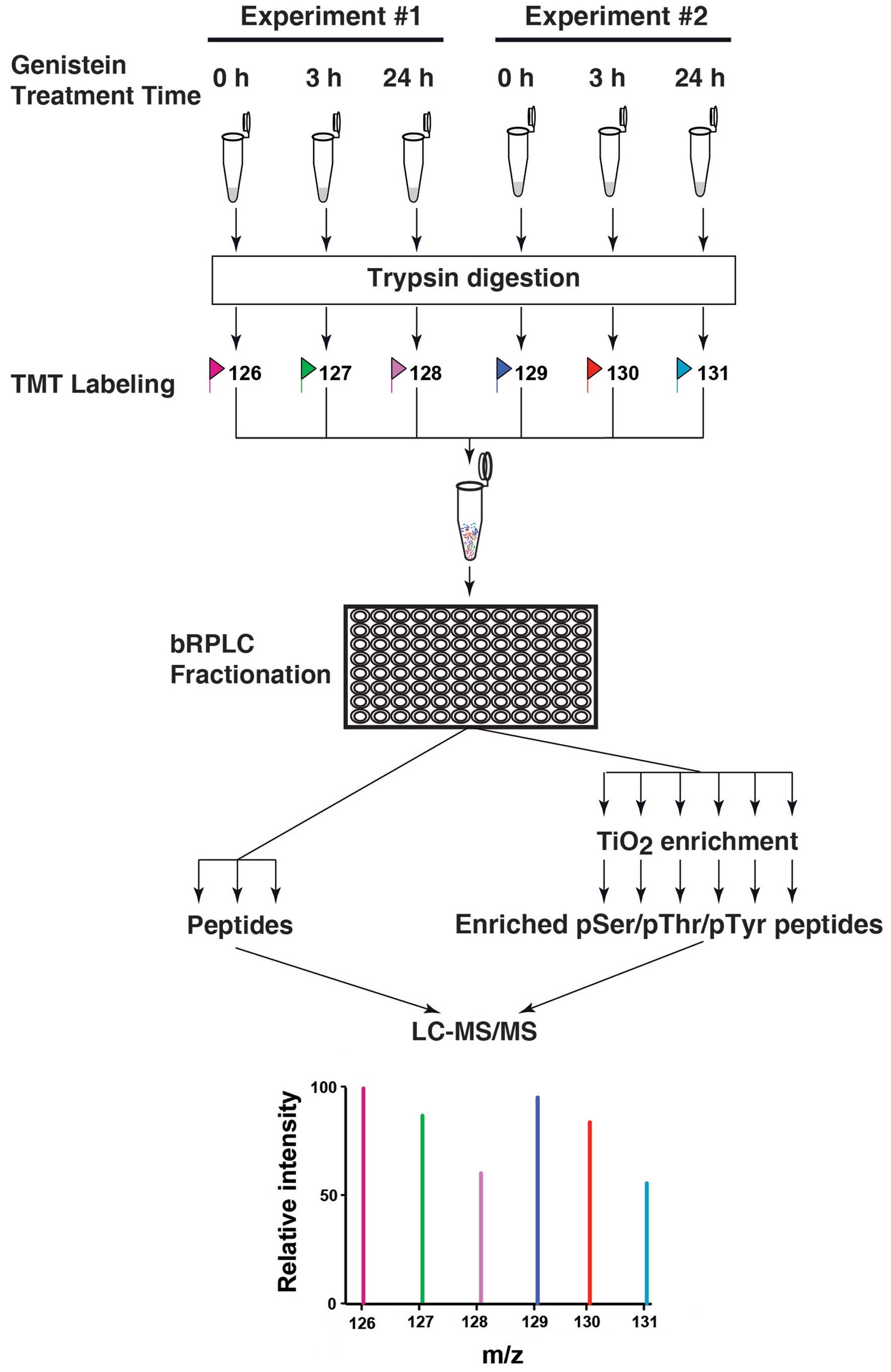
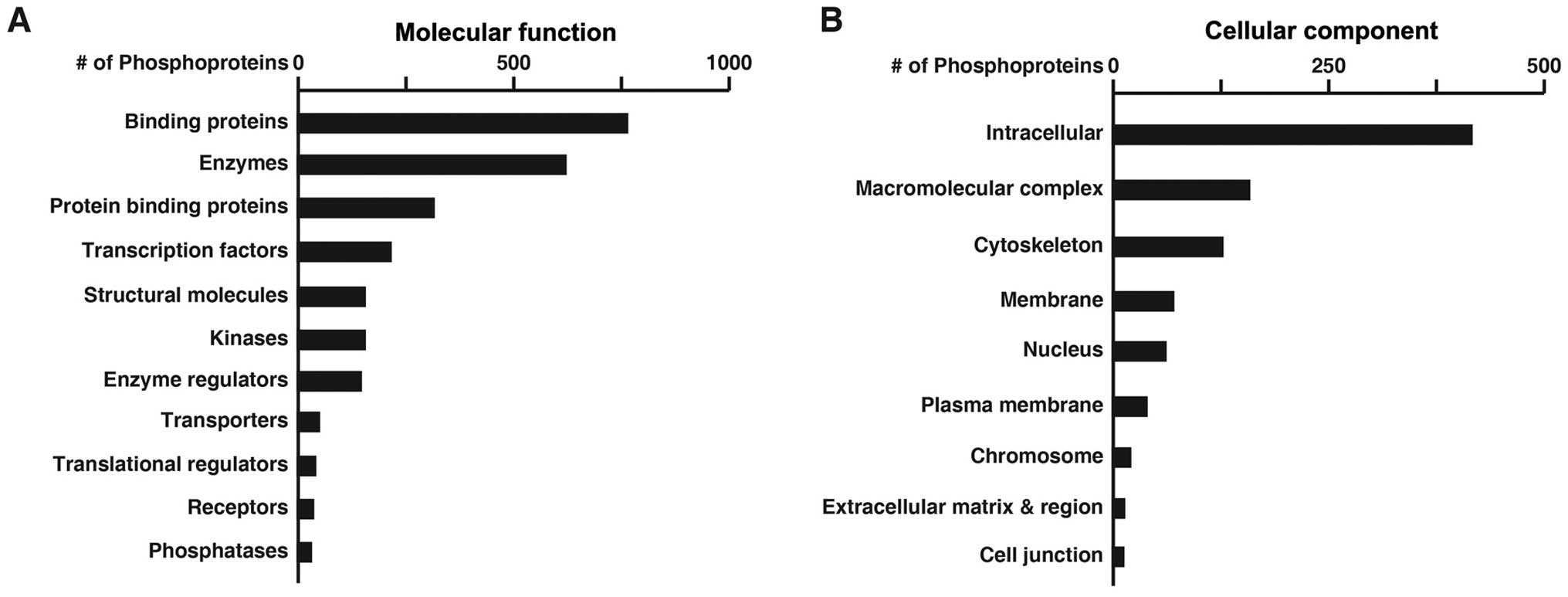
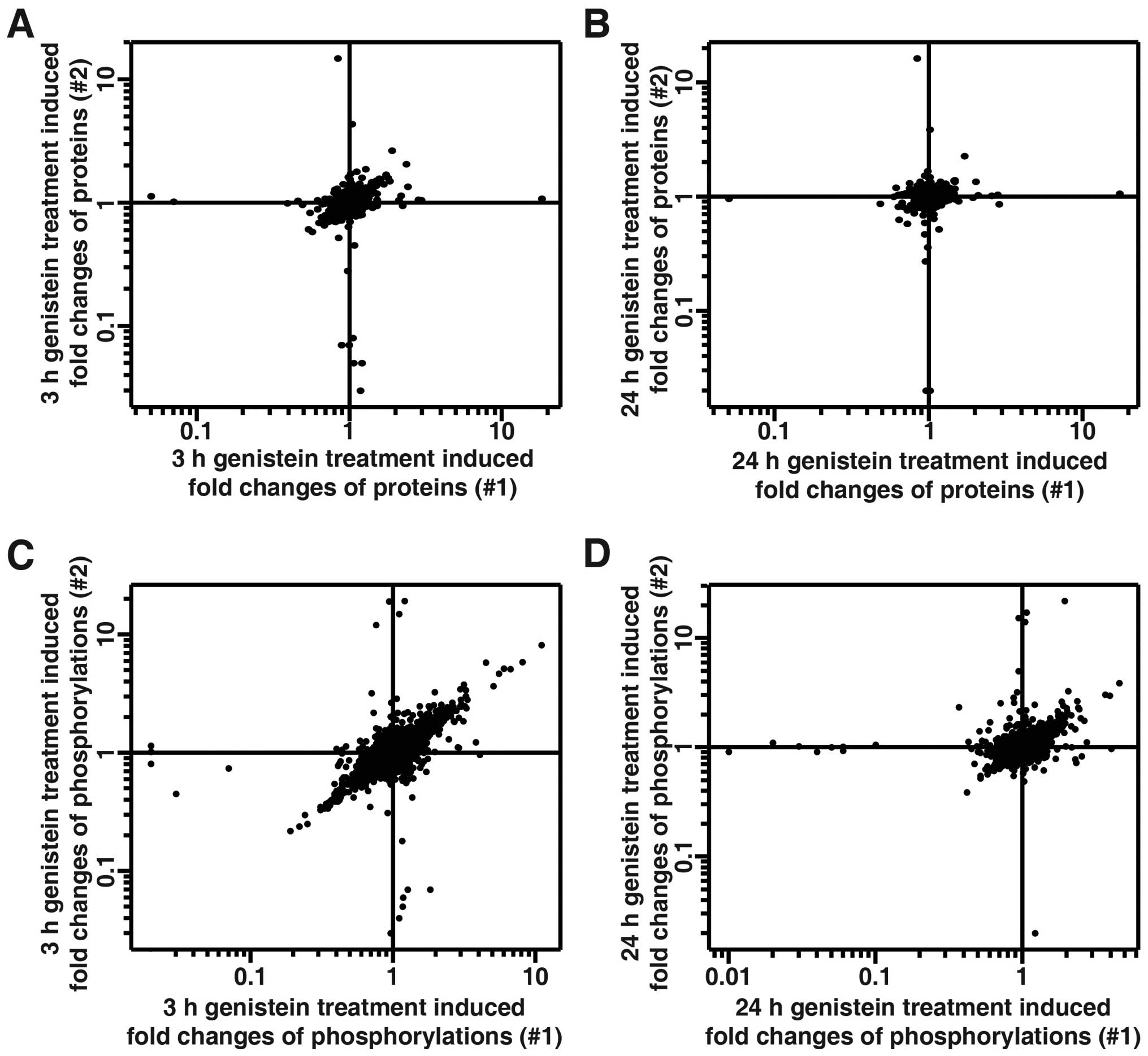
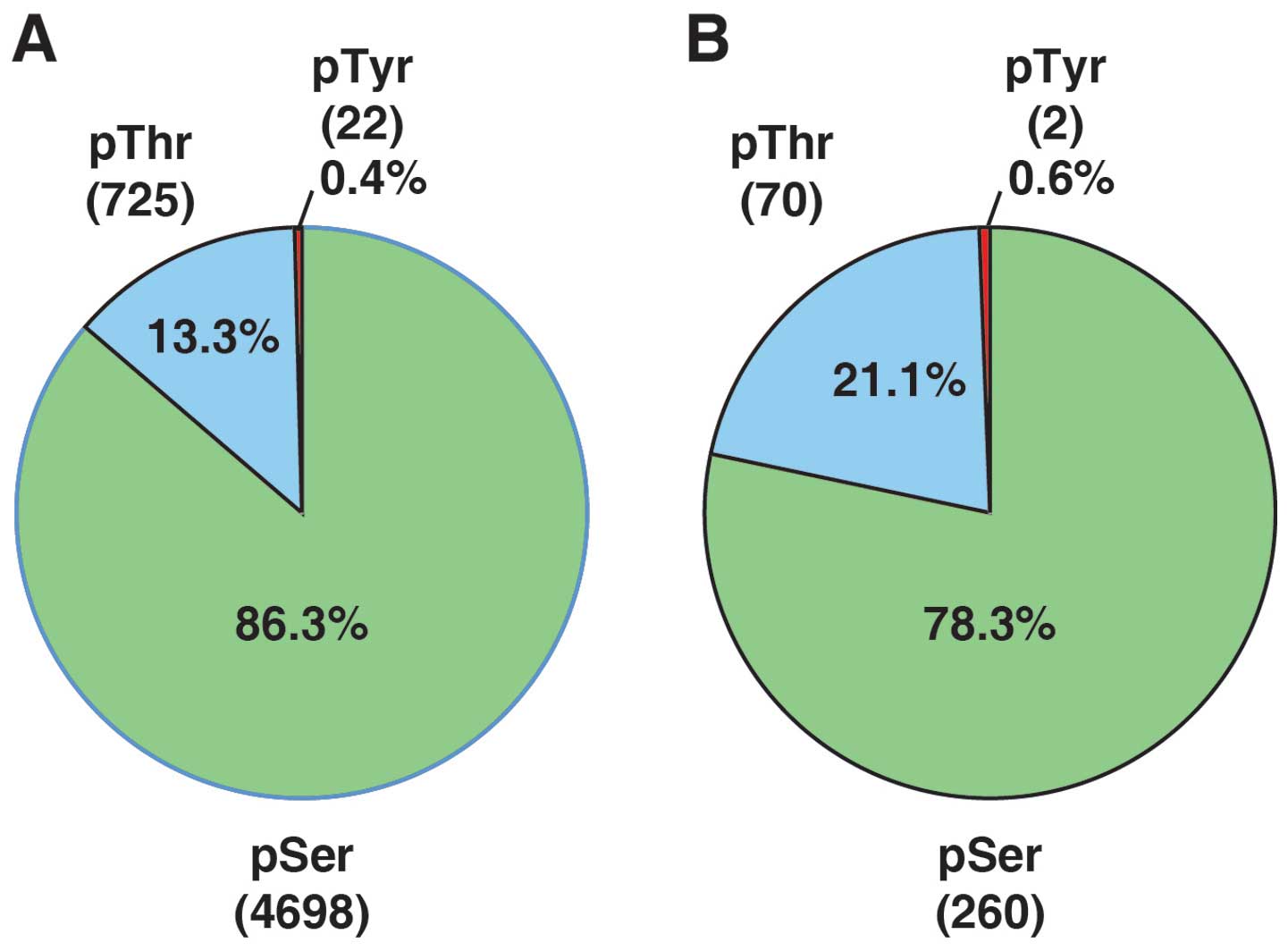
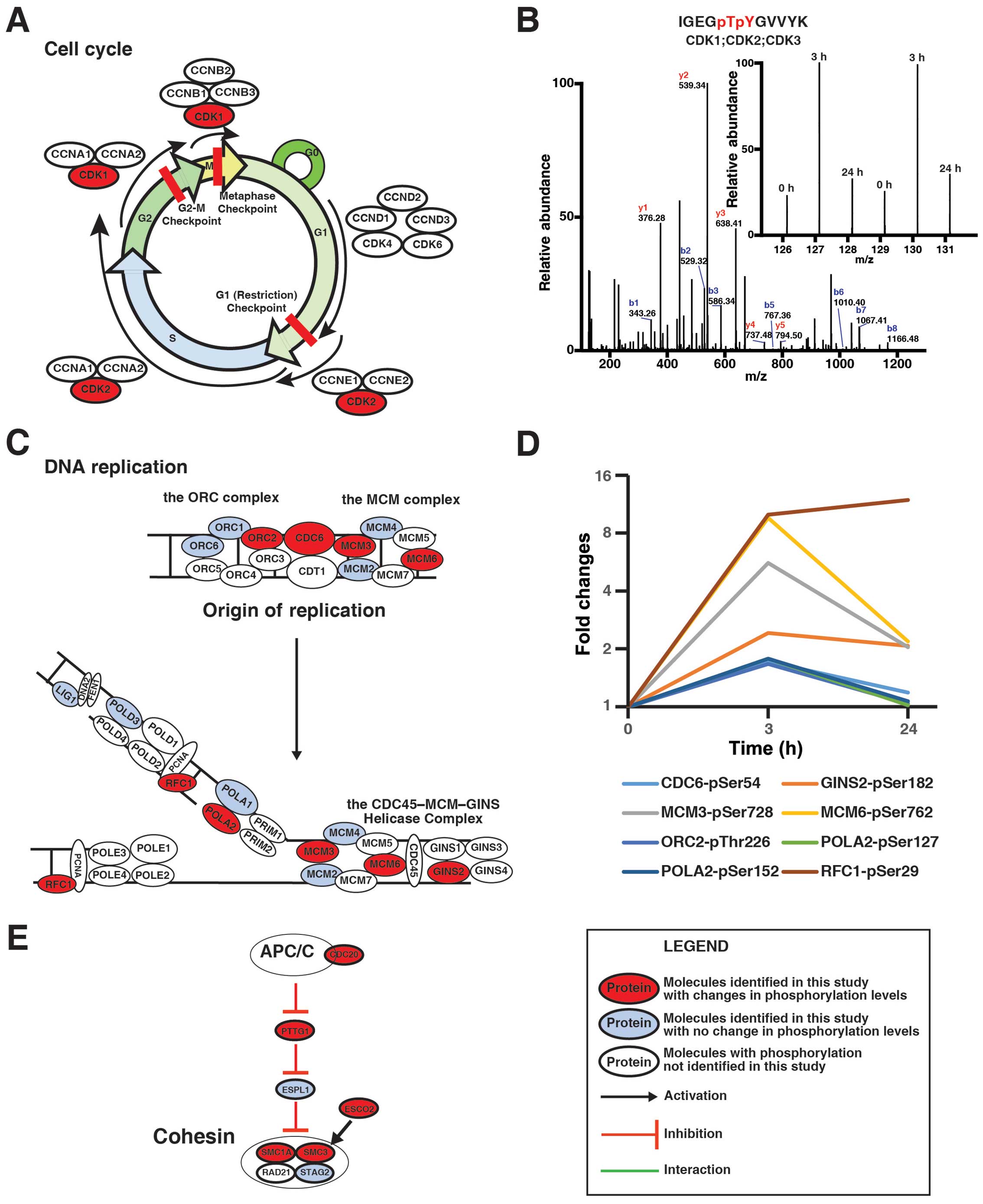
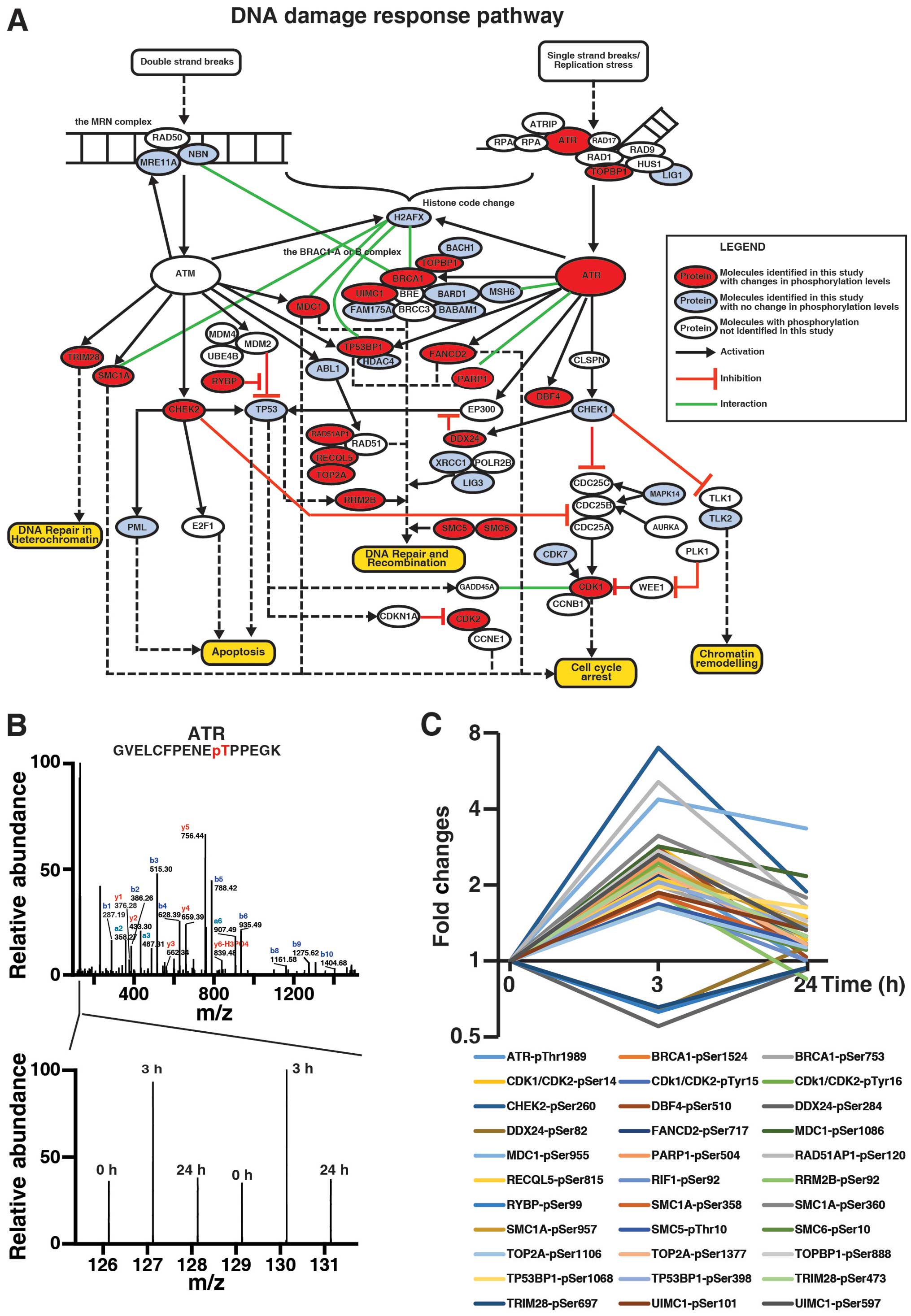
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Reis-Filho JS and Pusztai L: Gene
expression profiling in breast cancer: Classification,
prognostication, and prediction. Lancet. 378:1812–1823. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Perou CM and Børresen-Dale AL: Systems
biology and genomics of breast cancer. Cold Spring Harb Perspect
Biol. 3:a0032932011. View Article : Google Scholar
|
|
4
|
Adlercreutz CH, Goldin BR, Gorbach SL,
Höckerstedt KA, Watanabe S, Hämäläinen EK, Markkanen MH, Mäkelä TH,
Wähälä KT and Adlercreutz T: Soybean phytoestrogen intake and
cancer risk. J Nutr. 125(Suppl): S757–S770. 1995.
|
|
5
|
Messina MJ, Persky V, Setchell KD and
Barnes S: Soy intake and cancer risk: A review of the in vitro and
in vivo data. Nutr Cancer. 21:113–131. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nechuta SJ, Caan BJ, Chen WY, Lu W, Chen
Z, Kwan ML, Flatt SW, Zheng Y, Zheng W, Pierce JP, et al: Soy food
intake after diagnosis of breast cancer and survival: An in-depth
analysis of combined evidence from cohort studies of US and Chinese
women. Am J Clin Nutr. 96:123–132. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fioravanti L, Cappelletti V, Miodini P,
Ronchi E, Brivio M and Di Fronzo G: Genistein in the control of
breast cancer cell growth: Insights into the mechanism of action in
vitro. Cancer Lett. 130:143–152. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li Y, Upadhyay S, Bhuiyan M and Sarkar FH:
Induction of apoptosis in breast cancer cells MDA-MB-231 by
genistein. Oncogene. 18:3166–3172. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cappelletti V, Fioravanti L, Miodini P and
Di Fronzo G: Genistein blocks breast cancer cells in the G(2)M
phase of the cell cycle. J Cell Biochem. 79:594–600. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Horia E and Watkins BA: Complementary
actions of docosahexaenoic acid and genistein on COX-2, PGE2 and
invasiveness in MDA-MB-231 breast cancer cells. Carcinogenesis.
28:809–815. 2007. View Article : Google Scholar
|
|
11
|
Li Z, Li J, Mo B, Hu C, Liu H, Qi H, Wang
X and Xu J: Genistein induces G2/M cell cycle arrest via stable
activation of ERK1/2 pathway in MDA-MB-231 breast cancer cells.
Cell Biol Toxicol. 24:401–409. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li Z, Li J, Mo B, Hu C, Liu H, Qi H, Wang
X and Xu J: Genistein induces cell apoptosis in MDA-MB-231 breast
cancer cells via the mitogen-activated protein kinase pathway.
Toxicol In Vitro. 22:1749–1753. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gong L, Li Y, Nedeljkovic-Kurepa A and
Sarkar FH: Inactivation of NF-kappaB by genistein is mediated via
Akt signaling pathway in breast cancer cells. Oncogene.
22:4702–4709. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Thompson A, Schäfer J, Kuhn K, Kienle S,
Schwarz J, Schmidt G, Neumann T, Johnstone R, Mohammed AK and Hamon
C: Tandem mass tags: A novel quantification strategy for
comparative analysis of complex protein mixtures by MS/MS. Anal
Chem. 75:1895–1904. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ross PL, Huang YN, Marchese JN, Williamson
B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, et
al: Multiplexed protein quantitation in Saccharomyces cerevisiae
using amine-reactive isobaric tagging reagents. Mol Cell
Proteomics. 3:1154–1169. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nirujogi RS, Wright JD Jr, Manda SS, Zhong
J, Na CH, Meyerhoff J, Benton B, Jabbour R, Willis K, Kim MS, et
al: Phosphoproteomic analysis reveals compensatory effects in the
piriform cortex of VX nerve agent exposed rats. Proteomics.
15:487–499. 2015. View Article : Google Scholar
|
|
17
|
Roitinger E, Hofer M, Köcher T, Pichler P,
Novatchkova M, Yang J, Schlögelhofer P and Mechtler K: Quantitative
phosphoproteomics of the ataxia telangiectasia-mutated (ATM) and
ataxia telangiectasia-mutated and rad3-related (ATR) dependent DNA
damage response in Arabidopsis thaliana. Mol Cell Proteomics.
14:556–571. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wiśniewski JR, Zougman A, Nagaraj N and
Mann M: Universal sample preparation method for proteome analysis.
Nat Methods. 6:359–362. 2009. View Article : Google Scholar
|
|
19
|
Larsen MR, Thingholm TE, Jensen ON,
Roepstorff P and Jørgensen TJ: Highly selective enrichment of
phosphorylated peptides from peptide mixtures using titanium
dioxide micro-columns. Mol Cell Proteomics. 4:873–886. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rappsilber J, Ishihama Y and Mann M: Stop
and go extraction tips for matrix-assisted laser
desorption/ionization, nanoelectrospray, and LC/MS sample
pretreatment in proteomics. Anal Chem. 75:663–670. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cox J, Neuhauser N, Michalski A, Scheltema
RA, Olsen JV and Mann M: Andromeda: A peptide search engine
integrated into the MaxQuant environment. J Proteome Res.
10:1794–1805. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cox J, Matic I, Hilger M, Nagaraj N,
Selbach M, Olsen JV and Mann M: A practical guide to the MaxQuant
computational platform for SILAC-based quantitative proteomics. Nat
Protoc. 4:698–705. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cox J and Mann M: MaxQuant enables high
peptide identification rates, individualized p.p.b.-range mass
accuracies and proteome-wide protein quantification. Nat
Biotechnol. 26:1367–1372. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Vizcaíno JA, Deutsch EW, Wang R, Csordas
A, Reisinger F, Ríos D, Dianes JA, Sun Z, Farrah T, Bandeira N, et
al: ProteomeXchange provides globally coordinated proteomics data
submission and dissemination. Nat Biotechnol. 32:223–226. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mi H, Muruganujan A and Thomas PD: PANTHER
in 2013: Modeling the evolution of gene function, and other gene
attributes, in the context of phylogenetic trees. Nucleic Acids
Res. 41:D377–D386. 2013. View Article : Google Scholar :
|
|
26
|
Mi H, Muruganujan A, Casagrande JT and
Thomas PD: Large-scale gene function analysis with the PANTHER
classification system. Nat Protoc. 8:1551–1566. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Huang W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar :
|
|
28
|
Huang W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar
|
|
29
|
Pan H, Zhou W, He W, Liu X, Ding Q, Ling
L, Zha X and Wang S: Genistein inhibits MDA-MB-231 triple-negative
breast cancer cell growth by inhibiting NF-κB activity via the
Notch-1 pathway. Int J Mol Med. 30:337–343. 2012.PubMed/NCBI
|
|
30
|
Wang Z, Liang S, Lian X, Liu L, Zhao S,
Xuan Q, Guo L, Liu H, Yang Y, Dong T, et al: Identification of
proteins responsible for adriamycin resistance in breast cancer
cells using proteomics analysis. Sci Rep. 5:93012015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu S, Shiotani B, Lahiri M, Maréchal A,
Tse A, Leung CC, Glover JN, Yang XH and Zou L: ATR
autophosphorylation as a molecular switch for checkpoint
activation. Mol Cell. 43:192–202. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Vauzour D, Vafeiadou K, Rice-Evans C,
Cadenas E and Spencer JP: Inhibition of cellular proliferation by
the genistein metabolite 5,7,3′,4′-tetrahydroxyisoflavone is
mediated by DNA damage and activation of the ATR signalling
pathway. Arch Biochem Biophys. 468:159–166. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tuul M, Kitao H, Iimori M, Matsuoka K,
Kiyonari S, Saeki H, Oki E, Morita M and Maehara Y: Rad9, Rad17,
TopBP1 and claspin play essential roles in heat-induced activation
of ATR kinase and heat tolerance. PLoS One. 8:e553612013.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kumagai A, Lee J, Yoo HY and Dunphy WG:
TopBP1 activates the ATR-ATRIP complex. Cell. 124:943–955. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Foulkes WD, Smith IE and Reis-Filho JS:
Triple-negative breast cancer. N Engl J Med. 363:1938–1948. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tichy JR, Deal AM, Anders CK, Reeder-Hayes
K and Carey LA: Race, response to chemotherapy, and outcome within
clinical breast cancer subtypes. Breast Cancer Res Treat.
150:667–674. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Rajah TT, Peine KJ, Du N, Serret CA and
Drews NR: Physiological concentrations of genistein and
17β-estradiol inhibit MDA-MB-231 breast cancer cell growth by
increasing BAX/BCL-2 and reducing pERK1/2. Anticancer Res.
32:1181–1191. 2012.PubMed/NCBI
|
|
38
|
Santell RC, Kieu N and Helferich WG:
Genistein inhibits growth of estrogen-independent human breast
cancer cells in culture but not in athymic mice. J Nutr.
130:1665–1669. 2000.PubMed/NCBI
|
|
39
|
Krek W and Nigg EA: Differential
phosphorylation of vertebrate p34cdc2 kinase at the G1/S and G2/M
transitions of the cell cycle: Identification of major
phosphorylation sites. EMBO J. 10:305–316. 1991.PubMed/NCBI
|
|
40
|
Hagstrom KA and Meyer BJ: Condensin and
cohesin: More than chromosome compactor and glue. Nat Rev Genet.
4:520–534. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tang Z, Shu H, Oncel D, Chen S and Yu H:
Phosphorylation of Cdc20 by Bub1 provides a catalytic mechanism for
APC/C inhibition by the spindle checkpoint. Mol Cell. 16:387–397.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Whelan G, Kreidl E, Wutz G, Egner A,
Peters JM and Eichele G: Cohesin acetyltransferase Esco2 is a cell
viability factor and is required for cohesion in pericentric
heterochromatin. EMBO J. 31:71–82. 2012. View Article : Google Scholar :
|
|
43
|
Vega H, Waisfisz Q, Gordillo M, Sakai N,
Yanagihara I, Yamada M, van Gosliga D, Kayserili H, Xu C, Ozono K,
et al: Roberts syndrome is caused by mutations in ESCO2, a human
homolog of yeast ECO1 that is essential for the establishment of
sister chromatid cohesion. Nat Genet. 37:468–470. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Maiato H, Fairley EA, Rieder CL, Swedlow
JR, Sunkel CE and Earnshaw WC: Human CLASP1 is an outer kinetochore
component that regulates spindle microtubule dynamics. Cell.
113:891–904. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Maia AR, Garcia Z, Kabeche L, Barisic M,
Maffini S, Macedo-Ribeiro S, Cheeseman IM, Compton DA, Kaverina I
and Maiato H: Cdk1 and Plk1 mediate a CLASP2 phospho-switch that
stabilizes kinetochore-microtubule attachments. J Cell Biol.
199:285–301. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Liao H, Winkfein RJ, Mack G, Rattner JB
and Yen TJ: CENP-F is a protein of the nuclear matrix that
assembles onto kinetochores at late G2 and is rapidly degraded
after mitosis. J Cell Biol. 130:507–518. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Tadeu AM, Ribeiro S, Johnston J, Goldberg
I, Gerloff D and Earnshaw WC: CENP-V is required for centromere
organization, chromosome alignment and cytokinesis. EMBO J.
27:2510–2522. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lawo S, Bashkurov M, Mullin M, Ferreria
MG, Kittler R, Habermann B, Tagliaferro A, Poser I, Hutchins JR,
Hegemann B, et al: HAUS, the 8-subunit human Augmin complex,
regulates centrosome and spindle integrity. Curr Biol. 19:816–826.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Maddox PS, Hyndman F, Monen J, Oegema K
and Desai A: Functional genomics identifies a Myb domain-containing
protein family required for assembly of CENP-A chromatin. J Cell
Biol. 176:757–763. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Dunleavy EM, Roche D, Tagami H, Lacoste N,
Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y and
Almouzni-Pettinotti G: HJURP is a cell-cycle-dependent maintenance
and deposition factor of CENP-A at centromeres. Cell. 137:485–497.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Barnhart MC, Kuich PH, Stellfox ME, Ward
JA, Bassett EA, Black BE and Foltz DR: HJURP is a CENP-A chromatin
assembly factor sufficient to form a functional de novo
kinetochore. J Cell Biol. 194:229–243. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Barisic M, Sohm B, Mikolcevic P, Wandke C,
Rauch V, Ringer T, Hess M, Bonn G and Geley S: Spindly/CCDC99 is
required for efficient chromosome congression and mitotic
checkpoint regulation. Mol Biol Cell. 21:1968–1981. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Maréchal A and Zou L: DNA damage sensing
by the ATM and ATR kinases. Cold Spring Harb Perspect Biol.
5:52013. View Article : Google Scholar
|
|
54
|
Liu X, Sun C, Jin X, Li P, Ye F, Zhao T,
Gong L and Li Q: Genistein enhances the radiosensitivity of breast
cancer cells via G(2)/M cell cycle arrest and apoptosis. Molecules.
18:13200–13217. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Narod SA and Foulkes WD: BRCA1 and BRCA2:
1994 and beyond. Nat Rev Cancer. 4:665–676. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Venkitaraman AR: Cancer susceptibility and
the functions of BRCA1 and BRCA2. Cell. 108:171–182. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Wang B: BRCA1 tumor suppressor network:
Focusing on its tail. Cell Biosci. 2:62012. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wang B, Hurov K, Hofmann K and Elledge SJ:
NBA1, a new player in the Brca1 A complex, is required for DNA
damage resistance and checkpoint control. Genes Dev. 23:729–739.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Greenberg RA, Sobhian B, Pathania S,
Cantor SB, Nakatani Y and Livingston DM: Multifactorial
contributions to an acute DNA damage response by
BRCA1/BARD1-containing complexes. Genes Dev. 20:34–46. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Yu X and Chen J: DNA damage-induced cell
cycle checkpoint control requires CtIP, a phosphorylation-dependent
binding partner of BRCA1 C-terminal domains. Mol Cell Biol.
24:9478–9486. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Xu B, O'Donnell AH, Kim ST and Kastan MB:
Phosphorylation of serine 1387 in Brca1 is specifically required
for the Atm-mediated S-phase checkpoint after ionizing irradiation.
Cancer Res. 62:4588–4591. 2002.PubMed/NCBI
|
|
62
|
Cortez D, Wang Y, Qin J and Elledge SJ:
Requirement of ATM-dependent phosphorylation of brca1 in the DNA
damage response to double-strand breaks. Science. 286:1162–1166.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Kim H, Chen J and Yu X: Ubiquitin-binding
protein RAP80 mediates BRCA1-dependent DNA damage response.
Science. 316:1202–1205. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Sobhian B, Shao G, Lilli DR, Culhane AC,
Moreau LA, Xia B, Livingston DM and Greenberg RA: RAP80 targets
BRCA1 to specific ubiquitin structures at DNA damage sites.
Science. 316:1198–1202. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Ilves I, Tamberg N and Botchan MR:
Checkpoint kinase 2 (Chk2) inhibits the activity of the
Cdc45/MCM2-7/GINS (CMG) replicative helicase complex. Proc Natl
Acad Sci USA. 109:13163–13170. 2012. View Article : Google Scholar : PubMed/NCBI
|