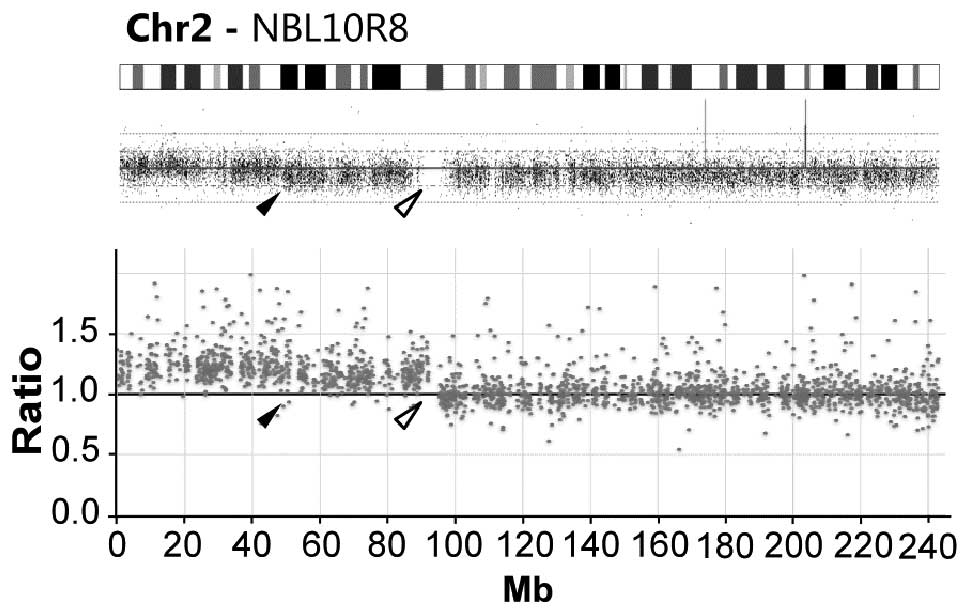
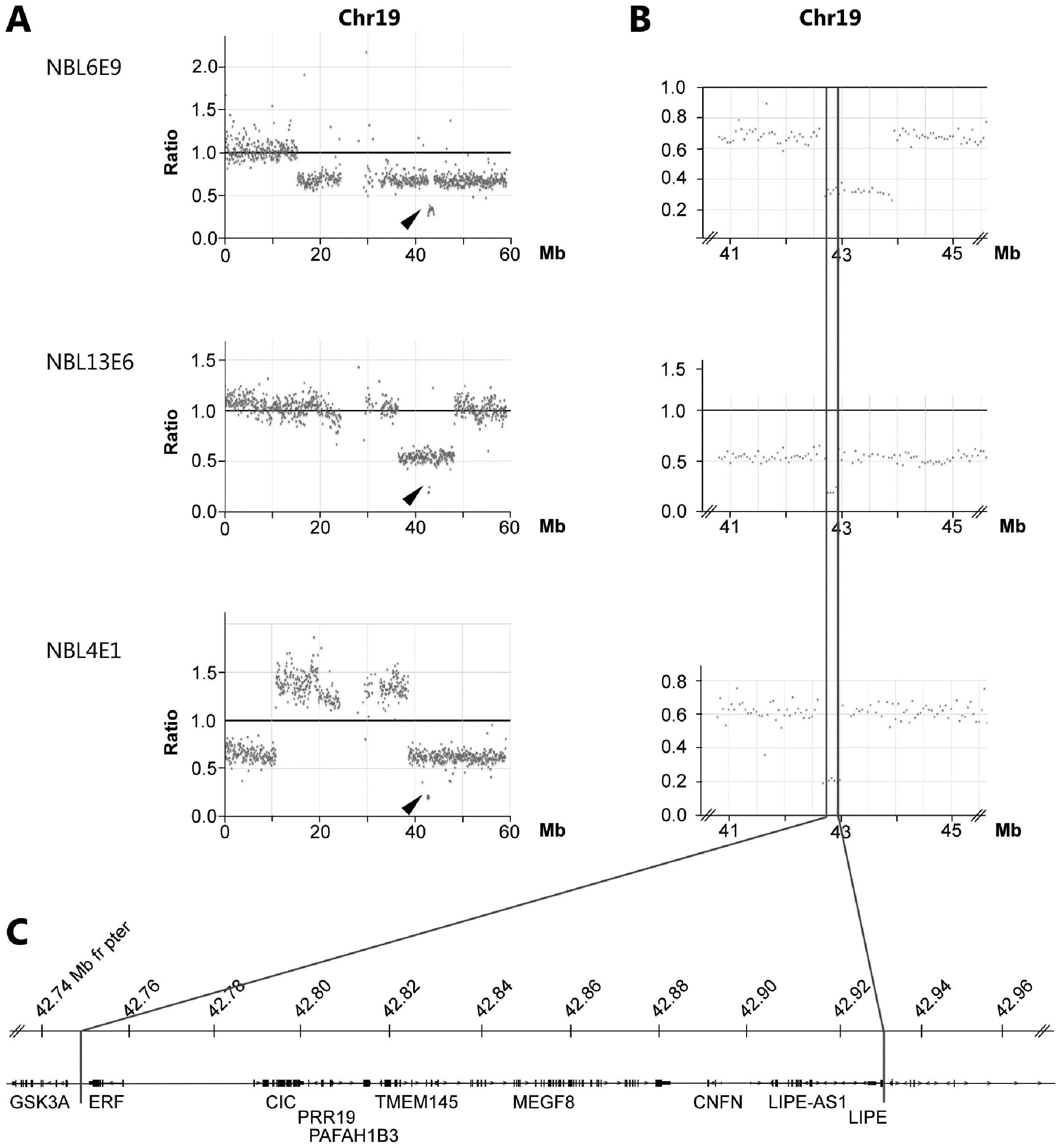
|
1
|
Cohn SL, Pearson AD, London WB, Monclair
T, Ambros PF, Brodeur GM, Faldum A, Hero B, Iehara T, Machin D, et
al: INRG Task Force: The International Neuroblastoma Risk Group
(INRG) classification system: An INRG Task Force report. J Clin
Oncol. 27:289–297. 2009. View Article : Google Scholar :
|
|
2
|
Carén H, Kryh H, Nethander M, Sjöberg RM,
Träger C, Nilsson S, Abrahamsson J, Kogner P and Martinsson T:
High-risk neuroblastoma tumors with 11q-deletion display a poor
prognostic, chromosome instability phenotype with later onset. Proc
Natl Acad Sci USA. 107:4323–4328. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Janoueix-Lerosey I, Schleiermacher G,
Michels E, Mosseri V, Ribeiro A, Lequin D, Vermeulen J, Couturier
J, Peuchmaur M, Valent A, et al: Overall genomic pattern is a
predictor of outcome in neuroblastoma. J Clin Oncol. 27:1026–1033.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cetinkaya C, Martinsson T, Sandgren J,
Träger C, Kogner P, Dumanski J, Díaz de Ståhl T and Hedborg F: Age
dependence of tumor genetics in unfavorable neuroblastoma: arrayCGH
profiles of 34 consecutive cases, using a Swedish 25-year
neuroblastoma cohort for validation. BMC Cancer. 13:2312013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Combaret V, Iacono I, Bréjon S,
Schleiermacher G, Pierron G, Couturier J, Bergeron C and Blay JY:
Analysis of genomic alterations in neuroblastoma by multiplex
ligation-dependent probe amplification and array comparative
genomic hybridization: A comparison of results. Cancer Genet.
205:657–664. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ambros IM, Brunner B, Aigner G, Bedwell C,
Beiske K, Bénard J, Bown N, Combaret V, Couturier J, Defferrari R,
et al: A multilocus technique for risk evaluation of patients with
neuroblastoma. Clin Cancer Res. 17:792–804. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sathirapongsasuti JF, Lee H, Horst BA,
Brunner G, Cochran AJ, Binder S, Quackenbush J and Nelson SF: Exome
sequencing-based copy-number variation and loss of heterozygosity
detection: Exome CNV. Bioinformatics. 27:2648–2654. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Krumm N, Sudmant PH, Ko A, O'Roak BJ,
Malig M, Coe BP, Quinlan AR, Nickerson DA and Eichler EE: NHLBI
Exome Sequencing Project: Copy number variation detection and
genotyping from exome sequence data. Genome Res. 22:1525–1532.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Valdés-Mas R, Bea S, Puente DA, López-Otín
C and Puente XS: Estimation of copy number alterations from exome
sequencing data. PLoS One. 7:e514222012. View Article : Google Scholar
|
|
10
|
Boeva V, Popova T, Bleakley K, Chiche P,
Cappo J, Schleiermacher G, Janoueix-Lerosey I, Delattre O and
Barillot E: Control-FREEC: A tool for assessing copy number and
allelic content using next-generation sequencing data.
Bioinformatics. 28:423–425. 2012. View Article : Google Scholar :
|
|
11
|
Carén H, Erichsen J, Olsson L, Enerbäck C,
Sjöberg RM, Abrahamsson J, Kogner P and Martinsson T:
High-resolution array copy number analyses for detection of
deletion, gain, amplification and copy-neutral LOH in primary
neuroblastoma tumors: Four cases of homozygous deletions of the
CDKN2A gene. BMC Genomics. 9:3532008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chang WCJ, Allaire JJ, Xie Y and McPehrson
J: Shiny: Web Application Framework for R. R package version
0.12.0. R package version 0120. 2015, https://cran.r-project.org/web/packages/shiny/index.html.
Accessed August 5, 2015
|
|
13
|
Hoefling H: A Path Algorithm for the Fused
Lasso Signal Approximator. J Comput Graph Stat. 19:984–1006. 2010.
View Article : Google Scholar
|
|
14
|
Molenaar JJ, Koster J, Zwijnenburg DA, van
Sluis P, Valentijn LJ, van der Ploeg I, Hamdi M, van Nes J,
Westerman BA, van Arkel J, et al: Sequencing of neuroblastoma
identifies chromothripsis and defects in neuritogenesis genes.
Nature. 483:589–593. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pandey GK, Mitra S, Subhash S, Hertwig F,
Kanduri M, Mishra K, Fransson S, Ganeshram A, Mondal T, Bandaru S,
et al: The risk-associated long noncoding RNA NBAT-1 controls
neuroblastoma progression by regulating cell proliferation and
neuronal differentiation. Cancer Cell. 26:722–737. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pugh TJ, Morozova O, Attiyeh EF,
Asgharzadeh S, Wei JS, Auclair D, Carter SL, Cibulskis K, Hanna M,
Kiezun A, et al: The genetic landscape of high-risk neuroblastoma.
Nat Genet. 45:279–284. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sausen M, Leary RJ, Jones S, Wu J,
Reynolds CP, Liu X, Blackford A, Parmigiani G, Diaz LA Jr,
Papadopoulos N, et al: Integrated genomic analyses identify ARID1A
and ARID1B alterations in the childhood cancer neuroblastoma. Nat
Genet. 45:12–17. 2013. View
Article : Google Scholar :
|
|
18
|
Boeva V, Zinovyev A, Bleakley K, Vert JP,
Janoueix-Lerosey I, Delattre O and Barillot E: Control-free calling
of copy number alterations in deep-sequencing data using GC-content
normalization. Bioinformatics. 27:268–269. 2011. View Article : Google Scholar :
|
|
19
|
Jiménez G, Shvartsman SY and Paroush Z:
The Capicua repressor - a general sensor of RTK signaling in
development and disease. J Cell Sci. 125:1383–1391. 2012.
View Article : Google Scholar
|
|
20
|
Eisenreich S, Abou-El-Ardat K, Szafranski
K, Campos Valenzuela JA, Rump A, Nigro JM, Bjerkvig R, Gerlach EM,
Hackmann K, Schröck E, et al: Novel CIC point mutations and an
exon-spanning, homozygous deletion identified in oligodendroglial
tumors by a comprehensive genomic approach including transcriptome
sequencing. PLoS One. 8:e766232013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sjöblom T, Jones S, Wood LD, Parsons DW,
Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, et al:
The consensus coding sequences of human breast and colorectal
cancers. Science. 314:268–274. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Choi N, Park J, Lee JS, Yoe J, Park GY,
Kim E, Jeon H, Cho YM, Roh TY and Lee Y:
miR-93/miR-106b/miR-375-CIC-CRABP1: A novel regulatory axis in
prostate cancer progression. Oncotarget. 6:23533–23547. 2015.
View Article : Google Scholar : PubMed/NCBI
|