Introduction
Colorectal cancer is one of the most common type of
cancers and is the fourth leading cause of cancer deaths in Korea
(1). In addition, the incidence
rates of colorectal cancer have continued to increase in both
genders from 1999 to 2010 (1).
Despite advances in surgical resection and systemic chemotherapies,
the most important factors contributing to progression and poor
prognosis of cancer are recurrence and metastasis (2–4).
However, the signaling pathways involved in tumor development,
progression, and metastasis are highly complex and require further
characterization. Therefore, it is essential to identify and
investigate signaling pathways involved in cancer recurrence or
metastasis of colorectal cancer.
Rho GTPases belong to the small GTPase family of
proteins (~21 kDa) that includes Ras, Rab, Arf, and Rho families
(5,6). These proteins have been implicated in
many important cancer-related processes in mammalian cells, such as
proliferation, migration, and survival. Mammalian cells express
more than 22 Rho GTPases, such as Rho isoforms (A, B and C), three
Rac isoforms (1, 2 and 3), Cdc42, among others (7,8). Rho
GTPases exist in two forms: the inactive, GDP-bound form, or the
active, GTP-bound form. This dynamic form allows these proteins to
function as molecular switches when activated by cell surface
receptors, leading to transcriptional activation, cytoskeleton
reorganization, and cell migration (9). Rho GTPases are highly expressed or
activated in multiple cancers (7);
RhoA inhibition using small interfering RNA has been shown to
reduce proliferation and tumor burden in vitro and in
vivo (10–12). Additionally, RhoA expression is
higher in tumor samples than in normal tissues (13). However, recently, it has been
suggested that RhoA acts as a tumor suppressor in colorectal
cancer, suppressing tumor progression and metastasis (14). Thus, it is still unclear whether
RhoA functions as a promoter or suppressor of colorectal
cancer.
Materials and methods
Ethics statement
All procedures performed in studies involving human
participants were in accordance with the ethical standards of
Ethics Committee of Soonchunhyang University Cheonan Hospital and
with the 1964 Helsinki declaration and its later amendments or
comparable ethical standards. Informed consent was obtained from
all individual participants included in the study.
Cell lines
Human colorectal cancer cell lines HCT116, HT29,
LoVo, SW480, SW620, colo201, colo205, and CaCO2 were
purchased from the Korean Cell Line Bank (KCLB). Cells were grown
in RPMI-1640 medium (Cellgro, USA) supplemented with 10% fetal
bovine serum (Equitech-bio, USA) and 1X penicillin/streptomycin
(Cellgro) at 37°C in a humidified atmosphere containing 5%
CO2.
Plasmid constructs and transfection
Short hairpin RNA (shRNA) constructs were designed
and cloned into the H1-shRNA vector (Genolution Pharmaceuticals
Inc.). The target sequence for RhoA is 5′-CAGAAAAGTGGACCCCAGAA-3′.
The sequence of nonsense shRNA against luciferase was provided by
Genolution Pharmaceuticals Inc. The plasmids were transfected into
HCT116 cells using Lipofectamine 2000 (Invitrogen, USA) according
to the manufacturer's instructions. Briefly, HCT116 cells were
cultured and transfected using Lipofectamine 2000. Medium was
changed 6 h after transfection, and transfected cells were selected
using Zeocin (Invitrogen). To evaluate transfection efficiency,
semi-quantitative reverse transcriptase-polymerase chain reaction
(RT-PCR) was used.
RNA extraction and RT-PCR
RNA was isolated using TRIzol (Invitrogen); equal
amounts of RNA were converted to cDNA using ReverTra
Ace® qPCR kit (Toyobo, Japan) according to the
manufacturer's instructions. To determine RhoA expression, PCR was
performed using the Maxime PCR PreMix kit (iNtRON, Korea). Primer
sequences are as follows: RhoA-F, 5′-CATCCGGAAGAAACTGGT-3′; RhoA-R,
5′-TCCCACAAAGCCAACTC-3′; GAPDH (glyceraldehyde-3-phosphate
dehydrogenase)-F, 5′-CTTAGCACCCCTGGCCAAG-3′; GAPDH-R,
5′-GATGTTCTGGAGAGCCCCG-3′; Zeocin-F, 5′-CGACGTGACCCTGTTCATCAG-3′;
Zeocin-R, 5′-GTTCGTGGACACGACCTCCGA-3′. The expected amplicon sizes
were 168 bp (RhoA), 121 bp (GAPDH), and 130 bp (Zeocin). The PCR
cycles consisted of a pre-denaturation step at 95°C for 10 min,
followed by 35 3-temperature cycles (95, 59.5 and 72°C) for 30 sec
each, and a final extension at 72°C for 5 min. The PCR products
were confirmed using the QIAxcel auto electrophoresis system
(Qiagen, USA).
Western blot analysis
Cell lysates were harvested using PRO-PREP (iNtRON,
Korea) for 30 min on ice and centrifuged at 13,000 rpm for 5 min at
4°C. Protein concentration of the supernatant was determined by BCA
assay (Thermo, USA). An equal amount of each protein extract (50
μg) was resolved using 10% polyacrylamide gel and
electro-transferred onto 0.2 μm polyvinylidene fluoride (PVDF)
membrane (Millipore, USA) using Trans-blot turbo (Bio-Rad
Laboratories, Inc., USA). Membranes were immunoblotted with either
1:1,000-diluted mouse anti-RhoA monoclonal antibody (Abnova,
Taiwan) or 1:5,000-diluted mouse anti-β-actin monoclonal antibody
(Sigma, USA) overnight at 4°C. Membranes were incubated with
1:10,000-diluted horseradish peroxidase-conjugated anti-mouse
immunoglobulin (Sigma) for 1 h at room temperature. The protein
signal was detected by enhanced chemiluminescence (Advansta, USA)
using the Molecular Imager ChemiDoc XRS+ System (Bio-Rad
Laboratories, Inc.).
Cell proliferation assay (MTT assay)
MTT assay was used to evaluate cell proliferation
after transfection. Cells (1.0×105 cells/well) were
seeded into a 96-well plate and incubated for an additional 24–72 h
post-transfection. After time-dependent incubation, the medium was
removed and the cells were washed with PBS. The cells were
incubated in a 5-mg/ml MTT (Sigma) solution for 4 h. Then, the
media was substituted with dimethyl sulfoxide (DMSO; Sigma) and
placed on the plate shaker for 15 min. Absorbance was read at 570
nm using a plate reader.
Migration assay (wound healing
assay)
Cell migration was analyzed in vitro using
the Culture insert system 24 (ibidi, Germany). The culture insert
was attached to the bottom of a 6-well plate, and 100 μl of media
containing 1.0×106 cells were seeded into each well of
the insert. The culture inserts were removed from the plate after
48 h, and cells were further cultured with fresh RPMI-1640 medium
contained 10% FBS. The cell gap was monitored every 12 h for 48 h.
Cell images were taken every 12 h with a phase contrast microscope,
AxionCam camera (Zeiss, Germany).
Invasion assay (Matrigel invasion
assay)
The transwell culture insert was pre-coated with 50
μl of Matrigel (BD, USA) according to the manufacturer's
instructions. Cells (5.0×106) were suspended in
serum-free RPMI-1640 medium and seeded into the pre-coated insert.
Eight hundred microliters of RPMI-1640 medium containing 10% FBS
was added outside the transwell culture insert. Cells were
incubated at 37°C for 24 h in a humidified atmosphere with 5%
CO2. After washing the transwell insert twice with PBS,
cells were fixed with 10% formaldehyde for 2 min. The cells were
permeabilized with methanol for 20 min and then stained with
methylgreen for 15 min. The transwell insert was washed twice with
PBS, wiped using cotton swab, and then observed using an inverted
microscope.
Immunofluorescent staining
Immunostaining was conducted as previously
described, with slight modification, to determine morphology change
after transfection (15). Briefly,
1×105 cells were seeded on a cover slip placed in a
6-well plate and incubated at 37°C in a humidified atmosphere with
5% CO2 for 24 h. The wells were washed twice with cold
PBS and fixed in cold methanol/acetone 1:1 (vol/vol) for 30 min at
−20°C. The methanol/acetone mixture was then removed and the wells
were dried for 15 min at room temperature. After rehydration with
PBS for 15 min at room temperature, cells were incubated with
blocking buffer (2 mg/ml BSA in PBS) for 1 h at room temperature,
and then rinsed with blocking buffer three times. Cells were
stained with 1 μg/ml DAPI for 2 min, sealed with clear nail polish,
and visualized using an Olympus FV10 confocal microscope.
Human colorectal carcinoma specimens
A total of 150 colorectal carcinoma tissue specimens
were obtained from Soonchunhyang University Cheonan Hospital,
Korea, where samples were collected from patients who underwent
surgery between 2002 and 2007. These tissues were formalin-fixed
and paraffin-embedded (FFPE). Clinicopathological data including
age, gender, TNM classification, and distal metastasis are shown at
Table I. Tumor stage was
identified according to the American Joint Committee on Cancer TNM
classification system. Sample collection for this study was
approved by the Ethics Committee of Soonchunhyang University,
Cheonan Hospital.
 | Table IClinicopathological features of
patient samples. |
Table I
Clinicopathological features of
patient samples.
| Clinicopathological
factors | N |
|---|
| Sex |
| Male | 93 |
| Female | 57 |
| pT stage |
| 1 | 4 |
| 2 | 22 |
| 3 | 108 |
| 4 | 16 |
| pN stage |
| 0 | 76 |
| 1 | 48 |
| 2 | 26 |
| Distal
metastasis |
| Negative | 144 |
| Positive | 6 |
| Vascular
invasion |
| Negative | 123 |
| Positive | 27 |
| Lymphatic
invasion |
| Negative | 118 |
| Positive | 32 |
| Clinical stage |
| I | 18 |
| II | 56 |
| III | 70 |
| IV | 6 |
Tissue microarray (TMA) and
immunohistochemistry
Immunohistochemical staining was performed using
tissue microarray (TMA) block sections to determine RhoA expression
in patient samples. The FFPE tumor tissues were re-embedded from
each FFPE block to the recipient block in duplicate. Each TMA block
contained 60 cores from 30 samples. For immunohistochemistry, 4-μm
sections were obtained using a microtome, deparaffinized in xylene,
and rehydrated in 100–70% alcohol series. Antigen retrieval was
achieved in citrate buffer (pH 6.0) using a microwave for 15 min.
To eliminate endogenous peroxidase activity, the sections were
incubated in peroxidase blocking solution (Dako, Denmark) for 30
min and then washed with phosphate-buffered saline containing 0.1%
Tween-20 (PBST). The sections were incubated with anti-mouse RhoA
antibody (Abnova, 1:500) for 2 h at room temperature, followed by
incubation in enhancer for 30 min and treatment with polymer for 1
h at room temperature. After washing with PBST, sections were
incubated with DAB, counterstained with hematoxylin, and observed
under a microscope.
IHC data analysis
The RhoA-stained tissue cores were examined by 2
independent observers (Chang-Jin Kim and Dongjun Jeong), and a
consensus score was determined for each specimen. A positive
reaction was scored into 4 grades, according to the intensity of
the staining: 0, 1+, 2+, and 3+. The percentages of RhoA-positive
cells were also scored into 4 categories: 0 (0%), 1 (1–30%), 2
(31–70%) and 3 (71–100%). The final score, calculated as the
product of the intensity score multiplied by the percentage score,
was classified as follows: 0 for negative; 1–3 for weak; 4–6 for
moderate; and 7–9 for strong. Samples with a final score ≤3 were
grouped together as RhoA expression negative while those with a
score ≥4 were grouped together as RhoA expression positive.
Statistical analysis
Statistical analysis was conducted using SPSS 19.0
(Chicago, IL, USA) program. The results of RT-PCR, western
blotting, and functional characterization of cells were analyzed
with one-way ANOVA test and Student's t-test. The relationship
between the result of immunohistochemistry and clinicopathological
data was analyzed with Chi-square test. Hazard ratio and 95%
confidence interval were evaluated using Cox regression models.
Kaplan-Meier method was used to analyze disease-free survival rate
using the log-rank test. A p-value of <0.05 was considered
statistically significant in all assessments.
Results
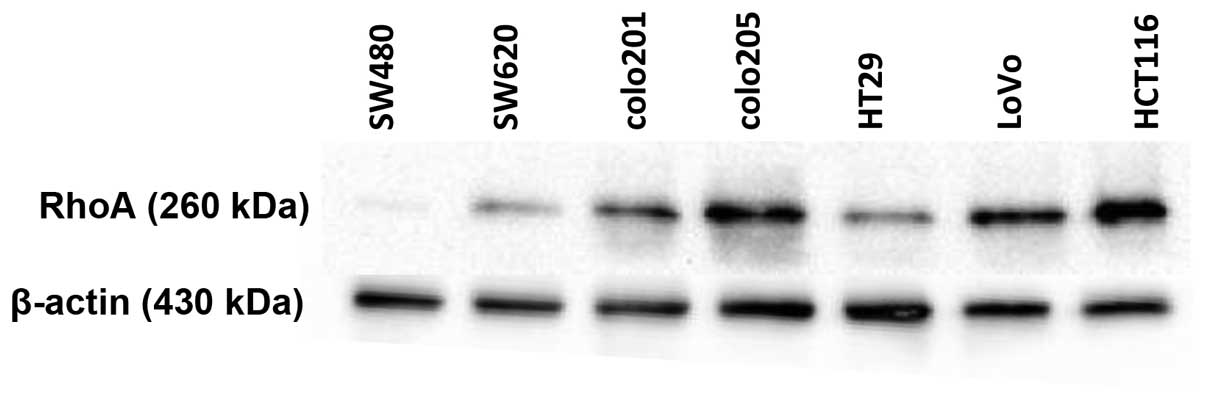
RhoA expression in various colorectal
carcinoma cell lines
As RhoA has been implicated in multiple cancers
including breast cancer, liver cancer, ovarian carcinoma, and
gastric carcinoma, we examined the expression of RhoA in both
non-metastatic and metastatic colorectal cancer cell lines. A
non-metastatic cancer cell line, SW480, expressed a very small
amount of RhoA, whereas many cancer cell lines including colo205,
LoVo, and HCT116 exhibited high levels of RhoA expression (Fig. 1). Notably, the metastatic cell line
HCT116, had the highest RhoA protein expression among the cell
lines tested. Therefore, we used the HCT116 cancer cell line for
further studies to determine the function of RhoA in the
carcinogenesis of colon cancer.
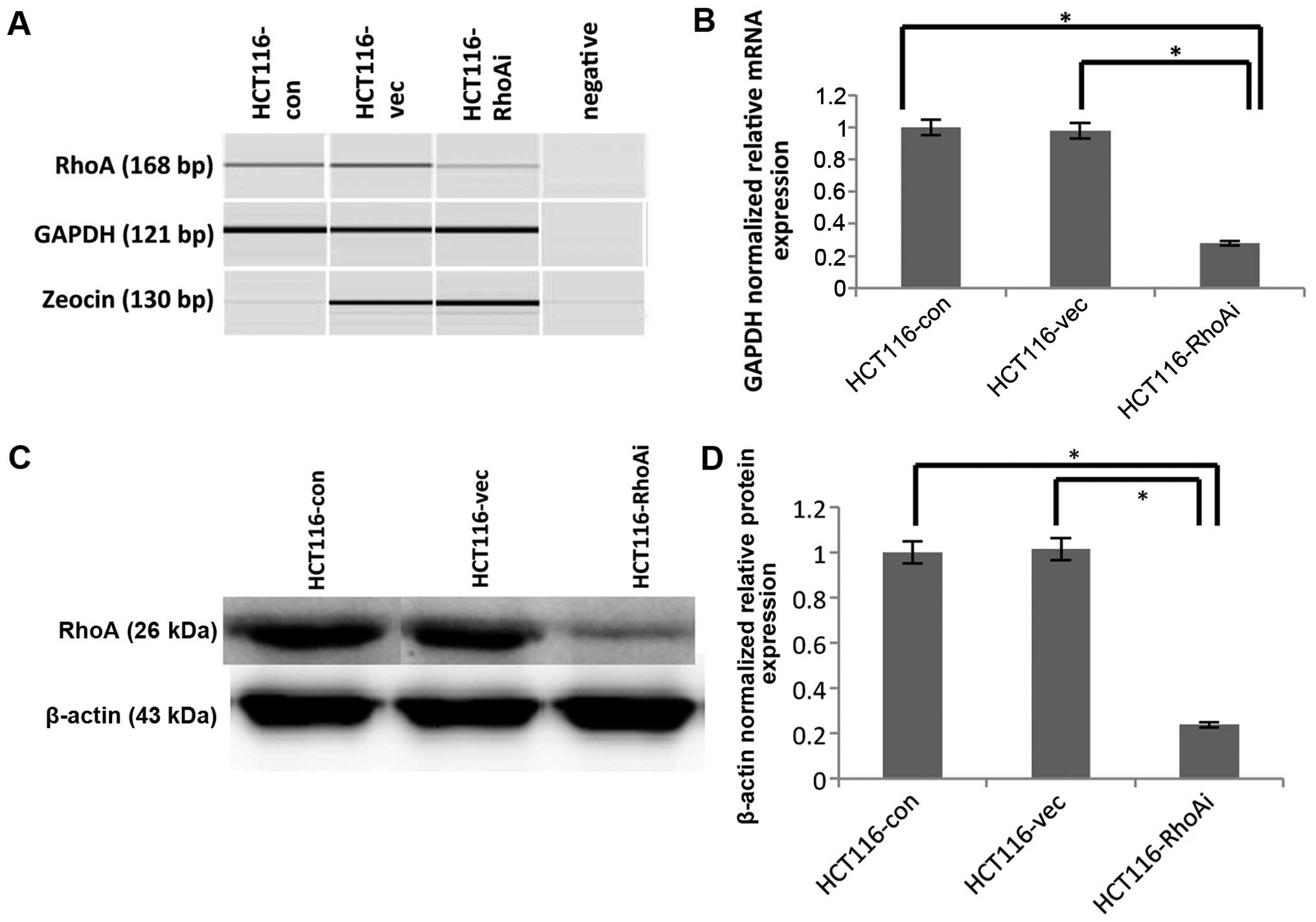
shRNA-mediated RhoA knockdown in the
HCT116 cell line
To determine the functional consequence of RhoA, we
used shRNA to knock down RhoA in HCT116 cells. The knockdown
efficiency of RhoA was confirmed using RT-PCR and immunoblotting
(Fig. 2). Specifically, HCT116
cells were transfected with either nonsense control (HCT116-vec) or
shRhoA (HCT116-RhoAi). RNA and protein lysates from each pair of
transfected cells were extracted and determined to assess RhoA
levels. To determine the knockdown efficiency at the RNA level,
RT-PCR was performed using cDNA from the samples. Amplicon size is
168 bp for RhoA, 121 bp for GAPDH, and 130 bp for zeocin. The PCR
products were analyzed with the auto electrophoresis system,
QIAxcel (Qiagen, USA). Zeocin was used to compare the transfection
efficiency between nonsense control and shRhoA, and indicated
similar transfection efficiencies (Fig. 2A). The expression of RhoA was
significantly downregulated in shRhoA-treated HCT116 cells
(HCT116-RhoAi) compared to untransfected (HCT116-con) and nonsense
control (HCT116-vec) cells (p=0.028 and p=0.035, respectively).
Specifically, the RhoA RNA level decreased by ~70% after shRhoA
transfection, and there was no difference between the untransfected
group (HCT116-con) and the control transfected group (HCT116-vec)
(Fig. 2B).
As changes at RNA level do not always correlate with
changes at the protein level, it is necessary to determine the
protein level after knockdown. To test this, we also determined
RhoA protein level after shRNA-mediated depletion. Protein lysates
from each pair of transfected cells were collected and immunobloted
with anti-RhoA Ab (Fig. 2C and D).
Protein expression also displayed the same pattern observed in RNA
after knockdown of RhoA. The protein level of RhoA was decreased by
70% in HCT116-RhoAi compared to untransfected cells or
vector-transfected cells (p<0.0001). These results led us to
confirm that RhoA was successfully knocked down.
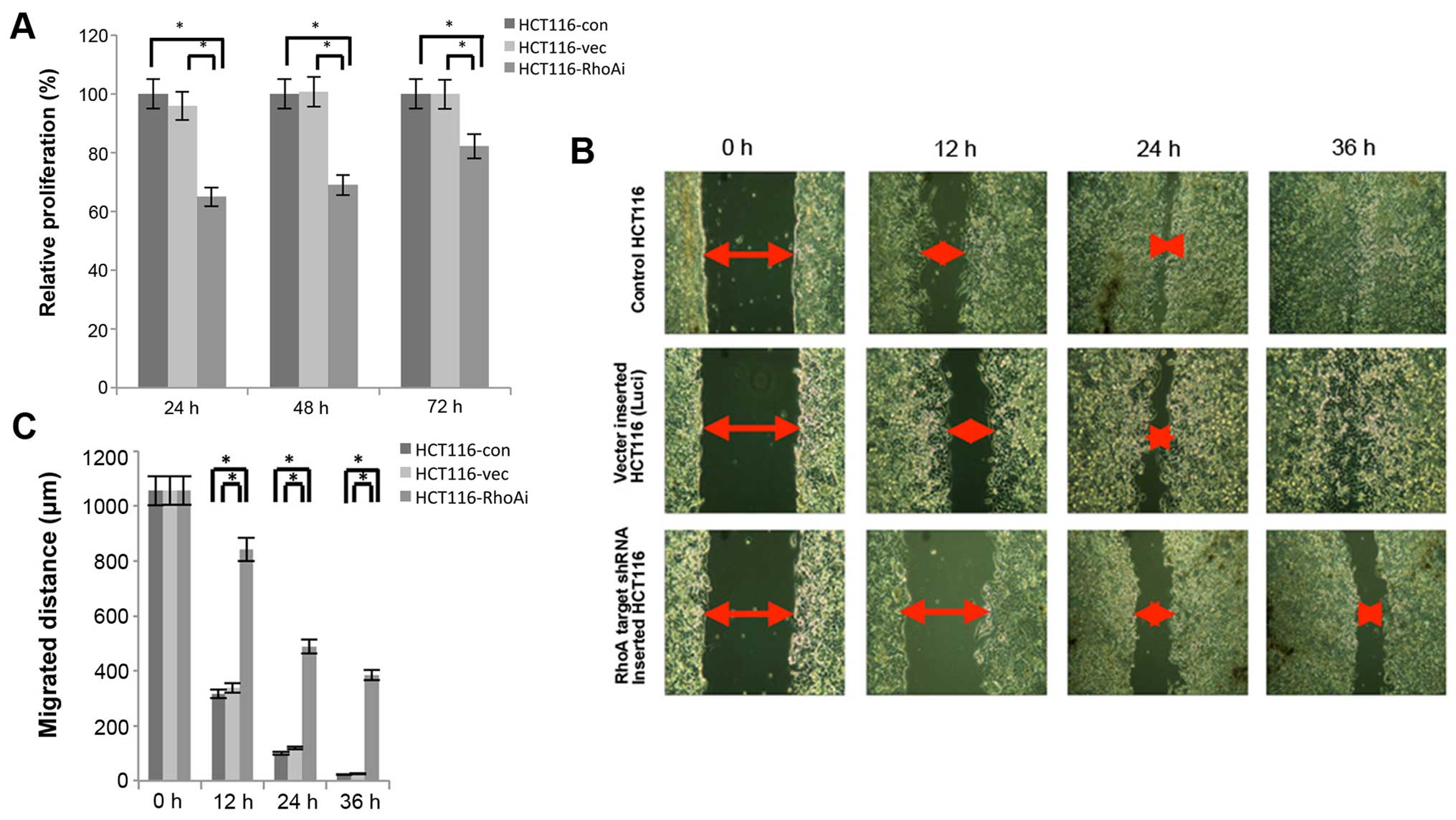
Knockdown of RhoA impairs the
proliferation of HCT116 cells
To functionally define the role of RhoA, we first
determined proliferation ability in the HCT116 cell line after RhoA
knockdown using an MTT assay. After transfection of either control
vector or shRhoA, cells were incubated for different time periods,
ranging from 24 to 72 h, to determine proliferation rate. As shown
in Fig. 3A, the proliferation of
shRhoA-transfected cells was lower than that of HCT116-con or
HCT116-vec cells (24 h, p<0.0001; 48 h, p<0.0001; 72 h,
p=0.002). The proliferation between HCT-con and HCT116-vec cells
was not significantly different. These results suggest that RhoA
plays a role in proliferation of colon cancer cells.
RhoA is essential for migration of HCT116
cells
In order to assess the effect of RhoA on migration
of HCT116 cells, migration ability was determined after knocking
down RhoA using a wound healing assay. Cells were seeded into
culture insert, which was attached to the bottom of a 6-well plate,
and the culture inserts were removed two days later. The cells were
further cultured and monitored every 12 h (Fig. 3B and C). Over time, cells in
control groups migrated gradually. However, the shRhoA-transfected
group displayed approximately a 2-fold delay in migration at 12 h
and a 3-fold delay at 24 h compared to control groups. More
significantly, while control groups showed almost no gaps at 36 h,
shRhoA group still displayed ~400-μm gaps, suggesting that
RhoA-deficient cells were significantly impaired in the ability to
migrate (Fig. 3C).
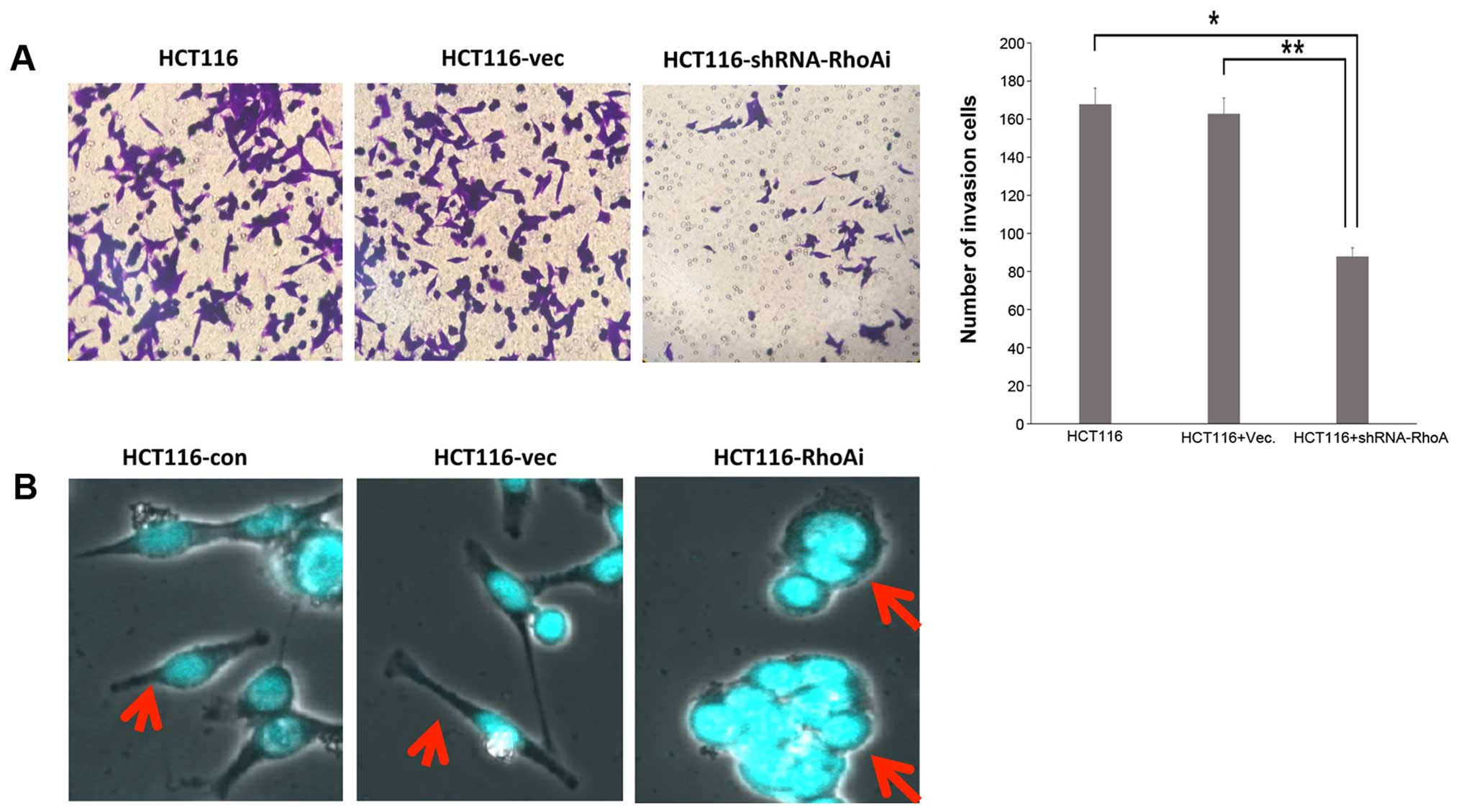
RhoA is required for invasion in HCT116
cells
As previous results showed that Rho is essential for
migration in HCT116 cells, we aimed to determine whether RhoA is
also required for invasion. To achieve this goal, HCT116 cells were
transfected and seeded on a Matrigel-coated insert. Cells were then
incubated for an additional 48 h after seeding, and cells were
imaged to assess cell invasion. HCT116 cells transfected with
shRhoA exhibited significantly less invasion than control cells
(Fig. 4A). Thus, we examined the
morphological characteristics of HCT116 cells following RhoA
knockdown. To assess cell morphology, nuclei of RhoA-depleted cells
were stained with DAPI and analyzed with a confocal microscope.
Vector-transfected or untransfected cells exhibited elongated
morphology with pseudopodia, whereas RhoA-depleted cells were more
rounded with less pseudopodia, and multi-nucleated in ~20% cells
tested (Fig. 4B). These results
indicate that RhoA is essential for cell migration and invasion of
colon cancer cells, two important factors contributing to mortality
caused by colon cancer.
RhoA expression is associated with a poor
prognosis in colorectal cancer
Based on the conclusion that RhoA is essential for
the migration and invasion in the HCT116 cell line, we then
analyzed RhoA expression in patient colorectal cancer samples to
define whether RhoA is relevant in human disease. The samples,
obtained from 150 patients who had undergone surgery, were stained
with RhoA antibody to determine RhoA expression. RhoA protein
stained primarily in the cytoplasm with a wide range of intensity;
RhoA stain intensity was graded from negative to strong expression
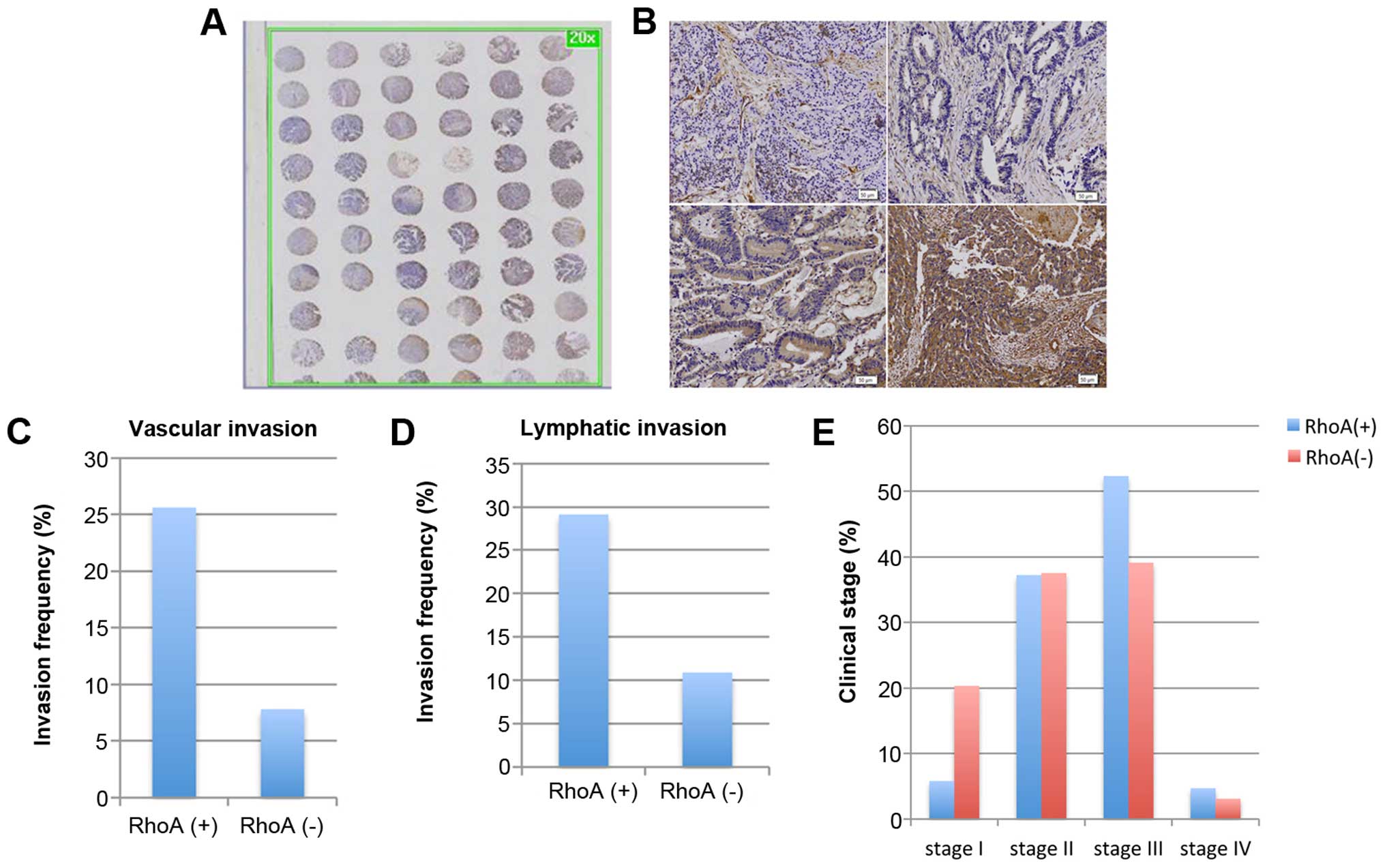
(Fig. 5A–C and Table II). RhoA expression was confirmed
to be positive in 86 cases out of 150 samples (57.3%). We further
correlated RhoA expression with several factors including age,
gender, pN stage, metastasis, and invasion, among others. RhoA
expression was not significantly correlated with age, gender, and
pN stage (Table III). However,
RhoA expression was significantly associated with pT stage,
vascular invasion, lymphatic invasion, and clinical stage (Fig. 5C–E and Table III), which is consistent with
in vitro data demonstrating defective migration and invasion
following RhoA knockdown (Figs. 3
and 4). Specifically, RhoA(+)
samples exhibited an approximately 3- to 4-fold higher invasion
rate of vascular invasion and lymphatic invasion compared to
RhoA(−) samples (vascular invasion, 25.6% of RhoA(+) versus 7.8% of
RhoA(−); lymphatic invasion, 29.1% of RhoA(+) versus 10.9% of
RhoA(−); Fig. 5C and D and
Table III). In addition, RhoA
expression was associated with clinical stages of colorectal
cancer; RhoA(−) specimens associated with lower clinical stages,
while RhoA(+) specimens had higher clinical stages [ clinical stage
I, 20.3% RhoA(−) versus 5.8% RhoA(+); clinical stage III, 39.1% of
RhoA(−) versus 52.3% of RhoA(+); Fig.
5E and Table III]. These
results indicate that, as tumors express more RhoA, the disease
becomes more invasive, progressing to a higher clinical stage.
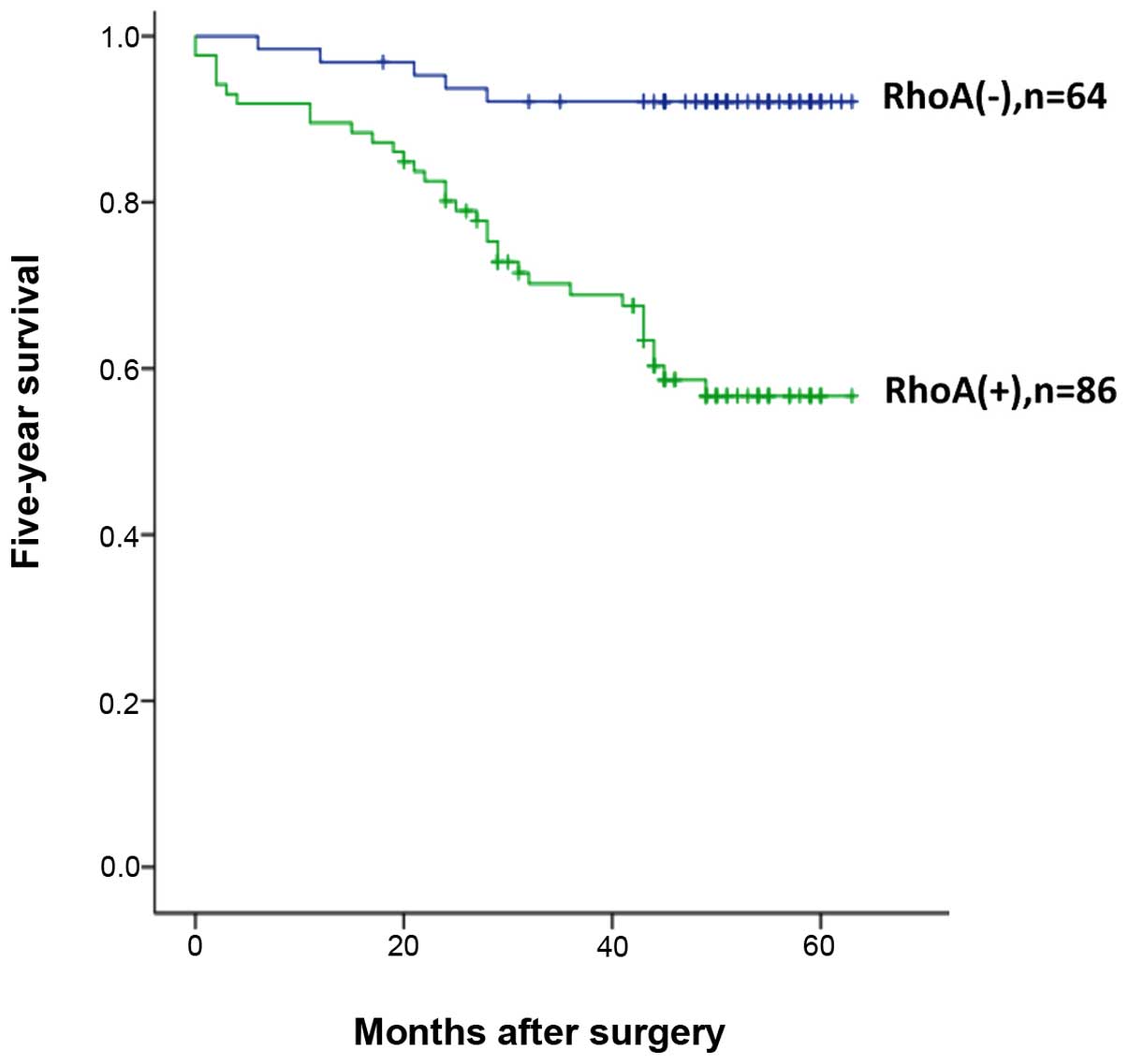
Finally, we correlated RhoA expression with 5-year survival rate.
Using univariate analysis, we found that RhoA expression was
significantly associated with patient survival rate. As shown in
Fig. 6, 92% of RhoA(−) patients
survived whereas only 56% of RhoA(+) patients survived five years
after surgery. These results indicate that RhoA is associated with
invasion and a poor prognosis in colorectal cancer, and could
serves as a promising therapeutic target for cancer therapy.
 | Table IIRhoA expression in colorectal
carcinoma tissue. |
Table II
RhoA expression in colorectal
carcinoma tissue.
| RhoA
expression | |
|---|
|
| |
|---|
| Negative | + | ++ | +++ | Positive rate
(%) |
|---|
| No. of cases | 64 | 45 | 34 | 7 | 57.3 |
 | Table IIIThe association of
clinicopathological features and RhoA expression in colorectal
carcinoma samples. |
Table III
The association of
clinicopathological features and RhoA expression in colorectal
carcinoma samples.
| RhoA | | |
|---|
|
| | |
|---|
| Clinicopathological
factors | Positive
(N=86) | Negative
(N=64) | Total | P-value |
|---|
| Age, years, mean
(SD) | 86 (62.01) | 64 (63.42) | | 0.479 |
| Sex, N (%) | | | | 0.259 |
| M | 50 (58.1) | 43 (67.2) | 93 (62.0) | |
| F | 36 (41.9) | 21 (32.8) | 57 (38.0) | |
| pT stage, N
(%) | | | | 0.027 |
| 1 | 0 (0) | 4 ( 6.3) | 4 (2.7) | |
| 2 | 12 (14.0) | 10 (15.6) | 22 (14.7) | |
| 3 | 61 (70.9) | 47 (73.4) | 108 (72.0) | |
| 4 | 13 (15.1) | 3 (4.7) | 16 (10.6) | |
| pN stage, N
(%) | | | | 0.245 |
| 0 | 39 (45.3) | 37 (57.8) | 76 (50.7) | |
| 1 | 29 (33.7) | 19 (29.7) | 48 (32.0) | |
| 2 | 18 (21.0) | 8 (12.5) | 26 (17.3) | |
| Distal metastasis,
N (%) | | | | 0.637 |
| (−) | 82 (95.3) | 62 (96.9) | 144 (96.0) | |
| (+) | 4 ( 4.7) | 2 (3.1) | 6 (4.0) | |
| Vascular invasion,
N (%) | | | | 0.005 |
| (−) | 64 (74.4) | 59 (92.2) | 123 (82.0) | |
| (+) | 22 (25.6) | 5 (7.8) | 27 (18.0) | |
| Lymphatic invasion,
N (%) | | | | 0.007 |
| (−) | 61 (70.9) | 57 (89.1) | 118 (78.7) | |
| (+) | 25 (29.1) | 7 (10.9) | 32 (21.3) | |
| Clinical stage, N
(%) | | | | 0.045 |
| I | 5 (5.8) | 13 (20.3) | 18 (12.0) | |
| II | 32 (37.2) | 24 (37.5) | 56 (37.3) | |
| III | 45 (52.3) | 25 (39.1) | 70 (46.7) | |
| IV | 4 (4.7) | 2 (3.1) | 6 (4.0) | |
Discussion
Rho GTPases are small proteins that function as
molecular switches in a wide range of systems to transduce signals
upon stimulation of cell surface receptors. Signaling through these
proteins leads to activation of many relevant pathways in cancer,
including cytoskeleton reorganization, proliferation,
differentiation, migration, and invasion.
In this study, we investigated the role of RhoA in
colorectal cancer using human cell lines as well as 150
patient-derived colorectal cancer samples. First, we found that
RhoA is highly expressed in colon cancer cell lines, especially in
a metastatic cell line SW620 compared to a non-metastatic cell line
SW480. To define the functional importance of RhoA, we generated an
shRNA construct against RhoA. We found that, although proliferation
of HCT116 cancer cells is only moderately impaired after RhoA
knockdown, migration and invasion were significantly reduced in
RhoA-depleted HCT116 cells compared to control cells. Furthermore,
RhoA(+) cells from colorectal patient samples were more enriched in
invasion of lymph node and blood vessels. Moreover, patients with
higher expression of RhoA had a significantly poorer 5-year
survival rate when followed up for five years after surgery. These
results demonstrate that RhoA is important in colorectal cancer,
and could be an interesting target for cancer therapeutics.
One of key factors contributing to the mortality in
colorectal cancer is metastatic spread of the disease, which is a
complex, multistage process. The signal transduction pathway
underlying metastasis is not fully known and requires extensive
study (16,17). Thus, it is crucial to study
signaling pathways contributing to invasion and metastasis in
colorectal cancer. In this regard, our results demonstrate that
RhoA is strongly associated with invasion and metastasis in
colorectal cancer. However, the data we show here is contradictory
to recent publications; these publications have shown that lower
RhoA expression is correlated with lymph node metastasis in
colorectal cancer patients, and that RhoA functions as a tumor
suppressor (14,18). Thus, it is important to examine and
understand these apparently discrepant results. One of main
differences is that we examined colorectal patient samples of
different clinical stages whereas Rodrigues et al (14) and Arango et al (18) analyzed only Dukes' stage C. Despite
these contradictory findings, many publications indicate that RhoA
does play a role in tumor growth and invasion of many types of
cancers (10,13,19,20).
In addition, RhoA-depleted cancer cells exhibit less proliferation
and smaller tumor sizes in vitro and in vivo
(21). Furthermore, RhoA is more
expressed in tumor samples compared to normal tissues (10,12,13).
Overall, it seems that RhoA functions either as a tumor suppressor
or activator in a context-dependent manner. As such, it is
essential that cancer therapies targeting RhoA should be approached
carefully.
In conclusion, our data provide clear evidence
implicating RhoA in migration, invasion, and poor prognosis in
colorectal cancer. Therefore, this signaling pathway could
ultimately serve as a promising target for the treatment and
prevention of colorectal cancer metastasis.
Acknowledgements
This study was supported by the Soonchunhyang
University Research Fund and a grant of the Korea Health Technology
R&D Project through the Korea Health Industry Development
Institute (KHID), funded by the Ministry of Health & Welfare,
Republic of Korea (grant no. HI15C1647).
References
|
1
|
Jung KW, Won YJ, Kong HJ, Oh CM, Seo HG
and Lee JS: Cancer statistics in Korea: Incidence, mortality,
survival and prevalence in 2010. Cancer Res Treat. 45:1–14. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wilke HJ and Van Cutsem E: Current
treatments and future perspectives in colorectal and gastric
cancer. Ann Oncol. 14(Suppl 2): pp. ii49–ii55. 2003, View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chan KM, Wu TH, Cheng CH, Lee WC, Chiang
JM, Chen JS and Wang JY: Prognostic significance of the number of
tumors and aggressive surgical approach in colorectal cancer
hepatic metastasis. World J Surg Oncol. 12:1552014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Swiderska M, Choromańska B, Dąbrowska E,
Konarzewska-Duchnowska E, Choromańska K, Szczurko G, Myśliwiec P,
Dadan J, Ladny JR and Zwierz K: The diagnostics of colorectal
cancer. Contemp Oncol (Pozn). 18:1–6. 2014.
|
|
5
|
Bai Y, Xiang X, Liang C and Shi L:
Regulating Rac in the nervous system: Molecular function and
disease implication of Rac GEFs and GAPs. BioMed Res Int.
2015:6324502015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Stankiewicz TR and Linseman DA: Rho family
GTPases: Key players in neuronal development, neuronal survival,
and neuro-degeneration. Front Cell Neurosci. 8:3142014. View Article : Google Scholar
|
|
7
|
Karlsson R, Pedersen ED, Wang Z and
Brakebusch C: Rho GTPase function in tumorigenesis. Biochim Biophys
Acta. 1796:91–98. 2009.PubMed/NCBI
|
|
8
|
Pai SY, Kim C and Williams DA: Rac GTPases
in human diseases. Dis Markers. 29:177–187. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jaffe AB and Hall A: Rho GTPases:
Biochemistry and biology. Annu Rev Cell Dev Biol. 21:247–269. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang H, Zhao G, Liu X, Sui A, Yang K, Yao
R, Wang Z and Shi Q: Silencing of RhoA and RhoC expression by RNA
interference suppresses human colorectal carcinoma growth in vivo.
J Exp Clin Cancer Res. 29:1232010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fan YM, Pang CP, Harvey AR and Cui Q:
Marked effect of RhoA-specific shRNA-producing plasmids on neurite
growth in PC12 cells. Neurosci Lett. 440:170–175. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu N, Bi F, Pan YL, Xue Y, Zhang X, Shi
YQ, Zhang YM, Du JP and Fan DM: The expression and possible
function of RhoA in human gastric cancer cell lines. Zhonghua Zhong
Liu Za Zhi. 26:26–29. 2004.(In Chinese). PubMed/NCBI
|
|
13
|
Takami Y, Higashi M, Kumagai S, Kuo PC,
Kawana H, Koda K, Miyazaki M and Harigaya K: The activity of RhoA
is correlated with lymph node metastasis in human colorectal
cancer. Dig Dis Sci. 53:467–473. 2008. View Article : Google Scholar
|
|
14
|
Rodrigues P, Macaya I, Bazzocco S,
Mazzolini R, Andretta E, Dopeso H, Mateo-Lozano S, Bilić J,
Cartón-García F, Nieto R, et al: RHOA inactivation enhances Wnt
signalling and promotes colorectal cancer. Nat Commun. 5:54582014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kodiha M, Umar R and Stochaj U: Optimized
immunofluorescence staining protocol to detect the nucleoporin
Nup98 in different subcellular compartments protocol exchage.
Protocol Exchange. 22–Jan;2009.Epub ahead of print. View Article : Google Scholar
|
|
16
|
Chambers AF, Groom AC and MacDonald IC:
Dissemination and growth of cancer cells in metastatic sites. Nat
Rev Cancer. 2:563–572. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lee JJ and Lotze MT: Molecular basis of
metastasis. N Engl J Med. 360:1679author reply 1679–1680.
2009.PubMed/NCBI
|
|
18
|
Arango D, Laiho P, Kokko A, Alhopuro P,
Sammalkorpi H, Salovaara R, Nicorici D, Hautaniemi S, Alazzouzi H,
Mecklin JP, et al: Gene-expression profiling predicts recurrence in
Dukes' C colorectal cancer. Gastroenterology. 129:874–884. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sun K, Duan X, Cai H, Liu X, Yang Y, Li M,
Zhang X and Wang J: Curcumin inhibits LPA-induced invasion by
attenuating RhoA/ROCK/MMPs pathway in MCF7 breast cancer cells.
Clin Exp Med. Jan 18–2015.Epub ahead of print. View Article : Google Scholar
|
|
20
|
Menhofer MH, Kubisch R, Schreiner L, Zorn
M, Foerster F, Mueller R, Raedler JO, Wagner E, Vollmar AM and
Zahler S: The actin targeting compound Chondramide inhibits breast
cancer metastasis via reduction of cellular contractility. PLoS
One. 9:e1125422014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang X, Zheng F, Zhang S and Lu J: Loss of
RhoA expression prevents proliferation and metastasis of SPCA1 lung
cancer cells in vitro. Biomed Pharmacother. 69:361–366. 2015.
View Article : Google Scholar : PubMed/NCBI
|