the
distribution of LMO1 and AR in the nucleus overlapped under DHT
treatment (Fig. 2, fourth column).
Together, the results suggest that LMO1, as part of an AR complex,
cotranslocates into the nucleus with AR in an androgen-dependent
manner, thus, probably modulating AR transcriptional activity.
Expression of LMO1 in clinical prostate
biopsies
To test the role of LMO1 expression in PCa
progression, we observed the expression of LMO1 in clinical
prostate biopsies including 64 cases of PCa and 33 cases of benign
prostatic hyperplasia (BPH) by immunohistochemical staining. The
representative images are shown in Fig. 3A. Higher staining of LMO1 was
observed in PCa tissues than in BPH tissues (Fig. 3B). Moreover, there was an increase
in LMO1 staining in poorly differentiated PCa (Gleason score 8–10)
compared with well and moderately differentiated PCa (Gleason score
2–7) (Fig. 3B). These data
indicate a connection between LMO1 expression and the development
of PCa.
LMO1 upregulates AR-mediated
transcriptional activation in a DHT-dependent manner
AR is an important transcription factor that
regulates expression of a variety of target genes that harbor the
androgen response element (ARE) in their promoters. To reveal the
functional role of LMO1 in AR signaling, we first assessed the
effect of its expression on the ARE luciferase activity via
luciferase assays in CV1, CWR22RV1 and PC-3 cells, respectively.
Cells were transfected with ARE-Luc, pRL-TK and LMO1. AR-deficient
CV1 and PC-3 cells were co-transfected with AR. LMO1 induced ARE
luciferase activity only in the presence of DHT stimulation. LMO1
upregulated ARE luciferase activity in a dose-dependent manner and
the increase reached a plateau at 300 ng LMO1 (Fig. 4A). LMO1 also increased ARE
luciferase activity in the presence of DHT in PC-3 and CWR22RV1
cells (Fig. 4B and C). On the
other hand, ARE luciferase activity was decreased when LMO1 was
silenced by siLMO1 in CWR22RV1 cells (Fig. 4D).
 | Figure 4LMO1 stimulate AR transactivation
DHT-dependently. (A–C) CV1, PC-3 or CWR22RV1 cells were transfected
with ARE-Luc, pRL-TK, together with the indicated expression
plasmids with or without DHT. AR-deficient CV1 and PC-3 cells were
co-transfected with AR. The ARE-promoter activities were measured
by luciferase assays. The expression of LMO1 was evaluated by
western blot assays. (D) The transcriptional activity of ARE was
estimated in CWR22RV1 cells transfected with ARE-Luc, pRL-TK,
together with LMO1-siRNA or control, with or without DHT. The
expression of LMO1 was evaluated by western blot assays. (E and F)
PC-3 or CWR22RV1 cells were transfected with PSA-Luc, pRL-TK
together with vector or Myc-LMO1 constructs, with or without DHT,
and then assayed for luciferase activity. Experiments were repeated
three times (n=3). *P<0.05, indicates significant
differences compared to the control group. |
Prostate-specific antigen (PSA) gene, another target
gene downstream of AR, is known to contain an ARE, and its
transcriptional level is regulated by AR (18). Furthermore, we checked PSA
luciferase and found LMO1 also promoted PSA luciferase activity in
CWR22RV1 and PC-3 cells, respectively (Fig. 4E and F).
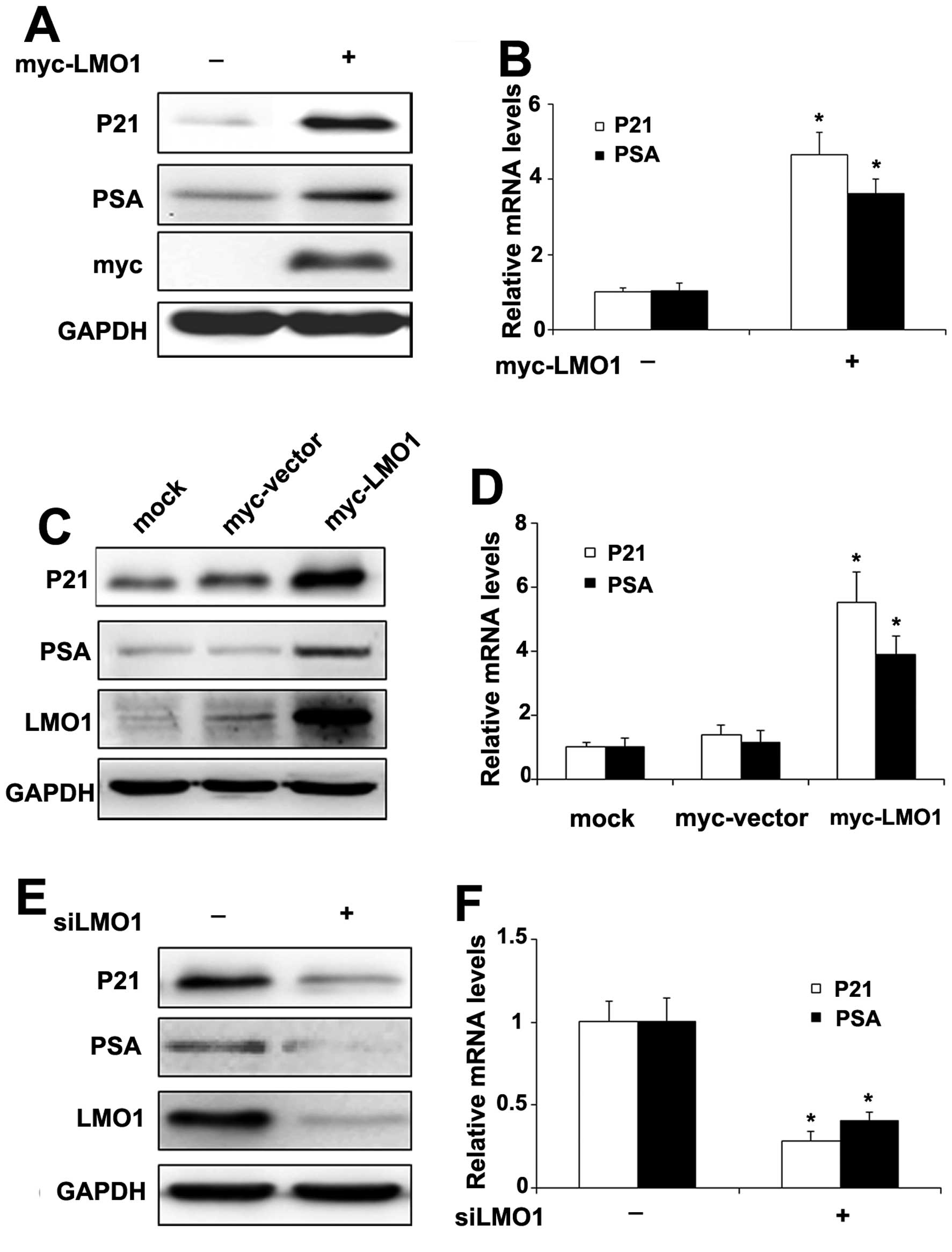
LMO1 facilitates the expression of
AR-targeted gene P21 and PSA
To further investigate the significance of LMO1
effect on AR signaling, we examined expression of the AR target
genes P21 (Waf1/Cip1) and PSA, which have been implicated in
prostate cancer cells (19,20).
When CWR22RV1 cells were transiently transfected with Myc-LMO1, the
protein (Fig. 5A) and mRNA levels
(Fig. 5B) of P21 and PSA
increased, respectively. Furthermore, the expression of protein
(Fig. 5C) and mRNA (Fig. 5D) of P21 and PSA were elevated,
respectively, in CWR22RV1 cells stably transfected with Myc-LMO1
compared to cells with the vector. On the other hand, when LMO1
levels were repressed by transfection with siLMO1, the protein
(Fig. 5E) and mRNA levels
(Fig. 5F) of P21 and PSA decreased
accordingly.
Discussion
One of the most troubling aspects of PCa progression
is that most cases will ultimately progress into a
hormone-refractory stage with high mortality rates after an initial
response to androgen-deprivation therapy. Despite advances in the
detection and treatment of PCa, current therapies remain limited
for the development of androgen-independent state (21). This highlights the need for a
better understanding of the mechanism of constitutive activation of
AR-mediated growth and subsequent androgen-independent receptor
activation involved in PCa progression, aiming to develop novel
targets for the diagnosis and treatment of the disease. Both the AR
and its coregulators have been extensively studied as hopeful
therapeutic targets for the treatment of PCa (22).
In the present study, we first provide evidence that
LMO1, which is overexpressed in human PCa tissue, is an
AR-interacting protein serving as a coactivator of AR to promote
the AR-mediated transactivation function and subsequently amplify
the expressions of P21 and PSA. These findings indicate that LMO1
is involved in PCa progression by acting as a coactivator of
AR.
LMO proteins are emerging as crucial molecules in a
wide variety of human cancers. All four LMO family members play key
roles in several cancers. LMO1 and LMO2 are involved in the onset
of T cell leukemia. LMO1 was identified near the break point of the
chromosomal translocation t(11;14) (p15;q11) in a T-ALL cell line
(8,23). LMO2 was subsequently found at the
junction of the chromosomal translocation t(11;14) (p13;q11) in
T-ALL (24,25). Even in the absence of chromosomal
lesions near LMO genes, overexpression of LMO1 or LMO2 is found in
~50% of human T-ALLs (26). The
roles of LMO1- and LMO2-transgenic mice during the development of
T-ALL have been more clearly defined (27,28).
Besides, LMO1 and LMO3 are implicated in human neuroblastoma.
Silence of LMO1 may suppress the growth of neuroblastoma cells with
high LMO1 expression, whereas overexpression of LMO1 in
neuroblastoma cells with low LMO1 expression promotes proliferation
(29). The overexpressing LMO3
shows a marked increase in cell growth and is indicative of poor
prognosis in neuroblastoma (30).
LMO4 seems to regulate progression of breast cancer by modulating
cell cycle arrest, cell proliferation, and apoptosis (31,32).
In the present study, the overexpression of LMO1 was
detected in human PCa tissues and it contributed to advanced stage
of PCa. Ma et al (33)
reported that LMO2 expression was significantly higher in PCa than
in normal epithelium and correlated positively with the level of
aggressiveness in human PCa. Yet no data on LMO1 expression in
human PCa are available to date. For the first time, our data
demonstrated that overexpression of LMO1 was found in human PCa
tissues, which also correlated with the level of cancer stage.
Moreover, there have been no data revealing the cellular location
of LMO1 in PCa cells. Our immunofluorescence analysis in the
present study demonstrated that LMO1 was distributed in both the
nucleus and the cytoplasm and then cotranslocated into the nucleus
with AR after DHT treatment.
AR cofactors that associate with AR and influence
its transcriptional activity could contribute to PCa progression.
Here, we first demonstrated LMO1 interacted with AR to form a
complex, then cotranslocated into the nucleus with AR in an
androgen-dependent manner and enhanced the AR transcriptional
activity. It is known that P21 and PSA are the target genes of AR
in prostate and their expressions are linked to the progression of
prostate cancer. P21, which belongs to a second family of
cyclin-dependent-kinase inhibitors, has various biological
functions such as cell cycle control, DNA repair and anti-apoptosis
(34,35). Androgens can upregulate the
expression of P21 at both the mRNA and protein levels in cell lines
(19,36). Increased expression of P21 is
associated with advanced PCa or cancer relapse in patients
(37,38). P21 protects cells against
p53-induced apoptosis, rescuing cells from a path of programmed
cell death to one of enhanced survival (39). PSA is highly expressed in prostate
gland and is one of the most reliable diagnostic markers for PCa
(40). Our results support that
LMO1 is also able to induce the expression of P21 and PSA in mRNA
and protein levels. These results imply that LMO1 acts as a
cofactor to modulate expression of some AR-target genes, thus
regulating PCa development and progression.
In summary, our results indicate that LMO1 binds to
AR and co-localize with AR in the nucleus, serving as a coactivator
of AR to promote the AR-mediated transactivation function and
subsequently induce the expression of P21 and PSA, downstream
targets of AR. In addition, the expression of LMO1 in PCa tissues
was significantly higher than that in BPH tissues and their
expression levels positively correlated with the grade of cancers.
This finding provides insights into the role of LMO1 in the
progression of PCa. Therefore, LMO1 could be a potential prognostic
marker and drug target for human prostate cancer.
Acknowledgements
We thank Dr Yue Zhao (China Medical University) for
providing the plasmids. We would like to express our gratitude to
all the subjects who participated in the present study.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yap TA, Zivi A, Omlin A and de Bono JS:
The changing therapeutic landscape of castration-resistant prostate
cancer. Nat Rev Clin Oncol. 8:597–610. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhou Y, Bolton EC and Jones JO: Androgens
and androgen receptor signaling in prostate tumorigenesis. J Mol
Endocrinol. 54:R15–R29. 2015. View Article : Google Scholar
|
|
4
|
Gelmann EP: Molecular biology of the
androgen receptor. J Clin Oncol. 20:3001–3015. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Heinlein CA and Chang C: Androgen receptor
in prostate cancer. Endocr Rev. 25:276–308. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Heemers HV and Tindall DJ: Androgen
receptor (AR) coregulators: A diversity of functions converging on
and regulating the AR transcriptional complex. Endocr Rev.
28:778–808. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chmelar R, Buchanan G, Need EF, Tilley W
and Greenberg NM: Androgen receptor coregulators and their
involvement in the development and progression of prostate cancer.
Int J Cancer. 120:719–733. 2007. View Article : Google Scholar
|
|
8
|
Boehm T, Foroni L, Kennedy M and Rabbitts
TH: The rhombotin gene belongs to a class of transcriptional
regulators with a potential novel protein dimerisation motif.
Oncogene. 5:1103–1105. 1990.PubMed/NCBI
|
|
9
|
Jurata LW and Gill GN: Structure and
function of LIM domains. Curr Top Microbiol Immunol. 228:75–113.
1998.
|
|
10
|
Dawid IB, Breen JJ and Toyama R: LIM
domains: Multiple roles as adapters and functional modifiers in
protein interactions. Trends Genet. 14:156–162. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Matthews JM, Lester K, Joseph S and Curtis
DJ: LIM-domain-only proteins in cancer. Nat Rev Cancer. 13:111–122.
2013. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rabbitts TH: LMO T-cell translocation
oncogenes typify genes activated by chromosomal translocations that
alter transcription and developmental processes. Genes Dev.
12:2651–2657. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu T, Li Y, Gu H, Zhu G, Li J, Cao L and
Li F: p21-Activated kinase 6 (PAK6) inhibits prostate cancer growth
via phosphorylation of androgen receptor and tumorigenic E3 ligase
murine double minute-2 (Mdm2). J Biol Chem. 288:3359–3369. 2013.
View Article : Google Scholar :
|
|
14
|
Chmelar R, Buchanan G, Need EF, Tilley W
and Greenberg NM: Downregulation of p21-activated kinase-1 inhibits
the growth of gastric cancer cells involving cyclin B1. Int J
Cancer. 125:2511–2519. 2009. View Article : Google Scholar
|
|
15
|
Berchuck A, Soisson AP, Clarke-Pearson DL,
Soper JT, Boyer CM, Kinney RB, McCarty KS Jr and Bast RC Jr:
Immunohistochemical expression of CA 125 in endometrial
adenocarcinoma: Correlation of antigen expression with metastatic
potential. Cancer Res. 49:2091–2095. 1989.PubMed/NCBI
|
|
16
|
Wang C, Li Y, Zhang H, Liu F, Cheng Z,
Wang D, Wang G, Xu H, Zhao Y, Cao L, et al: Oncogenic PAK4
regulates Smad2/3 axis involving gastric tumorigenesis. Oncogene.
33:3473–3484. 2014. View Article : Google Scholar
|
|
17
|
Li Y, Shao Y, Tong Y, Shen T, Zhang J, Li
Y, Gu H and Li F: Nucleo-cytoplasmic shuttling of PAK4 modulates
β-catenin intracellular translocation and signaling. Biochim
Biophys Acta. 1823:465–475. 2012. View Article : Google Scholar
|
|
18
|
Riegman PH, Vlietstra RJ, van der Korput
JA, Brinkmann AO and Trapman J: The promoter of the
prostate-specific antigen gene contains a functional androgen
responsive element. Mol Endocrinol. 5:1921–1930. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lu S, Liu M, Epner DE, Tsai SY and Tsai
MJ: Androgen regulation of the cyclin-dependent kinase inhibitor
p21 gene through an androgen response element in the proximal
promoter. Mol Endocrinol. 13:376–384. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cleutjens KB, van Eekelen CC, van der
Korput HA, Brinkmann AO and Trapman J: Two androgen response
regions cooperate in steroid hormone regulated activity of the
prostate-specific antigen promoter. J Biol Chem. 271:6379–6388.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Taplin ME and Balk SP: Androgen receptor:
A key molecule in the progression of prostate cancer to hormone
independence. J Cell Biochem. 91:483–490. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Debes JD and Tindall DJ: Mechanisms of
androgen-refractory prostate cancer. N Engl J Med. 351:1488–1490.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Boehm T, Baer R, Lavenir I, Forster A,
Waters JJ, Nacheva E and Rabbitts TH: The mechanism of chromosomal
translocation t(11;14) involving the T-cell receptor C delta locus
on human chromosome 14q11 and a transcribed region of chromosome
11p15. EMBO J. 7:385–394. 1988.PubMed/NCBI
|
|
24
|
Boehm T, Foroni L, Kaneko Y, Perutz MF and
Rabbitts TH: The rhombotin family of cysteine-rich LIM-domain
oncogenes: Distinct members are involved in T-cell translocations
to human chromosomes 11p15 and 11p13. Proc Natl Acad Sci USA.
88:4367–4371. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Van Vlierberghe P, van Grotel M, Beverloo
HB, Lee C, Helgason T, Buijs-Gladdines J, Passier M, van Wering ER,
Veerman AJ, Kamps WA, et al: The cryptic chromosomal deletion
del(11)(p12p13) as a new activation mechanism of LMO2 in pediatric
T-cell acute lymphoblastic leukemia. Blood. 108:3520–3529. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ferrando AA, Neuberg DS, Staunton J, Loh
ML, Huard C, Raimondi SC, Behm FG, Pui CH, Downing JR, Gilliland
DG, et al: Gene expression signatures define novel oncogenic
pathways in T cell acute lymphoblastic leukemia. Cancer Cell.
1:75–87. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tremblay M, Tremblay CS, Herblot S, Aplan
PD, Hébert J, Perreault C and Hoang T: Modeling T-cell acute
lymphoblastic leukemia induced by the SCL and LMO1 oncogenes. Genes
Dev. 24:1093–1105. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Neale GA, Rehg JE and Goorha RM:
Disruption of T-cell differentiation precedes T-cell tumor
formation in LMO-2 (rhombotin-2) transgenic mice. Leukemia.
11(Suppl 3): 289–290. 1997.PubMed/NCBI
|
|
29
|
Wang K, Diskin SJ, Zhang H, Attiyeh EF,
Winter C, Hou C, Schnepp RW, Diamond M, Bosse K, Mayes PA, et al:
Integrative genomics identifies LMO1 as a neuroblastoma oncogene.
Nature. 469:216–220. 2011. View Article : Google Scholar
|
|
30
|
Aoyama M, Ozaki T, Inuzuka H, Tomotsune D,
Hirato J, Okamoto Y, Tokita H, Ohira M and Nakagawara A: LMO3
interacts with neuronal transcription factor, HEN2, and acts as an
oncogene in neuroblastoma. Cancer Res. 65:4587–4597. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Montañez-Wiscovich ME, Seachrist DD,
Landis MD, Visvader J, Andersen B and Keri RA: LMO4 is an essential
mediator of ErbB2/HER2/Neu-induced breast cancer cell cycle
progression. Oncogene. 28:3608–3618. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang N, Lin KK, Lu Z, Lam KS, Newton R, Xu
X, Yu Z, Gill GN and Andersen B: The LIM-only factor LMO4 regulates
expression of the BMP7 gene through an HDAC2-dependent mechanism,
and controls cell proliferation and apoptosis of mammary epithelial
cells. Oncogene. 26:6431–6441. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ma S, Guan XY, Beh PS, Wong KY, Chan YP,
Yuen HF, Vielkind J and Chan KW: The significance of LMO2
expression in the progression of prostate cancer. J Pathol.
211:278–285. 2007. View Article : Google Scholar
|
|
34
|
Levine AJ: p53, the cellular gatekeeper
for growth and division. Cell. 88:323–331. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang J and Walsh K: Resistance to
apoptosis conferred by Cdk inhibitors during myocyte
differentiation. Science. 273:359–361. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lu S, Tsai SY and Tsai MJ: Molecular
mechanisms of androgen-independent growth of human prostate cancer
LNCaP-AI cells. Endocrinology. 140:5054–5059. 1999.PubMed/NCBI
|
|
37
|
Omar EA, Behlouli H, Chevalier S and
Aprikian AG: Relationship of p21(WAF-I) protein expression with
prognosis in advanced prostate cancer treated by androgen ablation.
Prostate. 49:191–199. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Fizazi K, Martinez LA, Sikes CR, Johnston
DA, Stephens LC, McDonnell TJ, Logothetis CJ, Trapman J, Pisters L,
et al: The association of p21(WAF-1/CIP1) with
progression to androgen-independent prostate cancer. Clin Cancer
Res. 8:775–781. 2002.PubMed/NCBI
|
|
39
|
Gorospe M, Cirielli C, Wang X, Seth P,
Capogrossi MC and Holbrook NJ: p21Waf1/Cip1 protects
against p53-mediated apoptosis of human melanoma cells. Oncogene.
14:929–935. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Gittes RF: Carcinoma of the prostate. N
Engl J Med. 324:236–245. 1991. View Article : Google Scholar : PubMed/NCBI
|